Self-Assembling Peptide RADA16 Nanofiber Scaffold Hydrogel-Wrapped Concentrated Growth Factors in Osteogenesis of MC3T3
Abstract
:1. Introduction
2. Materials and Methods
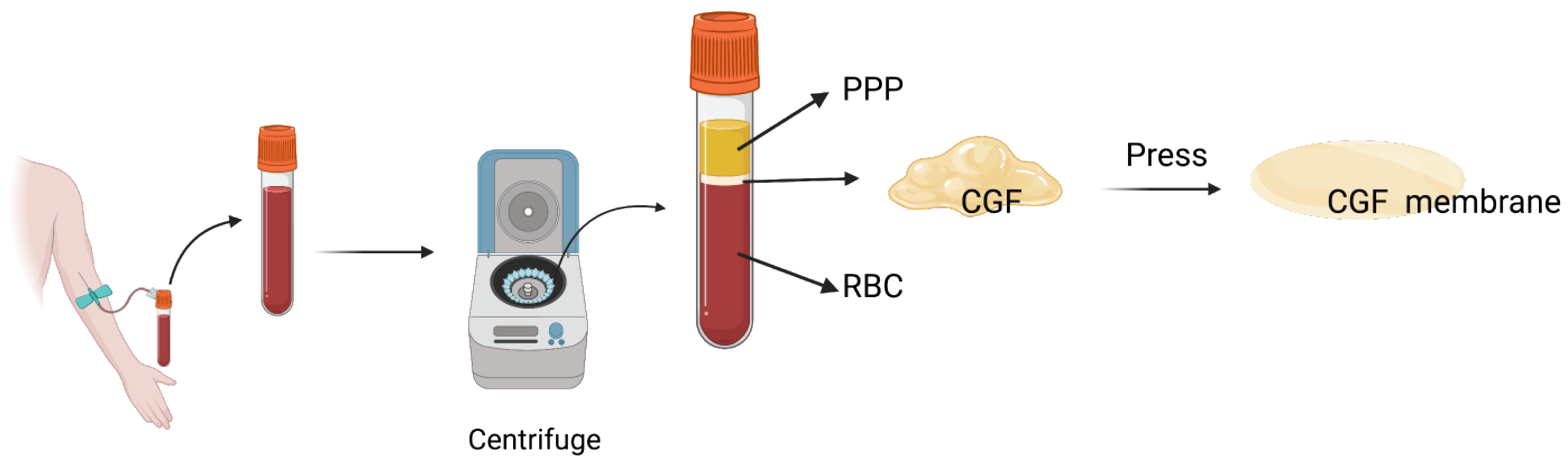
2.1. Blood Sample Centrifugation and CGF Preparation
2.2. RADA16 Synthesis
2.3. RADA16-CGF Fabrication
2.4. Scanning Electron Microscopy (SEM) Assay
2.5. Rheometry
2.6. ELISA Quantification for Growth Factors Released from RADA16-CGFs
2.7. Cells
2.8. Cell Culture Medium Preparation
2.9. Cell Adhesion Experiment
2.10. Cytotoxicity Experiment
2.11. Alkaline Phosphatase (ALP) Staining
2.12. Alizarin Red S Staining
2.13. Alp Gene Expression
2.14. Statistical Analysis
3. Results
3.1. Characterization of CGFs
3.2. Characterization of RADA16
3.3. Growth Factor Releasing Process of RADA16-CGF
3.4. Cell Proliferation of MC3T3 with RADA16-CGF
3.5. Effect of RADA16-CGFs on Mineralization Capability
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- McCrary, H.; Skirko, J.R. Bone Grafting of Alveolar Clefts. Oral Maxillofac. Surg. Clin. N. Am. 2021, 33, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Jeong, W. Secondary bone grafting for alveolar clefts: Surgical timing, graft materials, and evaluation methods. Arch. Craniofacial Surg. 2022, 23, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Tadic, D.; Epple, M. A thorough physicochemical characterisation of 14 calcium phosphate-based bone substitution materials in comparison to natural bone. Biomaterials 2004, 25, 987–994. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, P.; Li, D.J.; Auston, D.A.; Mir, H.S.; Yoon, R.S.; Koval, K.J. Autograft, Allograft, and Bone Graft Substitutes: Clinical Evidence and Indications for Use in the Setting of Orthopaedic Trauma Surgery. J. Orthop. Trauma 2019, 33, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Simonpieri, A.; Del Corso, M.; Sammartino, G.; Ehrenfest, D.M.D. The Relevance of Choukroun’s Platelet-Rich Fibrin and Metronidazole during Complex Maxillary Rehabilitations Using Bone Allograft. Part I: A New Grafting Protocol. Implant. Dent. 2009, 18, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Chen, J.; Liu, S.; Jin, Y. Stem cell-based bone and dental regeneration: A view of microenvironmental modulation. Int. J. Oral Sci. 2019, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Yu, Y.; Han, J.; Shi, D.; Sun, W.; Zhang, D.; Chen, L. Quantification of growth factors in advanced platelet-rich fibrin and concentrated growth factors and their clinical efficacy as adjunctive to the GTR procedure in periodontal intrabony defects. J. Periodontol. 2020, 91, 462–472. [Google Scholar] [CrossRef]
- Masuki, H.; Okudera, T.; Watanabe, T.; Suzuki, M.; Nishiyama, K.; Okudera, H.; Nakata, K.; Uematsu, K.; Su, C.-Y.; Kawase, T. Growth factor and pro-inflammatory cytokine contents in platelet-rich plasma (PRP), plasma rich in growth factors (PRGF), advanced platelet-rich fibrin (A-PRF), and concentrated growth factors (CGF). Int. J. Implant. Dent. 2016, 2, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Li, L.; Cai, W.; Jiang, B. The potential application of concentrated growth factor in regenerative endodontics. Int. Endod. J. 2019, 52, 646–655. [Google Scholar] [CrossRef]
- Boonyagul, S.; Banlunara, W.; Sangvanich, P.; Thunyakitpisal, P. Effect of acemannan, an extracted polysaccharide from Aloe vera, on BMSCs proliferation, differentiation, extracellular matrix synthesis, mineralization, and bone formation in a tooth extraction model. Odontology 2014, 102, 310–317. [Google Scholar] [CrossRef]
- Honda, H.; Tamai, N.; Naka, N.; Yoshikawa, H.; Myoui, A. Bone tissue engineering with bone marrow-derived stromal cells integrated with concentrated growth factor in Rattus norvegicus calvaria defect model. J. Artif. Organs 2013, 16, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Takeda, Y.; Katsutoshi, K.; Matsuzaka, K.; Inoue, T. The Effect of Concentrated Growth Factor on Rat Bone Marrow Cells In Vitro and on Calvarial Bone Healing In Vivo. Int. J. Oral Maxillofac. Implant. 2015, 30, 1187–1196. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-C.; Kim, S.-G.; Oh, J.-S.; You, J.-S.; Kim, J.-S.; Lim, S.-C.; Jeong, M.-A.; Kim, J.-S.; Jung, C.; Kwon, Y.-S.; et al. Early Bone Formation at a Femur Defect Using CGF and PRF Grafts in Adult Dogs: A Comparative Study. Implant. Dent. 2016, 25, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-H.; Kim, S.-H.; Sándor, G.K.; Kim, Y.-D. Comparison of platelet-rich plasma (PRP), platelet-rich fibrin (PRF), and concentrated growth factor (CGF) in rabbit-skull defect healing. Arch. Oral Biol. 2014, 59, 550–558. [Google Scholar] [CrossRef]
- Hong, S.; Chen, W.; Jiang, B. A Comparative Evaluation of Concentrated Growth Factor and Platelet-rich Fibrin on the Proliferation, Migration, and Differentiation of Human Stem Cells of the Apical Papilla. J. Endod. 2018, 44, 977–983. [Google Scholar] [CrossRef]
- Yu, B.; Wang, Z. Effect of concentrated growth factors on beagle periodontal ligament stem cells in vitro. Mol. Med. Rep. 2014, 9, 235–242. [Google Scholar] [CrossRef]
- Calabriso, N.; Stanca, E.; Rochira, A.; Damiano, F.; Giannotti, L.; Stanca, B.D.C.; Massaro, M.; Scoditti, E.; Demitri, C.; Nitti, P.; et al. Angiogenic Properties of Concentrated Growth Factors (CGFs): The Role of Soluble Factors and Cellular Components. Pharmaceutics 2021, 13, 635. [Google Scholar] [CrossRef]
- Wang, L.; Wan, M.; Li, Z.; Zhong, N.; Liang, D.; Ge, L. A comparative study of the effects of concentrated growth factors in two different forms on osteogenesis in vitro. Mol. Med. Rep. 2019, 20, 1039–1048. [Google Scholar] [CrossRef]
- Yokoi, H.; Kinoshita, T.; Zhang, S. Dynamic reassembly of peptide RADA16 nanofiber scaffold. Proc. Natl. Acad. Sci. USA 2005, 102, 8414–8419. [Google Scholar] [CrossRef]
- Kulkarni, D.; Musale, S.; Panzade, P.; Paiva-Santos, A.C.; Sonwane, P.; Madibone, M.; Choundhe, P.; Giram, P.; Cavalu, S. Surface Functionalization of Nanofibers: The Multifaceted Approach for Advanced Biomedical Applications. Nanomaterials 2022, 12, 3899. [Google Scholar] [CrossRef]
- Semino, C.E. Self-assembling Peptides: From Bio-inspired Materials to Bone Regeneration. J. Dent. Res. 2008, 87, 606–616. [Google Scholar] [CrossRef] [PubMed]
- Koutsopoulos, S. Self-assembling peptide nanofiber hydrogels in tissue engineering and regenerative medicine: Progress, design guidelines, and applications. J. Biomed. Mater. Res. Part A 2016, 104, 1002–1016. [Google Scholar] [CrossRef]
- Horii, A.; Wang, X.; Gelain, F.; Zhang, S. Biological Designer Self-Assembling Peptide Nanofiber Scaffolds Significantly Enhance Osteoblast Proliferation, Differentiation and 3-D Migration. PLoS ONE 2007, 2, e190. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Hong, N.; Liu, H.; Wang, J.; Li, Y.; Wu, S. Differentiated adipose-derived stem cell cocultures for bone regeneration in RADA16-I in vitro. J. Cell. Physiol. 2018, 233, 9458–9472. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Guo, A.; Li, K.; Tao, B.; Lei, D.; Deng, Z. Effects of a novel self-assembling peptide scaffold on bone regeneration and controlled release of two growth factors. J. Biomed. Mater. Res. Part A 2022, 110, 943–953. [Google Scholar] [CrossRef]
- Takeuchi, T.; Bizenjima, T.; Ishii, Y.; Imamura, K.; Suzuki, E.; Seshima, F.; Saito, A. Enhanced healing of surgical periodontal defects in rats following application of a self-assembling peptide nanofibre hydrogel. J. Clin. Periodontol. 2016, 43, 279–288. [Google Scholar] [CrossRef]
- Yu, M.; Wang, X.; Liu, Y.; Qiao, J. Cytokine release kinetics of concentrated growth factors in different scaffolds. Clin. Oral Investig. 2019, 23, 1663–1671. [Google Scholar] [CrossRef]
- Campbell, S.; Lees, C.; Moscoso, G.; Hall, P. Ultrasound antenatal diagnosis of cleft palate by a new technique: The 3D ‘reverse face’ view. Ultrasound Obstet. Gynecol. 2005, 25, 12–18. [Google Scholar] [CrossRef]
- Sakkas, A.; Wilde, F.; Heufelder, M.; Winter, K.; Schramm, A. Autogenous bone grafts in oral implantology—Is it still a “gold standard”? A consecutive review of 279 patients with 456 clinical procedures. Int. J. Implant. Dent. 2017, 3, 1–17. [Google Scholar] [CrossRef]
- Tapety, F.I.; Amizuka, N.; Uoshima, K.; Nomura, S.; Maeda, T. A histological evaluation of the involvement of Bio-OssR in osteoblastic differentiation and matrix synthesis. Clin. Oral Implant. Res. 2004, 15, 315–324. [Google Scholar] [CrossRef]
- Burnouf, T.; Goubran, H.A.; Chen, T.-M.; Ou, K.-L.; El-Ekiaby, M.; Radosevic, M. Blood-derived biomaterials and platelet growth factors in regenerative medicine. Blood Rev. 2013, 27, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zheng, J.; Zheng, Q.; Wu, Y.; Wu, B.; Huang, S.; Fang, W.; Guo, X. FGL-functionalized self-assembling nanofiber hydrogel as a scaffold for spinal cord-derived neural stem cells. Mater. Sci. Eng. C 2015, 46, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Olsen, B.R. The roles of vascular endothelial growth factor in bone repair and regeneration. Bone 2016, 91, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Kaigler, D.; Avila, G.; Wisner-Lynch, L.; Nevins, M.L.; Nevins, M.; Rasperini, G.; E Lynch, S.; Giannobile, W.V. Platelet-derived growth factor applications in periodontal and peri-implant bone regeneration. Expert Opin. Biol. Ther. 2011, 11, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Keppler, L.; Rutkowski, J. A Review of Platelet Derived Growth Factor Playing Pivotal Role in Bone Regeneration. J. Oral Implant. 2014, 40, 330–340. [Google Scholar] [CrossRef]
- Gelain, F.; Unsworth, L.D.; Zhang, S. Slow and sustained release of active cytokines from self-assembling peptide scaffolds. J. Control. Release 2010, 145, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Zhou, A.; Chen, S.; He, B.; Zhao, W.; Chen, X.; Jiang, D. Controlled release of TGF-beta 1 from RADA self-assembling peptide hydrogel scaffolds. Drug Des. Devel. Ther. 2016, 10, 3043–3051. [Google Scholar] [CrossRef]
- Hunt, N.; Grover, L.M. Cell encapsulation using biopolymer gels for regenerative medicine. Biotechnol. Lett. 2010, 32, 733–742. [Google Scholar] [CrossRef]
- Wang, R.; Wang, Z.; Guo, Y.; Li, H.; Chen, Z. Design of a RADA16-based self-assembling peptide nanofiber scaffold for biomedical applications. J. Biomater. Sci. Polym. Ed. 2019, 30, 713–736. [Google Scholar] [CrossRef]
- Sankar, S.; O’neill, K.; D’arc, M.B.; Rebeca, F.; Buffier, M.; Aleksi, E.; Fan, M.; Matsuda, N.; Gil, E.S.; Spirio, L. Clinical Use of the Self-Assembling Peptide RADA16: A Review of Current and Future Trends in Biomedicine. Front. Bioeng. Biotechnol. 2021, 9, 679525. [Google Scholar] [CrossRef]
- Tabatabaei, F.; Aghamohammadi, Z.; Tayebi, L. In vitro and in vivo effects of concentrated growth factor on cells and tissues. J. Biomed. Mater. Res. Part A 2020, 108, 1338–1350. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, R.; Chen, J.; Wang, D.; Xu, Y.; Ou, G. Self-Assembling Peptide RADA16 Nanofiber Scaffold Hydrogel-Wrapped Concentrated Growth Factors in Osteogenesis of MC3T3. J. Funct. Biomater. 2023, 14, 260. https://doi.org/10.3390/jfb14050260
Yang R, Chen J, Wang D, Xu Y, Ou G. Self-Assembling Peptide RADA16 Nanofiber Scaffold Hydrogel-Wrapped Concentrated Growth Factors in Osteogenesis of MC3T3. Journal of Functional Biomaterials. 2023; 14(5):260. https://doi.org/10.3390/jfb14050260
Chicago/Turabian StyleYang, Renjie, Jiali Chen, Dingjie Wang, Yichen Xu, and Guomin Ou. 2023. "Self-Assembling Peptide RADA16 Nanofiber Scaffold Hydrogel-Wrapped Concentrated Growth Factors in Osteogenesis of MC3T3" Journal of Functional Biomaterials 14, no. 5: 260. https://doi.org/10.3390/jfb14050260
APA StyleYang, R., Chen, J., Wang, D., Xu, Y., & Ou, G. (2023). Self-Assembling Peptide RADA16 Nanofiber Scaffold Hydrogel-Wrapped Concentrated Growth Factors in Osteogenesis of MC3T3. Journal of Functional Biomaterials, 14(5), 260. https://doi.org/10.3390/jfb14050260






