Functional L-Arginine Derivative as an Efficient Vector for Intracellular Protein Delivery for Potential Cancer Therapy
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis
2.3. Preparation and Characterization of DRC/Protein Nanoformulations
2.4. Protein Loading Efficiency
2.5. The Stability of Nanoformulations
2.6. Synthesis of FITC-Labeled BSA (BSA-FITC) and FITC-Labeled Saporin (SA-FITC)
2.7. Cell Culture and Cellular Uptake
2.8. Cytotoxicity Assay
2.9. In Vitro Anticancer Activity
2.10. Statistical Analysis
3. Results and Discussion
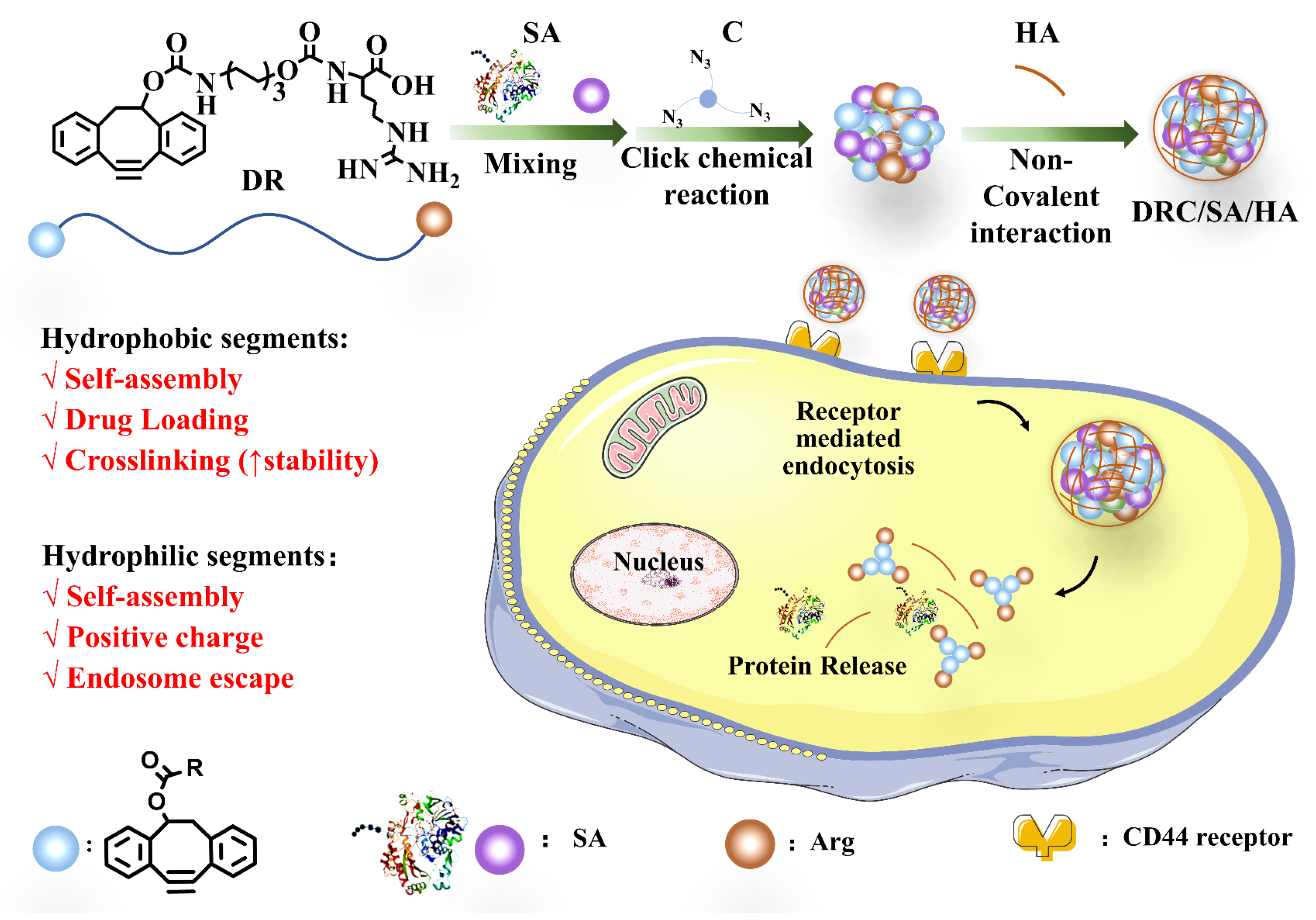
3.1. Synthesis the of Vector and Crosslinker
3.2. Preparation and Characterization of Various Nanoformulations
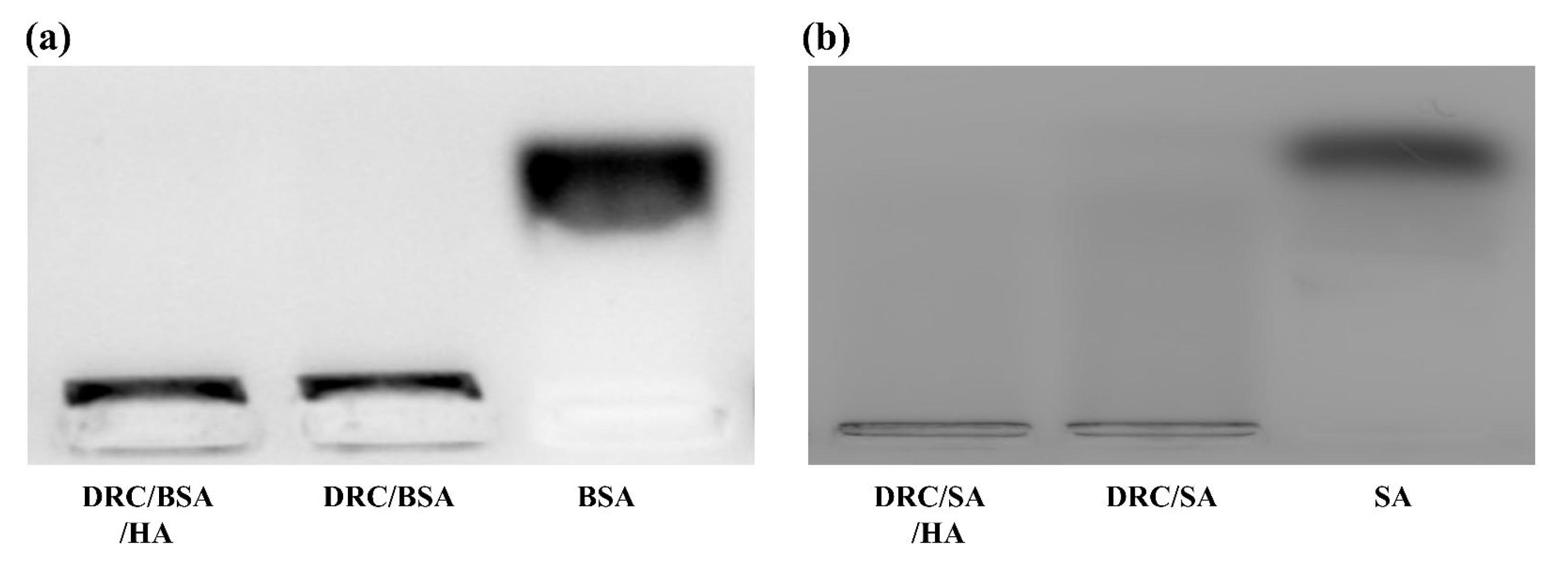
3.3. Stability of the Protein-Loaded Nanoparticles
3.4. Cellular Uptake of Proteins
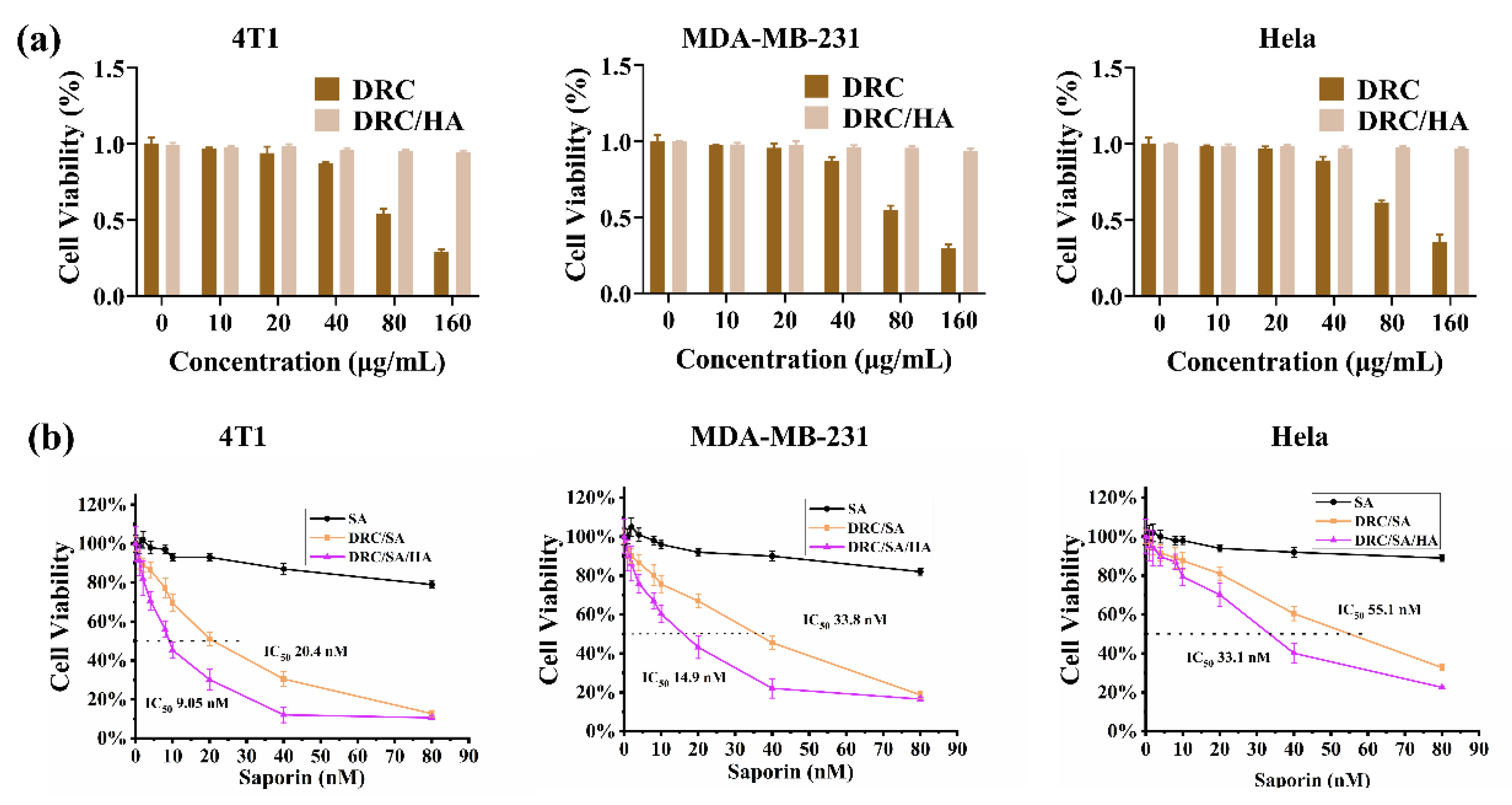
3.5. Cytotoxicity Evaluation of DRC and Anticancer Ability of DRC-Based Nanoformulations Containing Saporin
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hafeez, U.; Gan, H.K.; Scott, A.M. Monoclonal antibodies as immunomodulatory therapy against cancer and autoimmune diseases. Curr. Opin. Pharmacol. 2018, 41, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Urquhart, L. Top product forecasts for 2022. Nat. Rev. Drug Discov. 2022, 21, 11. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Yu, C.; Wei, J.; Li, L.; Zhang, C.; Wu, Q.; Liu, J.; Yao, S.Q.; Huang, W. Rational Design of Nanocarriers for Intracellular Protein Delivery. Adv. Mater. 2019, 31, e1902791. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Alberti, K.; Sun, S.; Arellano, C.L.; Xu, Q. Combinatorially Designed Lipid-like Nanoparticles for Intracellular Delivery of Cytotoxic Protein for Cancer Therapy. Angew. Chem. Int. Edit. 2014, 53, 2893–2898. [Google Scholar] [CrossRef]
- Nischan, N.; Herce, H.D.; Natale, F.; Bohlke, N.; Budisa, N.; Cardoso, M.C.; Hackenberger, C.P.R. Covalent Attachment of Cyclic TAT Peptides to GFP Results in Protein Delivery into Live Cells with Immediate Bioavailability. Angew. Chem. Int. Edit. 2015, 54, 1950–1953. [Google Scholar] [CrossRef] [PubMed]
- Brodin, J.D.; Sprangers, A.J.; McMillan, J.R.; Mirkin, C.A. DNA-Mediated Cellular Delivery of Functional Enzymes. J. Am. Chem. Soc. 2015, 137, 14838–14841. [Google Scholar] [CrossRef]
- Yu, M.; Wu, J.; Shi, J.; Farokhzad, O.C. Nanotechnology for protein delivery: Overview and perspectives. J. Control Release 2016, 240, 24–37. [Google Scholar] [CrossRef]
- Cao, Y.; Ma, E.; Cestellos-Blanco, S.; Zhang, B.; Qiu, R.; Su, Y.; Doudna, J.A.; Yang, P. Nontoxic nanopore electroporation for effective intracellular delivery of biological macromolecules. Proc. Natl. Acad. Sci. USA 2019, 116, 7899–7904. [Google Scholar] [CrossRef]
- Stewart, M.P.; Langer, R.; Jensen, K.F. Intracellular Delivery by Membrane Disruption: Mechanisms, Strategies, and Concepts. Chem. Rev. 2018, 118, 7409–7531. [Google Scholar] [CrossRef]
- Mogaki, R.; Hashim, P.K.; Okuro, K.; Aida, T. Guanidinium-based “molecular glues” for modulation of biomolecular functions. Chem. Soc. Rev. 2017, 46, 6480–6491. [Google Scholar] [CrossRef]
- Guidotti, G.; Brambilla, L.; Rossi, D. Cell-Penetrating Peptides: From Basic Research to Clinics. Trends Pharmacol. Sci. 2017, 38, 406–424. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Monteiro da Silva, G.; Deatherage, C.; Burd, C.; DiMaio, D. Cell-Penetrating Peptide Mediates Intracellular Membrane Passage of Human Papillomavirus L2 Protein to Trigger Retrograde Trafficking. Cell 2018, 174, 1465–1476. [Google Scholar] [CrossRef] [PubMed]
- Mout, R.; Ray, M.; Tay, T.; Sasaki, K.; Yesilbag Tonga, G.; Rotello, V.M. General Strategy for Direct Cytosolic Protein Delivery via Protein–Nanoparticle Co-engineering. ACS Nano 2017, 11, 6416–6421. [Google Scholar] [CrossRef]
- Schneider, A.F.L.; Kithil, M.; Cardoso, M.C.; Lehmann, M.; Hackenberger, C.P.R. Cellular uptake of large biomolecules enabled by cell-surface-reactive cell-penetrating peptide additives. Nat. Chem. 2021, 13, 530–539. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Zeng, K.; Zhang, Y.; Shao, J.; Yan, J.; Liao, J.-Y.; Wang, W.; Dai, X.; Weng, Q.; Yao, S.Q.; et al. In vivo targeted delivery of antibodies into cancer cells with pH-responsive cell-penetrating poly(disulfide)s. ChemComm. 2022, 58, 1314–1317. [Google Scholar] [CrossRef]
- Laurent, Q.; Martinent, R.; Lim, B.; Pham, A.-T.; Kato, T.; López-Andarias, J.; Sakai, N.; Matile, S. Thiol-Mediated Uptake. JACS Au 2021, 1, 710–728. [Google Scholar] [CrossRef]
- Zhang, R.; Nie, T.; Fang, Y.; Huang, H.; Wu, J. Poly(disulfide)s: From Synthesis to Drug Delivery. Biomacromolecules 2022, 23, 1–19. [Google Scholar] [CrossRef]
- Wang, H.; Hu, Y.; Wang, Y.; Lu, J.; Lu, H. Doxorubicin@ PEPylated interferon-polydisulfide: A multi-responsive nanoparticle for enhanced chemo–protein combination therapy. Giant 2021, 5, 100040. [Google Scholar] [CrossRef]
- Chen, N.X.; He, Y.; Zang, M.M.; Zhang, Y.X.; Lu, H.Y.; Zhao, Q.F.; Wang, S.L.; Gao, Y.K. Approaches and materials for endocytosis-independent intracellular delivery of protein. Biomaterials 2022, 286, 121567. [Google Scholar] [CrossRef]
- Xu, X.; Shen, S.; Mo, R. Bioresponsive nanogels for protein delivery. View 2022, 3, 20200136. [Google Scholar] [CrossRef]
- Kube, S.; Hersch, N.; Naumovska, E.; Gensch, T.; Hendriks, J.; Franzen, A.; Landvogt, L.; Siebrasse, J.-P.; Kubitscheck, U.; Hoffmann, B.; et al. Fusogenic Liposomes as Nanocarriers for the Delivery of Intracellular Proteins. Langmuir 2017, 33, 1051–1059. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Gao, X.; Liu, J.; Zheng, Q.; Li, Y.; Yu, P.; Wang, M.; Mao, L. Biodegradable Metal–Organic-Frameworks-Mediated Protein Delivery Enables Intracellular Cascade Biocatalysis and Pyroptosis In Vivo. ACS Appl. Mater. Interfaces 2022, 14, 47472–47481. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; He, B.; Yu, J.; Wang, Y.; Wang, C.; Zhang, S.; Wang, H.; Hu, J.; Zhang, Q.; Cheng, Y. Fluoropolymers for intracellular and in vivo protein delivery. Biomaterials 2018, 182, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-W.; Luther, D.C.; Goswami, R.; Jeon, T.; Clark, V.; Elia, J.; Gopalakrishnan, S.; Rotello, V.M. Direct Cytosolic Delivery of Proteins through Coengineering of Proteins and Polymeric Delivery Vehicles. J. Am. Chem. Soc. 2020, 142, 4349–4355. [Google Scholar] [CrossRef]
- Qian, L.; Fu, J.; Yuan, P.; Du, S.; Huang, W.; Li, L.; Yao, S.Q. Intracellular Delivery of Native Proteins Facilitated by Cell-Penetrating Poly(disulfide)s. Angew. Chem. Int. Ed. 2018, 130, 1548–1552. [Google Scholar] [CrossRef]
- Xu, J.; Li, Z.; Fan, Q.; Lv, J.; Li, Y.; Cheng, Y. Dynamic Polymer Amphiphiles for Efficient Intracellular and In Vivo Protein Delivery. Adv. Mater. 2021, 33, 2104355. [Google Scholar] [CrossRef]
- Kern, H.B.; Srinivasan, S.; Convertine, A.J.; Hockenbery, D.; Press, O.W.; Stayton, P.S. Enzyme-Cleavable Polymeric Micelles for the Intracellular Delivery of Proapoptotic Peptides. Mol. Pharm. 2017, 14, 1450–1459. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Gao, W. In Situ Growth of Self-Assembled Protein–Polymer Nanovesicles for Enhanced Intracellular Protein Delivery. ACS Appl. Mater. Interfaces 2017, 9, 2023–2028. [Google Scholar] [CrossRef]
- Dutta, K.; Hu, D.; Zhao, B.; Ribbe, A.E.; Zhuang, J.; Thayumanavan, S. Templated Self-Assembly of a Covalent Polymer Network for Intracellular Protein Delivery and Traceless Release. J. Am. Chem. Soc. 2017, 139, 5676–5679. [Google Scholar] [CrossRef]
- Garcia-Seisdedos, H.; Empereur-Mot, C.; Elad, N.; Levy, E.D. Proteins evolve on the edge of supramolecular self-assembly. Nature 2017, 548, 244–247. [Google Scholar] [CrossRef]
- Levy, E.D.; Erba, E.B.; Robinson, C.V.; Teichmann, S.A. Assembly reflects evolution of protein complexes. Nature 2008, 453, 1262–1265. [Google Scholar] [CrossRef]
- Zhang, N.; Zhao, F.; Zou, Q.; Li, Y.; Ma, G.; Yan, X. Multitriggered Tumor-Responsive Drug Delivery Vehicles Based on Protein and Polypeptide Coassembly for Enhanced Photodynamic Tumor Ablation. Small 2016, 12, 5936–5943. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.; Yu, X.; Shen, B.; Sun, L. Peptide Self-Assembled Nanostructures for Drug Delivery Applications. J. Nanomater. 2017, 2017, 4562474. [Google Scholar] [CrossRef]
- Gu, W.; Liu, T.; Fan, D.; Zhang, J.; Xia, Y.; Meng, F.; Xu, Y.; Cornelissen, J.J.L.M.; Liu, Z.; Zhong, Z. A6 peptide-tagged, ultra-small and reduction-sensitive polymersomal vincristine sulfate as a smart and specific treatment for CD44+ acute myeloid leukemia. J. Control Release 2021, 329, 706–716. [Google Scholar] [CrossRef] [PubMed]
- McManus, J.J.; Charbonneau, P.; Zaccarelli, E.; Asherie, N. The physics of protein self-assembly. Curr. Opin. Colloid Interface Sci. 2016, 22, 73–79. [Google Scholar] [CrossRef]
- Wang, F.; Gnewou, O.; Wang, S.; Osinski, T.; Zuo, X.; Egelman, E.H.; Conticello, V.P. Deterministic chaos in the self-assembly of β sheet nanotubes from an amphipathic oligopeptide. Matter 2021, 4, 3217–3231. [Google Scholar] [CrossRef] [PubMed]
- Guilbaud-Chéreau, C.; Dinesh, B.; Schurhammer, R.; Collin, D.; Bianco, A.; Ménard-Moyon, C. Protected Amino Acid–Based Hydrogels Incorporating Carbon Nanomaterials for Near-Infrared Irradiation-Triggered Drug Release. ACS Appl. Mater. Interfaces 2019, 11, 13147–13157. [Google Scholar] [CrossRef]
- Li, Y.; Zou, Q.; Yuan, C.; Li, S.; Xing, R.; Yan, X. Amino Acid Coordination Driven Self-Assembly for Enhancing both the Biological Stability and Tumor Accumulation of Curcumin. Angew. Chem. Int. Edit. 2018, 57, 17084–17088. [Google Scholar] [CrossRef]
- Cheng, G.; Han, X.; Zheng, S.Y. Magnetically Driven Nanotransporter-Assisted Intracellular Delivery and Autonomous Release of Proteins. ACS Appl. Mater. Interfaces 2020, 12, 41096–41104. [Google Scholar] [CrossRef]
- Barrios, A.; Estrada, M.; Moon, J.H. Carbamoylated Guanidine-Containing Polymers for Non-Covalent Functional Protein Delivery in Serum-Containing Media. Angew Chem. Int. Ed. 2022, 61, e202116722. [Google Scholar] [CrossRef]
- Chen, J.; He, H.; Deng, C.; Yin, L.; Zhong, Z. Saporin-loaded CD44 and EGFR dual-targeted nanogels for potent inhibition of metastatic breast cancer in vivo. Int. J. Pharm. 2019, 560, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Song, T.; Gao, Y.; Song, M.; Qian, J.; Zhang, H.; Zhou, J.; Ding, Y. Fluoropolymers-mediated efficient biomacromolecule drug delivery. Med. Drug Discov. 2022, 14, 100123. [Google Scholar] [CrossRef]
- Yu, S.; Yang, H.; Li, T.; Pan, H.; Ren, S.; Luo, G.; Jiang, J.; Yu, L.; Chen, B.; Zhang, Y.; et al. Efficient intracellular delivery of proteins by a multifunctional chimaeric peptide in vitro and in vivo. Nat. Commun. 2021, 12, 5131. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Zhou, S.; Tang, B.; Xu, H.; Wu, X.; Li, N.; Zan, X.; Geng, W. Efficient delivery of cytosolic proteins by protein-hexahistidine-metal co-assemblies. Acta Biomater. 2021, 129, 199–208. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, X.; Qu, Y.; Xiong, S.; Jiang, Z.; Tang, Y.; Yan, F.; Deng, Y.; Sun, Y. Functional L-Arginine Derivative as an Efficient Vector for Intracellular Protein Delivery for Potential Cancer Therapy. J. Funct. Biomater. 2023, 14, 301. https://doi.org/10.3390/jfb14060301
He X, Qu Y, Xiong S, Jiang Z, Tang Y, Yan F, Deng Y, Sun Y. Functional L-Arginine Derivative as an Efficient Vector for Intracellular Protein Delivery for Potential Cancer Therapy. Journal of Functional Biomaterials. 2023; 14(6):301. https://doi.org/10.3390/jfb14060301
Chicago/Turabian StyleHe, Xiao, Yannv Qu, Su Xiong, Zhiru Jiang, Yaqin Tang, Fei Yan, Yuanfei Deng, and Yansun Sun. 2023. "Functional L-Arginine Derivative as an Efficient Vector for Intracellular Protein Delivery for Potential Cancer Therapy" Journal of Functional Biomaterials 14, no. 6: 301. https://doi.org/10.3390/jfb14060301
APA StyleHe, X., Qu, Y., Xiong, S., Jiang, Z., Tang, Y., Yan, F., Deng, Y., & Sun, Y. (2023). Functional L-Arginine Derivative as an Efficient Vector for Intracellular Protein Delivery for Potential Cancer Therapy. Journal of Functional Biomaterials, 14(6), 301. https://doi.org/10.3390/jfb14060301









