Unveiling Nanoparticles: Recent Approaches in Studying the Internalization Pattern of Iron Oxide Nanoparticles in Mono- and Multicellular Biological Structures
Abstract
1. Introduction
2. Internalization Mechanism of Iron Oxide Nanoparticles
3. Investigations of the Iron Oxide Nanoparticle Internalization Process in Conventional 2D Cell Cultures
3.1. Optical and Fluorescence Microscopy
3.2. Electron Microscopy
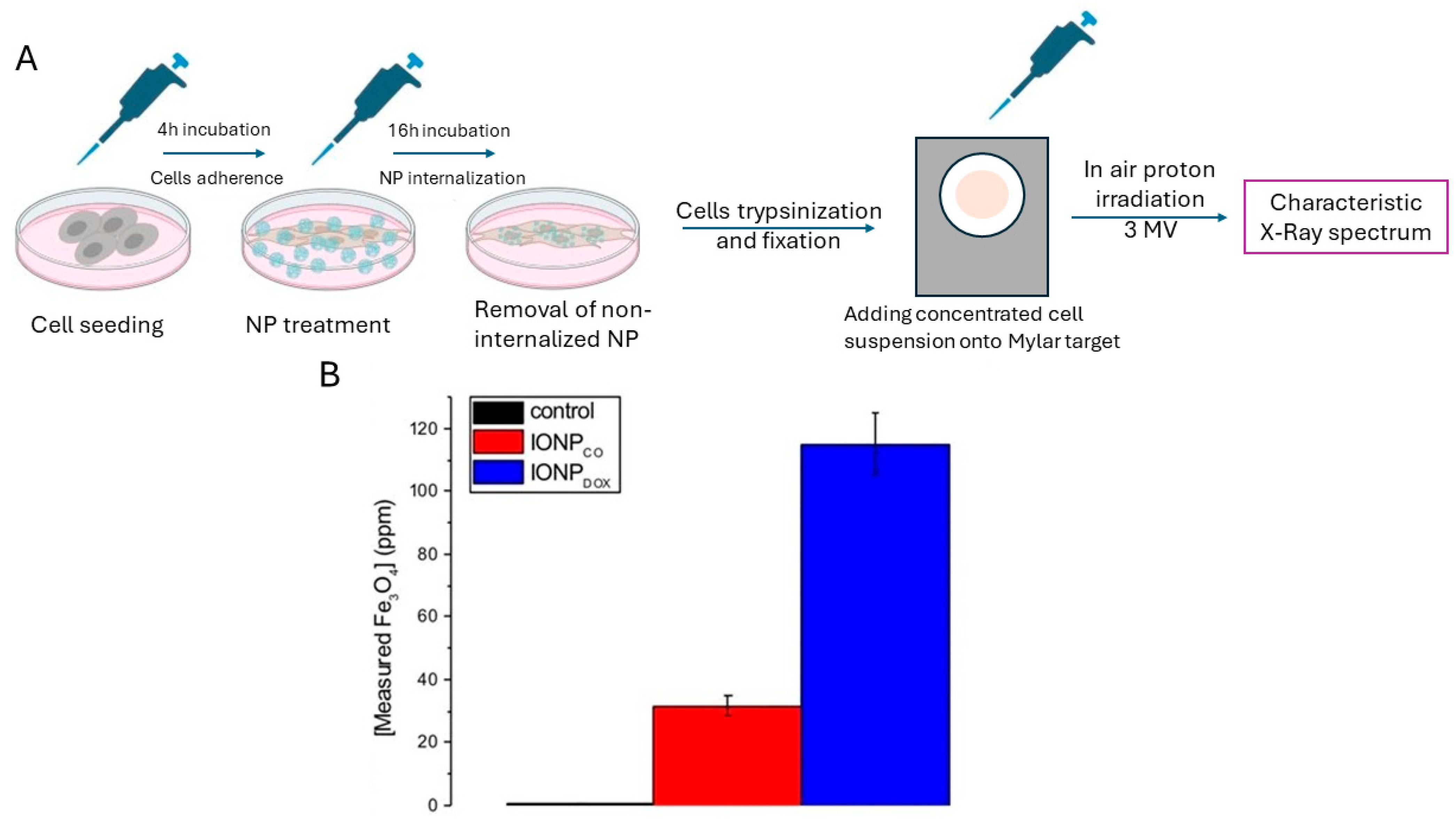
3.3. Quantitative Spectroscopic Determinations
4. Immunohistological Studies for Complex Biological Structures
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Murugan, K.; Choonara, Y.E.; Kumar, P.; Bijukumar, D.; du Toit, L.C.; Pillay, V. Parameters and characteristics governing cellular internalization and trans-barrier trafficking of nanostructures. Int. J. Nanomed. 2015, 10, 2191–2206. [Google Scholar] [CrossRef]
- Valdiglesias, V.; Kiliç, G.; Costa, C.; Fernández-Bertólez, N.; Pásaro, E.; Teixeira, J.P.; Laffon, B. Effects of iron oxide nanoparticles: Cytotoxicity, genotoxicity, developmental toxicity, and neurotoxicity. Environ. Mol. Mutagen. 2015, 56, 125–148. [Google Scholar] [CrossRef]
- Guggenheim, E.J.; Rappoport, J.Z.; Lynch, I. Mechanisms for cellular uptake of nanosized clinical MRI contrast agents. Nanotoxicology 2020, 14, 504–532. [Google Scholar] [CrossRef]
- Montizaan, D. Unravelling the Mechanisms of Recognition and Internalization of Nanoparticles by Cells. Ph.D. Thesis, University of Groningen, Groningen, The Netherlands, 2020. [Google Scholar] [CrossRef]
- Rueda-Gensini, L.; Cifuentes, J.; Castellanos, M.C.; Puentes, P.R.; Serna, J.A.; Muñoz-Camargo, C.; Juan, C. Tailoring iron oxide nanoparticles for efficient cellular internalization and endosomal escape. Nanomaterials 2020, 10, 1816. [Google Scholar] [CrossRef]
- Reimer, P.; Balzer, T. Ferucarbotran (Resovist): A new clinically approved RES-specific contrast agent for contrast-enhanced MRI of the liver: Properties, clinical development, and applications. Eur. Radiol. 2003, 13, 1266–1276. [Google Scholar] [CrossRef]
- Feridex Prescribing Information. Available online: https://www.drugs.com/pro/feridex.html (accessed on 16 March 2024).
- Geppert, M.; Himly, M. Iron Oxide Nanoparticles in Bioimaging—An Immune Perspective. Front. Immunol. 2021, 12, 688927. [Google Scholar] [CrossRef]
- Pharmacosmos Products. Available online: https://www.pharmacosmos.com/pharmacosmos/products (accessed on 16 March 2024).
- Go Beyond Efficacy with Ferahme for Iron Deficiency Anemia. Available online: https://www.feraheme.com/home/ (accessed on 16 March 2024).
- Jain, T.K.; Reddy, M.K.; Morales, M.A.; Leslie-Pelecky, D.L.; Labhasetwar, V. Biodistribution, clearance, and biocompatibility of iron oxide magnetic nanoparticles in rats. Mol. Pharm. 2008, 5, 316–327. [Google Scholar] [CrossRef]
- Zhang, T.; Xu, Q.; Huang, T.; Ling, D.; Gao, J. New Insights into Biocompatible Iron Oxide Nanoparticles: A Potential Booster of Gene Delivery to Stem Cells. Small 2020, 16, 2001588. [Google Scholar] [CrossRef]
- Mulens-Arias, V.; Rojas, J.M.; Barber, D.F. The Intrinsic Biological Identities of Iron Oxide Nanoparticles and Their Coatings: Unexplored Territory for Combinatorial Therapies. Nanomaterials 2020, 10, 837. [Google Scholar] [CrossRef]
- Roemhild, K.; von Maltzahn, F.; Weiskirchen, R.; Knüchel, R.; von Stillfried, S.; Lammers, T. Iron metabolism: Pathophysiology and pharmacology. Trends Pharmacol. Sci. 2021, 42, 640–656. [Google Scholar] [CrossRef]
- Poller, W.C.; Pieber, M.; Boehm-Sturm, P.; Ramberger, E.; Karampelas, V.; Möller, K.; Schleicher, M.; Wiekhorst, F.; Löwa, N.; Wagner, S.; et al. Very small superparamagnetic iron oxide nanoparticles: Long-term fate and metabolic processing in atherosclerotic mice. Nanomedicine 2018, 14, 2575–2586. [Google Scholar] [CrossRef]
- Gu, J.; Xu, H.; Han, Y.; Dai, W.; Hao, W.; Wang, C.; Gu, N.; Xu, H.; Cao, J. The internalization pathway, metabolic fate and biological effect of superparamagnetic iron oxide nanoparticles in the macrophage-like RAW264.7 cell. Sci. China Life Sci. 2011, 54, 793–805. [Google Scholar] [CrossRef]
- Santoyo Salazar, J.; Perez, L.; de Abril, O.; Truong Phuoc, L.; Ihiawakrim, D.; Vazquez, M.; Greneche, J.M.; Begin-Colin, S.; Pourroy, G. Magnetic Iron Oxide Nanoparticles in 10−40 nm Range: Composition in Terms of Magnetite/Maghemite Ratio and Effect on the Magnetic Properties. Chem. Mater. 2011, 23, 1379–1386. [Google Scholar] [CrossRef]
- Chen, L.; Xie, J.; Wu, H.; Li, J.; Wang, Z.; Song, L.; Zang, F.; Ma, M.; Gu, N.; Zhang, Y. Precise Study on Size-Dependent Properties of Magnetic Iron Oxide Nanoparticles for In Vivo Magnetic Resonance Imaging. J. Nanomater. 2018, 2018, 3743164. [Google Scholar] [CrossRef]
- Augustine, R.; Hasan, A.; Primavera, R.; Joyce, R.; Thakor, A.S.; Kevadiya, B.D. Cellular uptake and retention of nanoparticles: Insights on particle properties and interaction with cellular components. Mater. Today Commun. 2020, 25, 101692. [Google Scholar] [CrossRef]
- Rennick, J.J.; Johnston, A.P.R.; Parton, R.G. Key principles and methods for studying the endocytosis of biological and nanoparticle therapeutics. Nat. Nanotechnol. 2021, 16, 266–276. [Google Scholar] [CrossRef]
- Michael, H.; Wojciech, P. Ross Histology: A Text and Atlas, 7th ed.; Hipocrate: Bucharest, Romania, 2020; ISBN 9786069457580. [Google Scholar]
- Popescu, R.C.; Savu, D.; Dorobantu, I.; Vasile, B.S.; Hosser, H.; Boldeiu, A.; Temelie, M.; Straticiuc, M.; Iancu, D.A.; Andronescu, E.; et al. Efficient uptake and retention of iron oxide-based nanoparticles in HeLa cells leads to an effective intracellular delivery of doxorubicin. Sci. Rep. 2020, 10, 10530. [Google Scholar] [CrossRef]
- Mazumdar, S.; Chitkara, D.; Mittal, A. Exploration and insights into the cellular internalization and intracellular fate of amphiphilic polymeric nanocarriers. Acta Pharm. Sin. B 2021, 11, 903–924. [Google Scholar] [CrossRef]
- Malatesta, M. Transmission Electron Microscopy as a Powerful Tool to Investigate the Interaction of Nanoparticles with Subcellular Structures. Int. J. Mol. Sci. 2021, 22, 2789. [Google Scholar] [CrossRef]
- Brown, A.P.; Brydson, R.M.D.; Hondow, N.S. Measuring in vitro cellular uptake of nanoparticles by transmission electron microscopy. J. Phys. Conf. Ser. 2014, 522, 3–7. [Google Scholar] [CrossRef]
- Vtyurina, N.; Åberg, C.; Salvati, A. Imaging of nanoparticle uptake and kinetics of intracellular trafficking in individual cells. Nanoscale 2021, 13, 10436–10446. [Google Scholar] [CrossRef]
- Claudia, M.; Kristin, Ö.; Jennifer, O.; Eva, R.; Eleonore, F. Comparison of fluorescence-based methods to determine nanoparticle uptake by phagocytes and non-phagocytic cells in vitro. Toxicology 2017, 378, 25–36. [Google Scholar] [CrossRef]
- Popescu, R.C.; Straticiuc, M.; Mustăciosu, C.; Temelie, M.; Trușcă, R.; Vasile, B.Ș.; Boldeiu, A.; Mirea, D.; Andrei, R.F.; Cenușă, C.; et al. Enhanced internalization of nanoparticles following ionizing radiation leads to mitotic catastrophe in MG-63 human osteosarcoma cells. Int. J. Mol. Sci. 2020, 21, 7220. [Google Scholar] [CrossRef]
- Liu, Y.; Cheng, W.; Xin, H.; Liu, R.; Wang, Q.; Cai, W.; Peng, X.; Yang, F.; Xin, H. Nanoparticles advanced from preclinical studies to clinical trials for lung cancer therapy. Cancer Nanotechnol. 2023, 14, 28. [Google Scholar] [CrossRef]
- Mundekkad, D.; Cho, W.C. Nanoparticles in Clinical Translation for Cancer Therapy. Int. J. Mol. Sci. 2022, 23, 1685. [Google Scholar] [CrossRef]
- Cancer Nano-Therapies in the Clinic and Clinical Trials. Available online: https://www.cancer.gov/nano/cancer-nanotechnology/current-treatments (accessed on 16 March 2024).
- Popescu, R.C.; Savu, D.I.; Bierbaum, M.; Grbenicek, A.; Schneider, F.; Hosser, H.; Vasile, B.Ș.; Andronescu, E.; Wenz, F.; Giordano, F.A.; et al. Intracellular delivery of doxorubicin by iron oxide-based nano-constructs increases clonogenic inactivation of ionizing radiation in hela cells. Int. J. Mol. Sci. 2021, 22, 6778. [Google Scholar] [CrossRef]
- Popescu, R.C.; Kopatz, V.; Andronescu, E.; Savu, D.I.; Doerr, W. Nanoparticle-Mediated Drug Delivery of Doxorubicin Induces a Differentiated Clonogenic Inactivation in 3D Tumor Spheroids In Vitro. Int. J. Mol. Sci. 2023, 24, 2198. [Google Scholar] [CrossRef]
- Tudor, M.; Popescu, R.C.; Negoita, R.D.; Gilbert, A.; Ilisanu, M.A.; Temelie, M.; Dinischiotu, A.; Chevalier, F.; Mihailescu, M.; Savu, D.I. In vitro hyperspectral biomarkers of human chondrosarcoma cells in nanoparticle-mediated radiosensitization using carbon ions. Sci. Rep. 2023, 13, 14878. [Google Scholar] [CrossRef]
- Gutiérrez, L.; de la Cueva, L.; Moros, M.; Mazarío, E.; de Bernardo, S.; de la Fuente, J.M.; Morales, M.P.; Salas, G. Aggregation effects on the magnetic properties of iron oxide colloids. Nanotechnology 2019, 30, 112001. [Google Scholar] [CrossRef]
- Nguyen, T.-N.; Tran, Q.-H.; Terki, F.; Charnay, C.; Dumail, X.; Reibel, C.; Cazals, G.; Valette, G.; Jay-Allemand, C.; Bidel, L.P.R. Aggregation of magnetic nanoparticles functionalized with trans-resveratrol in aqueous solution. Discov. Nano 2023, 18, 64. [Google Scholar] [CrossRef]
- Portilla, Y.; Fernández-Afonso, Y.; Pérez-Yagüe, S.; Mulens-Arias, V.; Morales, M.P.; Gutiérrez, L.; Barber, D.F. Different coatings on magnetic nanoparticles dictate their degradation kinetics in vivo for 15 months after intravenous administration in mice. J. Nanobiotechnol. 2022, 20, 543. [Google Scholar] [CrossRef]
- Strączek, T.; Fiejdasz, S.; Rybicki, D.; Goc, K.; Przewoźnik, J.; Mazur, W.; Nowakowska, M.; Zapotoczny, S.; Rumian, S.; Kapusta, C. Dynamics of Superparamagnetic Iron Oxide Nanoparticles with Various Polymeric Coatings. Materials 2019, 12, 1793. [Google Scholar] [CrossRef]
- Khandhar, A.P.; Ferguson, R.M.; Arami, H.; Kemp, S.J.; Krishnan, K.M. Tuning surface coatings of optimized magnetite nanoparticle tracers for in vivo Magnetic Particle Imaging. IEEE Trans. Magn. 2015, 51, 5300304. [Google Scholar] [CrossRef]
- Nelson, N.; Port, J.; Pandey, M. Use of Superparamagnetic Iron Oxide Nanoparticles (SPIONs) via Multiple Imaging Modalities and Modifications to Reduce Cytotoxicity: An Educational Review. J. Nanotheranostics 2020, 1, 105–135. [Google Scholar] [CrossRef]
- Chen, W.; Yang, Z. Human gastric carcinoma cells targeting peptide-functionalized iron oxide nanoparticles delivery for magnetic resonance imaging. Process Biochem. 2020, 99, 171–178. [Google Scholar] [CrossRef]
- Xia, Y.; Padmanabhan, P.; Vijayaragavan, V.; Murukeshan, V.M.; Gulyás, B. Amyloid Beta42 (Aβ42) Peptide Functionalized Iron Oxide Nanoparticles for Specific Targeting of SH-SY5Y Neuroblastoma Cells. J. Nanosci. Nanotechnol. 2021, 21, 5044–5050. [Google Scholar] [CrossRef]
- Bychkova, A.V.; Yakunina, M.N.; Lopukhova, M.V.; Degtyarev, Y.N.; Motyakin, M.V.; Pokrovsky, V.S.; Kovarski, A.L.; Gorobets, M.G.; Retivov, V.M.; Khachatryan, D.S. Albumin-Functionalized Iron Oxide Nanoparticles for Theranostics: Engineering and Long-Term In Situ Imaging. Pharmaceutics 2022, 14, 2771. [Google Scholar] [CrossRef]
- Singh, A.; Bajpai, J.; Bajpai, A.K.; Mongre, R.K.; Lee, M.-S. Encapsulation of cytarabine into casein coated iron oxide nanoparticles (CCIONPs) and study of in vitro drug release and anticancer activities. J. Drug Deliv. Sci. Technol. 2020, 55, 101396. [Google Scholar] [CrossRef]
- Zhong, J.; Zheng, C.; Gao, H.; Tong, W.; Hui, H.; Tian, J. Noninvasive imaging of the lung NETosis by anti-Ly6G iron oxide nanoparticles. Heliyon 2022, 8, e10043. [Google Scholar] [CrossRef]
- Choi, W.I.; Lee, J.H.; Kim, J.-Y.; Heo, S.U.; Jeong, Y.Y.; Kim, Y.H.; Tae, G. Targeted antitumor efficacy and imaging via multifunctional nano-carrier conjugated with anti-HER2 trastuzumab. Nanomed. Nanotechnol. Biol. Med. 2015, 11, 359–368. [Google Scholar] [CrossRef]
- Francia, V.; Montizaan, D.; Salvati, A. Interactions at the cell membrane and pathways of internalization of nano-sized materials for nanomedicine. Beilstein J. Nanotechnol. 2020, 11, 338–353. [Google Scholar] [CrossRef]
- Auría-Soro, C.; Nesma, T.; Juanes-Velasco, P.; Landeira-Viñuela, A.; Fidalgo-Gomez, H.; Acebes-Fernandez, V.; Gongora, R.; Almendral Parra, M.J.; Manzano-Roman, R.; Fuentes, M. Interactions of nanoparticles and biosystems: Microenvironment of nanoparticles and biomolecules in nanomedicine. Nanomaterials 2019, 9, 1365. [Google Scholar] [CrossRef]
- Biorender. Available online: https://www.biorender.com (accessed on 15 March 2024).
- Foroozandeh, P.; Aziz, A.A. Insight into Cellular Uptake and Intracellular Trafficking of Nanoparticles. Nanoscale Res. Lett. 2018, 13, 339. [Google Scholar] [CrossRef]
- Behzadi, S.; Serpooshan, V.; Tao, W.; Hamaly, M.A.; Alkawareek, M.Y.; Dreaden, E.C.; Brown, D.; Alkilany, A.M.; Farokhzad, O.C.; Mahmoudi, M. Cellular uptake of nanoparticles: Journey inside the cell. Chem. Soc. Rev. 2017, 46, 4218–4244. [Google Scholar] [CrossRef]
- Jiang, W.; Xie, H.; Ghoorah, D.; Shang, Y.; Shi, H.; Liu, F.; Yang, X.; Xu, H. Conjugation of functionalized SPIONs with transferrin for targeting and imaging brain glial tumors in rat model. PLoS ONE 2012, 7, e37376. [Google Scholar] [CrossRef]
- Chen, C.C.V.; Ku, M.C.; Jayaseema, D.M.; Lai, J.S.; Hueng, D.Y.; Chang, C. Simple SPION Incubation as an Efficient Intracellular Labeling Method for Tracking Neural Progenitor Cells Using MRI. PLoS ONE 2013, 8, e56125. [Google Scholar] [CrossRef][Green Version]
- Gharib, A.; Faezizadeh, Z.; Mesbah-Namin, S.A.R.; Saravani, R. Experimental treatment of breast cancer-bearing BALB/c mice by artemisinin and transferrin-loaded magnetic nanoliposomes. Pharmacogn. Mag. 2015, 11 (Suppl. S1), S117–S122. [Google Scholar] [CrossRef]
- Luther, E.M.; Petters, C.; Bulcke, F.; Kaltz, A.; Thiel, K.; Bickmeyer, U.; Dringen, R. Endocytotic uptake of iron oxide nanoparticles by cultured brain microglial cells. Acta Biomater. 2013, 9, 8454–8465. [Google Scholar] [CrossRef]
- Mazzolini, J.; Weber, R.J.M.; Chen, H.-S.; Khan, A.; Guggenheim, E.; Shaw, R.K.; Chipman, J.K.; Viant, M.R.; Rappoport, J.Z. Protein Corona Modulates Uptake and Toxicity of Nanoceria via Clathrin-Mediated Endocytosis. Biol. Bull. 2016, 231, 40–60. [Google Scholar] [CrossRef]
- Svitkova, B.; Zavisova, V.; Nemethova, V.; Koneracka, M.; Kretova, M.; Razga, F.; Ursinyova, M.; Gabelova, A. Differences in surface chemistry of iron oxide nanoparticles result in different routes of internalization. Beilstein J. Nanotechnol. 2021, 12, 270–281. [Google Scholar] [CrossRef]
- Bohmer, N.; Jordan, A. Caveolin-1 and CDC42 mediated endocytosis of silica-coated iron oxide nanoparticles in HeLa cells. Beilstein J. Nanotechnol. 2015, 6, 167–176. [Google Scholar] [CrossRef]
- Gandek, T.B.; van der Koog, L.; Nagelkerke, A. A Comparison of Cellular Uptake Mechanisms, Delivery Efficacy, and Intracellular Fate between Liposomes and Extracellular Vesicles. Adv. Healthc. Mater. 2003, 12, e2300319. [Google Scholar] [CrossRef]
- Portilla, Y.; Mulens-Arias, V.; Paradela, A.; Ramos-Fernández, A.; Pérez-Yagüe, S.; Morales, M.P.; Barber, D.F. The surface coating of iron oxide nanoparticles drives their intracellular trafficking and degradation in endolysosomes differently depending on the cell type. Biomaterials 2022, 281, 121365. [Google Scholar] [CrossRef]
- Tahara, T.; Shibata, T.; Nakamura, M.; Yamashita, H.; Yoshioka, D.; Okubo, M.; Maruyama, N.; Kamano, T.; Kamiya, Y.; Nakagawa, Y.; et al. Effect of MDR1 gene promoter methylation in patients with ulcerative colitis. Int. J. Mol. Med. 2009, 23, 521–527. [Google Scholar] [CrossRef]
- Zhao, K.; Ruan, L.; Liu, X.; Wu, L.; Cao, J.; Shen, S. Iron oxide nanoparticles served as the primary carrier to increase drug loading in macrophages. Biomed. Mater. 2023, 18, 15018. [Google Scholar] [CrossRef]
- Calero, M.; Gutiérrez, L.; Salas, G.; Luengo, Y.; Lázaro, A.; Acedo, P.; Morales, M.P.; Miranda, R.; Villanueva, A. Efficient and safe internalization of magnetic iron oxide nanoparticles: Two fundamental requirements for biomedical applications. Nanomed. Nanotechnol. Biol. Med. 2014, 10, 733–743. [Google Scholar] [CrossRef]
- Sun, Z.; Worden, M.; Wroczynskyj, Y.; Manna, P.K.; Thliveris, J.A.; van Lierop, J.; Hegmann, T.; Miller, D.W. Differential internalization of brick shaped iron oxide nanoparticles by endothelial cells. J. Mater. Chem. B 2016, 4, 5913–5920. [Google Scholar] [CrossRef]
- Freis, B.; Ramirez, M.D.L.A.; Kiefer, C.; Harlepp, S.; Iacovita, C.; Henoumont, C.; Affolter-Zbaraszczuk, C.; Meyer, F.; Mertz, D.; Boos, A.; et al. Effect of the Size and Shape of Dendronized Iron Oxide Nanoparticles Bearing a Targeting Ligand on MRI, Magnetic Hyperthermia, and Photothermia Properties—From Suspension to In Vitro Studies. Pharmaceutics 2023, 15, 1104. [Google Scholar] [CrossRef]
- Lei, W.; Min, W.; Hui, D.; Yun, L.; An, X. Effect of Surface Modification on Cellular Internalization of Fe3O4 Nanoparticles in Strong Static Magnetic Field. J. Nanosci. Nanotechnol. 2015, 15, 5184–5192. [Google Scholar] [CrossRef]
- Kenzaoui, B.H.; Vilà, M.R.; Miquel, J.M.; Cengelli, F.; Juillerat-Jeanneret, L. Evaluation of uptake and transport of cationic and anionic ultrasmall iron oxide nanoparticles by human colon cells. Int. J. Nanomed. 2012, 7, 1275–1286. [Google Scholar] [CrossRef]
- Zhang, W.; Cao, S.; Liang, S.; Tan, C.H.; Luo, B.; Xu, X.; Saw, P.E. Differently Charged Super-Paramagnetic Iron Oxide Nanoparticles Preferentially Induced M1-Like Phenotype of Macrophages. Front. Bioeng. Biotechnol. 2020, 8, 537. [Google Scholar] [CrossRef]
- Mahaling, B.; Katti, D.S. Physicochemical properties of core–shell type nanoparticles govern their spatiotemporal biodistribution in the eye. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 2149–2160. [Google Scholar] [CrossRef]
- Kim, E.; Kim, J.M.; Kim, L.; Choi, S.J.; Park, I.S.; Han, J.Y.; Chu, Y.C.; Choi, E.S.; Na, K.; Hong, S. The effect of neutral-surface iron oxide nanoparticles on cellular uptake and signaling pathways. Int. J. Nanomed. 2016, 11, 4595–4607. [Google Scholar] [CrossRef]
- Eshaghi, B.; Alsharif, N.; An, X.; Akiyama, H.; Brown, K.A.; Gummuluru, S.; Reinhard, B.M. Stiffness of HIV-1 Mimicking Polymer Nanoparticles Modulates Ganglioside-Mediated Cellular Uptake and Trafficking. Adv. Sci. 2020, 7, 2000649. [Google Scholar] [CrossRef]
- Perez, J.E.; Fage, F.; Pereira, D.; Abou-Hassan, A.; Asnacios, S.; Asnacios, A.; Wilhelm, C. Transient cell stiffening triggered by magnetic nanoparticle exposure. J. Nanobiotechnology 2021, 19, 117. [Google Scholar] [CrossRef]
- Kunzmann, A.; Andersson, B.; Vogt, C.; Feliu, N.; Ye, F.; Gabrielsson, S.; Toprak, M.S.; Buerki-Thurnherr, T.; Laurent, S.; Vahter, M.; et al. Efficient internalization of silica-coated iron oxide nanoparticles of different sizes by primary human macrophages and dendritic cells. Toxicol. Appl. Pharmacol. 2011, 253, 81–93. [Google Scholar] [CrossRef]
- Lee, S.H.; Park, D.J.; Yun, W.S.; Park, J.-E.; Choi, J.S.; Key, J.; Seo, Y.J. Endocytic trafficking of polymeric clustered superparamagnetic iron oxide nanoparticles in mesenchymal stem cells. J. Control. Release 2020, 326, 408–418. [Google Scholar] [CrossRef]
- Diaz-Diestra, D.M.; Palacios-Hernandez, T.; Liu, Y.; Smith, D.E.; Nguyen, A.K.; Todorov, T.; Gray, P.J.; Zheng, J.; Skoog, S.A.; Goering, P.L. Impact of Surface Chemistry of Ultrasmall Superparamagnetic Iron Oxide Nanoparticles on Protein Corona Formation and Endothelial Cell Uptake, Toxicity, and Barrier Function. Toxicol. Sci. 2022, 188, 261–275. [Google Scholar] [CrossRef]
- Wilson, L.; Brundle, R.; Wilson, G. Encyclopedia of Materials Characterization: Surfaces, Interfaces, Thin Films; Butterworth-Heinemann: Oxford, UK, 1992; ISBN 978-0-08-052360-6. [Google Scholar]
- Altammar, K.A. A review on nanoparticles: Characteristics, synthesis, applications, and challenges. Front. Microbiol. 2023, 14, 1155622. [Google Scholar] [CrossRef]
- Torres-Rivero, K.; Bastos-Arrieta, J.; Fiol, N.; Florido, A. Metal and Metal Oxide Nanoparticles: An Integrated Perspective of the Green Synthesis Methods by Natural Products and Waste Valorization: Applications and Challenges, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2021; Volume 94. [Google Scholar] [CrossRef]
- Arshad, F.; Hassan, I.U.; Naikoo, G.A. Chapter 3-Characterization of nanomaterials. In Woodhead Publishing Series in Biomaterials; Woodhead Publishing: Sawston, UK, 2023; pp. 37–61. [Google Scholar] [CrossRef]
- Ionescu, I.C. Surface microscopy study of metallic and ceramic orthodontic brackets. Rev. ROMÂNÅ Stomatol. 2015, LXI, 20–24. Available online: https://rjs.com.ro/articles/2015.1/RJS_2015_1_Art-03.pdf (accessed on 13 March 2024). [CrossRef]
- Lee, B.; Yoon, S.; Lee, J.W.; Kim, Y.; Chang, J.; Yun, J.; Ro, J.C.; Lee, J.-S.; Lee, J.H. Statistical Characterization of the Morphologies of Nanoparticles through Machine Learning Based Electron Microscopy Image Analysis. ACS Nano 2020, 14, 17125–17133. [Google Scholar] [CrossRef]
- Diaspro, A.; Bianchini, P.; Cella Zanacchi, F.; Usai, C. Optical Fluorescence Microscopy BT-Encyclopedia of Biophysics. In Encyclopedia of Biophysics; Roberts, G.C.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 1790–1792. [Google Scholar] [CrossRef]
- Alessio, P.; Aoki, P.H.B.; Furini, L.N.; Aliaga, A.E.; Leopoldo Constantino, C.J. 3-Spectroscopic Techniques for Characterization of Nanomaterials. In Nanocharacterization Techniques; William Andrew Publishing: New York, USA, 2017; pp. 65–98. [Google Scholar] [CrossRef]
- Jayawardena, H.S.N.; Liyanage, S.H.; Rathnayake, K.; Patel, U.; Yan, M. Analytical Methods for Characterization of Nanomaterial Surfaces. Anal. Chem. 2021, 93, 1889–1911. [Google Scholar] [CrossRef]
- Sanità, G.; Carrese, B.; Lamberti, A. Nanoparticle Surface Functionalization: How to Improve Biocompatibility and Cellular Internalization. Front. Mol. Biosci. 2020, 7, 587012. [Google Scholar] [CrossRef]
- Jagadeesh, P.; Rangappa, S.M.; Siengchin, S. Advanced characterization techniques for nanostructured materials in biomedical applications. Adv. Ind. Eng. Polym. Res. 2024, 7, 122–143. [Google Scholar] [CrossRef]
- Nicholas, T.P.; Haick, A.K.; Bammler, T.K.; Workman, T.W.; Kavanagh, T.J.; Faustman, E.M.; Gharib, S.A.; Altemeier, W.A. The Effects of Genotype × Phenotype Interactions on Transcriptional Response to Silver Nanoparticle Toxicity in Organotypic Cultures of Murine Tracheal Epithelial Cells. Toxicol. Sci. 2020, 173, 131–143. [Google Scholar] [CrossRef]
- Iglesias, O. Time-Dependent Phenomena in Nanoparticle Assemblies; Pan Stanford Publishing: Singapore, 2014; pp. 91–128. [Google Scholar] [CrossRef]
- Baabu, P.R.S.; Kumar, H.K.; Gumpu, M.B.; Babu, K.J.; Kulandaisamy, A.J.; Rayappan, J.B.B. Iron Oxide Nanoparticles: A Review on the Province of Its Compounds, Properties and Biological Applications. Materials 2023, 16, 59. [Google Scholar] [CrossRef]
- Penczek, P.A. Resolution measures in molecular electron microscopy. Methods Enzymol. 2010, 482, 73–100. [Google Scholar] [CrossRef]
- Microscope Resolution: Concepts, Factors and Calculation. Available online: https://www.leica-microsystems.com/science-lab/life-science/microscope-resolution-concepts-factors-and-calculation/ (accessed on 16 March 2024).
- Costanzo, M.; Vurro, F.; Cisterna, B.; Boschi, F.; Marengo, A.; Montanari, E.; Di Meo, C.; Matricardi, P.; Berlier, G.; Stella, B.; et al. Uptake and intracellular fate of biocompatible nanocarriers in cycling and noncycling cells. Nanomedicine 2019, 14, 301–316. [Google Scholar] [CrossRef]
- Burger, N.; Biswas, A.; Barzan, D.; Kirchner, A.; Hosser, H.; Hausmann, M.; Hildenbrand, G.; Herskind, C.; Wenz, F.; Veldwijk, M.R. A method for the efficient cellular uptake and retention of small modified gold nanoparticles for the radiosensitization of cells. Nanomedicine 2014, 10, 1365–1373. [Google Scholar] [CrossRef]
- Fouquet, C.; Gilles, J.-F.; Heck, N.; Dos Santos, M.; Schwartzmann, R.; Cannaya, V.; Morel, M.-P.; Davidson, R.S.; Trembleau, A.; Bolte, S. Improving axial resolution in confocal microscopy with new high refractive index mounting media. PLoS ONE 2015, 10, e0121096. [Google Scholar] [CrossRef]
- Reclusa, P.; Verstraelen, P.; Taverna, S.; Gunasekaran, M.; Pucci, M.; Pintelon, I.; Claes, N.; de Miguel-Pérez, D.; Alessandro, R.; Bals, S.; et al. Improving extracellular vesicles visualization: From static to motion. Sci. Rep. 2020, 10, 6494. [Google Scholar] [CrossRef]
- Simonsen, J.B.; Kromann, E.B. Pitfalls and opportunities in quantitative fluorescence-based nanomedicine studies—A commentary. J. Control. Release 2021, 335, 660–667. [Google Scholar] [CrossRef]
- Torrano, A.A.; Bräuchle, C. Precise quantification of silica and ceria nanoparticle uptake revealed by 3D fluorescence microscopy. Beilstein J. Nanotechnol. 2014, 5, 1616–1624. [Google Scholar] [CrossRef]
- Katebi, S.; Esmaeili, A.; Ghaedi, K.; Zarrabi, A. Superparamagnetic iron oxide nanoparticles combined with NGF and quercetin promote neuronal branching morphogenesis of PC12 cells. Int. J. Nanomed. 2019, 14, 2157–2169. [Google Scholar] [CrossRef]
- Reynders, H.; Van Zundert, I.; Silva, R.; Carlier, B.; Deschaume, O.; Bartic, C.; Rocha, S.; Basov, S.; Van Bael, M.J.; Himmelreich, U.; et al. Label-Free Iron Oxide Nanoparticles as Multimodal Contrast Agents in Cells Using Multi-Photon and Magnetic Resonance Imaging. Int. J. Nanomed. 2021, 16, 8375–8389. [Google Scholar] [CrossRef]
- Nowak-Jary, J.; Machnicka, B. Pharmacokinetics of magnetic iron oxide nanoparticles for medical applications. J. Nanobiotechnol. 2022, 20, 305. [Google Scholar] [CrossRef]
- Denora, N.; Lee, C.; Iacobazzi, R.M.; Choi, J.Y.; Song, I.H.; Yoo, J.S.; Piao, Y.; Lopalco, A.; Leonetti, F.; Lee, B.C.; et al. TSPO-targeted NIR-fluorescent ultra-small iron oxide nanoparticles for glioblastoma imaging. Eur. J. Pharm. Sci. 2019, 139, 105047. [Google Scholar] [CrossRef]
- Sun, M.; Sun, B.; Liu, Y.; Shen, Q.-D.; Jiang, S. Dual-Color Fluorescence Imaging of Magnetic Nanoparticles in Live Cancer Cells Using Conjugated Polymer Probes. Sci. Rep. 2016, 6, 22368. [Google Scholar] [CrossRef]
- Liu, M.; Li, Q.; Liang, L.; Li, J.; Wang, K.; Li, J.; Lv, M.; Chen, N.; Song, H.; Lee, J.; et al. Real-Time visualization of clustering and intracellular transport of gold nanoparticles by correlative imaging. Nat. Commun. 2017, 8, 15646. [Google Scholar] [CrossRef]
- Durymanov, M.; Permyakova, A.; Sene, S.; Guo, A.; Kroll, C.; Giménez-Marqués, M.; Serre, C.; Reineke, J. Cellular Uptake, Intracellular Trafficking, and Stability of Biocompatible Metal-Organic Framework (MOF) Particles in Kupffer Cells. Mol. Pharm. 2019, 16, 2315–2325. [Google Scholar] [CrossRef]
- Hoechst 33342 Solution. Available online: https://www.thermofisher.com/order/catalog/product/62249 (accessed on 16 March 2024).
- Mihailescu, M.; Miclea, L.C.; Pleava, A.M.; Tarba, N.; Scarlat, E.N.; Negoita, R.D.; Moisescu, M.G.; Savopol, T. Method for nanoparticles uptake evaluation based on double labeled fluorescent cells scanned in enhanced darkfield microscopy. Biomed. Opt. Express 2023, 14, 2796–2810. [Google Scholar] [CrossRef]
- Fakhrullin, R.; Nigamatzyanova, L.; Fakhrullina, G. Dark-field/hyperspectral microscopy for detecting nanoscale particles in environmental nanotoxicology research. Sci. Total Environ. 2021, 772, 145478. [Google Scholar] [CrossRef]
- Wang, B.; Sun, D.; Zhang, C.; Wang, K.; Bai, J. Dark-field microscopy for characterization of single molecule dynamics in vitro and in vivo. Anal. Methods 2019, 11, 2778–2784. [Google Scholar] [CrossRef]
- Su, L.; Zhang, B.; Huang, Y.; Fan, Z.; Zhao, Y. Enhanced cellular uptake of iron oxide nanoparticles modified with 1,2-dimyristoyl-sn-glycero-3-phosphocholine. RSC Adv. 2017, 7, 38001–38007. [Google Scholar] [CrossRef]
- Solorio-Rodríguez, A.; Escamilla-Rivera, V.; Uribe-Ramírez, M.; González-Pozos, S.; Hernández-Soto, J.; Rafael-Vázquez, L.; De Vizcaya-Ruiz, A. In vitro cytotoxicity study of superparamagnetic iron oxide and silica nanoparticles on pneumocyte organelles. Toxicol. Vitr. 2021, 72, 105071. [Google Scholar] [CrossRef]
- Popescu, R.C.; Andronescu, E.; Vasile, B.Ș.; Truşcă, R.; Boldeiu, A.; Mogoantă, L.; Mogoșanu, G.D.; Temelie, M.; Radu, M.; Grumezescu, A.M.; et al. Fabrication and Cytotoxicity of Gemcitabine-Functionalized Magnetite Nanoparticles. Molecules 2017, 22, 1080. [Google Scholar] [CrossRef]
- Knight, L.C.; Romano, J.E.; Krynska, B.; Faro, S.; Mohamed, F.B. Binding and Internalization of Iron Oxide Nanoparticles Targeted To Nuclear Oncoprotein. J. Mol. Biomark. Diagn. 2010, 1, 10000102. [Google Scholar] [CrossRef]
- Karageorgou, M.A.; Bouziotis, P.; Stiliaris, E.; Stamopoulos, D. Radiolabeled Iron Oxide Nanoparticles as Dual Modality Contrast Agents in SPECT/MRI and PET/MRI. Nanomaterials 2023, 13, 503. [Google Scholar] [CrossRef]
- Salvanou, E.-A.; Kolokithas-Ntoukas, A.; Liolios, C.; Xanthopoulos, S.; Paravatou-Petsotas, M.; Tsoukalas, C.; Avgoustakis, K.; Bouziotis, P. Preliminary Evaluation of Iron Oxide Nanoparticles Radiolabeled with 68Ga and 177Lu as Potential Theranostic Agents. Nanomaterials 2022, 12, 2490. [Google Scholar] [CrossRef]
- Turiel-Fernández, D.; Gutiérrez-Romero, L.; Corte-Rodriguez, M.; Bettmer, J.; Montes-Bayón, M. Ultrasmall iron oxide nanoparticles cisplatin (IV) prodrug nanoconjugate: ICP-MS based strategies to evaluate the formation and drug delivery capabilities in single cells. Anal. Chim. Acta 2021, 1159, 338356. [Google Scholar] [CrossRef]
- Zhang, Q.; Rajan, S.S.; Tyner, K.M.; Casey, B.J.; Dugard, C.K.; Jones, Y.; Paredes, A.M.; Clingman, C.S.; Howard, P.C.; Goering, P.L. Effects of iron oxide nanoparticles on biological responses and MR imaging properties in human mammary healthy and breast cancer epithelial cells: Human breast epithelial cell responses to iron oxide nanoparticles. J. Biomed. Mater. Res. Part B Appl. Biomater. 2015, 104, 1032–1042. [Google Scholar] [CrossRef]
- Comparison of ICP-OES and ICP-MS for Trace Element Analysis. Available online: https://www.thermofisher.com/ro/en/home/industrial/environmental/environmental-learning-center/contaminant-analysis-information/metal-analysis/comparison-icp-oes-icp-ms-trace-element-analysis.html (accessed on 16 March 2024).
- David, C.; Gonzalez de Vega, R. Facets of ICP-MS and their potential in the medical sciences-Part 2: Nanomedicine, immunochemistry, mass cytometry, and bioassays. Anal. Bioanal. Chem. 2022, 414, 7363–7386. [Google Scholar] [CrossRef]
- Fernández-Trujillo, S.; Jiménez-Moreno, M.; Rodríguez-Fariñas, N.; Rodríguez Martín-Doimeadios, R.C. Critical evaluation of the potential of ICP-MS-based systems in toxicological studies of metallic nanoparticles. Anal. Bioanal. Chem. 2024, 416, 2657–2676. [Google Scholar] [CrossRef]
- Theiner, S.; Loehr, K.; Koellensperger, G.; Mueller, L.; Jakubowski, N. Single-cell analysis by use of ICP-MS. J. Anal. At. Spectrom. 2020, 35, 1784–1813. [Google Scholar] [CrossRef]
- Yin, Q.; Pan, A.; Chen, B.; Wang, Z.; Tang, M.; Yan, Y.; Wang, Y.; Xia, H.; Chen, W.; Du, H.; et al. Quantitative imaging of intracellular nanoparticle exposure enables prediction of nanotherapeutic efficacy. Nat. Commun. 2021, 12, 2385. [Google Scholar] [CrossRef]
- Burducea, I.; Straticiuc, M.; Ghita, D.G.; Moșu, D.V.; Călinescu, C.I.; Podaru, N.; Mous, D.; Ursu, I.; Zamfir, N. A new ion beam facility based on a 3 MV TandetronTM at IFIN-HH, Romania. Nucl. Instrum. Methods Phys. Res. Sect. B-Beam Interact. Mater. At. 2015, 359, 12–19. [Google Scholar] [CrossRef]
- Salvati, A.; Nelissen, I.; Haase, A.; Åberg, C.; Moya, S.; Jacobs, U.; Alnasser, F.; Bewersdorff, T.; Deville, S.; Luch, A.; et al. Quantitative measurement of nanoparticle uptake by flow cytometry illustrated by an interlaboratory comparison of the uptake of labelled polystyrene nanoparticles. NanoImpact 2018, 9, 42–50. [Google Scholar] [CrossRef]
- Lozano, O.; Toussaint, O.; Dogné, J.M.; Lucas, S. The use of PIXE for engineered nanomaterials quantification in complex matrices. J. Phys. Conf. Ser. 2013, 429, 12010. [Google Scholar] [CrossRef]
- Chelarescu, E.; Dulama, I.; Gheboianu, A.; Bucurică, I.; Pacesila, D. PIXE Analytical Method Applied in the Study of Environment Samples used as Bioindicators. Rom. J. Phys. 2016, 61, 1369–1379. Available online: https://rjp.nipne.ro/2016_61_7-8/RomJPhys.61.p1369.pdf (accessed on 14 March 2024).
- Preoteasa, E.A.; Scafes, A.C.; Preoteasa, E.S.; Straja, D.; Iancu, D. Potential of in-Air Pixe for the Elemental Analysis of Dental Composites. Rom. J. Phys. 2022, 67, 1–23. Available online: https://rjp.nipne.ro/2022_67_5-6/RomJPhys.67.701.pdf (accessed on 14 March 2024).
- Costa, J.P.d.C.d.; Assis, M.; Teodoro, V.; Rodrigues, A.; Foggi, C.C.d.; San-Miguel, M.A.; Carmo, J.P.P.d.; Andrés, J.; Longo, E. Electron beam irradiation for the formation of thick Ag film on Ag3PO4. RSC Adv. 2020, 10, 21745–21753. [Google Scholar] [CrossRef]
- Ng, C.T.; Yong, L.Q.; Hande, M.P.; Ong, C.N.; Yu, L.E.; Bay, B.H.; Baeg, G.H. Zinc oxide nanoparticles exhibit cytotoxicity and genotoxicity through oxidative stress responses in human lung fibroblasts and Drosophila melanogaster. Int. J. Nanomed. 2017, 12, 1621–1637. [Google Scholar] [CrossRef]
- Hamzeh, M.; Sunahara, G.I. In vitro cytotoxicity and genotoxicity studies of titanium dioxide (TiO2) nanoparticles in Chinese hamster lung fibroblast cells. Toxicol. Vitr. 2013, 27, 864–873. [Google Scholar] [CrossRef]
- Takamiya, A.S.; Monteiro, D.R.; Bernabé, D.G.; Gorup, L.F.; Camargo, E.R.; Gomes-Filho, J.E.; Oliveira, S.H.P.; Barbosa, D.B. In Vitro and In Vivo Toxicity Evaluation of Colloidal Silver Nanoparticles Used in Endodontic Treatments. J. Endod. 2016, 42, 953–960. [Google Scholar] [CrossRef]
- Fraga, S.; Faria, H.; Soares, M.E.; Duarte, J.A.; Soares, L.; Pereira, E.; Costa-Pereira, C.; Teixeira, J.P.; Bastos, M.L.; Carmo, H. Influence of the surface coating on the cytotoxicity, genotoxicity and uptake of gold nanoparticles in human HepG2 cells. J. Appl. Toxicol. 2013, 33, 1111–1119. [Google Scholar] [CrossRef]
- Youhannayee, M.; Nakhaei-Rad, S.; Haghighi, F.; Klauke, K.; Janiak, C.; Ahmadian, M.R.; Rabenalt, R.; Albers, P.; Getzlaff, M. Physical characterization and uptake of iron oxide nanoparticles of different prostate cancer cells. J. Magn. Magn. Mater. 2019, 473, 205–214. [Google Scholar] [CrossRef]
- Xia, Q.; Zhang, Y.; Li, Z.; Hou, X.; Feng, N. Red blood cell membrane-camouflaged nanoparticles: A novel drug delivery system for antitumor application. Acta Pharm. Sin. B 2019, 9, 675–689. [Google Scholar] [CrossRef]
- Suh, K.S.; Lee, Y.S.; Seo, S.H.; Kim, Y.S.; Choi, E.M. Gold nanoparticles attenuates antimycin A-induced mitochondrial dysfunction in MC3T3-E1 osteoblastic cells. Biol. Trace Elem. Res. 2013, 153, 428–436. [Google Scholar] [CrossRef]
- Pan, L.; Pei, X.; He, R.; Wan, Q.; Wang, J. Multiwall carbon nanotubes/polycaprolactone composites for bone tissue engineering application. Colloids Surf. B Biointerfaces 2012, 93, 226–234. [Google Scholar] [CrossRef]
- Chen, Y.; Li, N.; Yang, Y.; Liu, Y. A dual targeting cyclodextrin/gold nanoparticle conjugate as a scaffold for solubilization and delivery of paclitaxel. RSC Adv. 2015, 5, 8938–8941. [Google Scholar] [CrossRef]
- Cacciatore, I.; Ciulla, M.; Fornasari, E.; Marinelli, L.; Di Stefano, A. Solid lipid nanoparticles as a drug delivery system for the treatment of neurodegenerative diseases. Expert Opin. Drug Deliv. 2016, 13, 1121–1131. [Google Scholar] [CrossRef]
- Assa, F.; Jafarizadeh-Malmiri, H.; Ajamein, H.; Vaghari, H.; Anarjan, N.; Ahmadi, O.; Berenjian, A. Chitosan magnetic nanoparticles for drug delivery systems. Crit. Rev. Biotechnol. 2017, 37, 492–509. [Google Scholar] [CrossRef]
- Cao, S.; Deng, Y.; Zhang, L.; Aleahmad, M. Chitosan nanoparticles, as biological macromolecule-based drug delivery systems to improve the healing potential of artificial neural guidance channels: A review. Int. J. Biol. Macromol. 2022, 201, 569–579. [Google Scholar] [CrossRef]
- Ozcelikkale, A.; Moon, H.-R.; Linnes, M.; Han, B. In vitro microfluidic models of tumor microenvironment to screen transport of drugs and nanoparticles. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2017, 9, e1460. [Google Scholar] [CrossRef]
- Asghar, W.; El Assal, R.; Shafiee, H.; Pitteri, S.; Paulmurugan, R.; Demirci, U. Engineering cancer microenvironments for in vitro 3-D tumor models. Mater. Today 2015, 18, 539–553. [Google Scholar] [CrossRef]
- Adityan, S.; Tran, M.; Bhavsar, C.; Wu, S.Y. Nano-therapeutics for modulating the tumour microenvironment: Design, development, and clinical translation. J. Control. Release 2020, 327, 512–532. [Google Scholar] [CrossRef]
- Chaicharoenaudomrung, N.; Kunhorm, P.; Noisa, P. Three-dimensional cell culture systems as an in vitro platform for cancer and stem cell modeling. World J. Stem Cells 2019, 11, 1065–1083. [Google Scholar] [CrossRef]
- Badr-Eldin, S.M.; Aldawsari, H.M.; Kotta, S.; Deb, P.K.; Venugopala, K.N. Three-Dimensional In Vitro Cell Culture Models for Efficient Drug Discovery: Progress So Far and Future Prospects. Pharmaceuticals 2022, 15, 926. [Google Scholar] [CrossRef]
- Florczyk, S.J.; Kievit, F.M.; Wang, K.; Erickson, A.E.; Ellenbogen, R.G.; Zhang, M. 3D Porous Chitosan-Alginate Scaffolds Promote Proliferation and Enrichment of Cancer Stem-Like Cells. J. Mater. Chem. B 2016, 4, 6326–6334. [Google Scholar] [CrossRef]
- Choi, Y.-J.; Jun, Y.-J.; Kim, D.Y.; Yi, H.-G.; Chae, S.-H.; Kang, J.; Lee, J.; Gao, G.; Kong, J.-S.; Jang, J.; et al. A 3D cell printed muscle construct with tissue-derived bioink for the treatment of volumetric muscle loss. Biomaterials 2019, 206, 160–169. [Google Scholar] [CrossRef]
- Thoma, C.R.; Zimmermann, M.; Agarkova, I.; Kelm, J.M.; Krek, W. 3D cell culture systems modeling tumor growth determinants in cancer target discovery. Adv. Drug Deliv. Rev. 2014, 69–70, 29–41. [Google Scholar] [CrossRef]
- Jubelin, C.; Muñoz-Garcia, J.; Griscom, L.; Cochonneau, D.; Ollivier, E.; Heymann, M.-F.; Vallette, F.M.; Oliver, L.; Heymann, D. Three-dimensional in vitro culture models in oncology research. Cell Biosci. 2022, 12, 155. [Google Scholar] [CrossRef]
- Jubelin, C.; Muñoz-Garcia, J.; Cochonneau, D.; Ollivier, E.; Vallette, F.; Heymann, M. Technical report: Liquid overlay technique allows the generation of homogeneous osteosarcoma, glioblastoma, lung and prostate adenocarcinoma spheroids that can be used for drug cytotoxicity measurements. Front. Bioeng. Biotechnol. 2023, 11, 1260049. [Google Scholar] [CrossRef]
- Van Zundert, I.; Fortuni, B.; Rocha, S. From 2D to 3D cancer cell models—The enigmas of drug delivery research. Nanomaterials 2020, 10, 2236. [Google Scholar] [CrossRef]
- Sokolova, V.; Ebel, J.-F.; Kollenda, S.; Klein, K.; Kruse, B.; Veltkamp, C.; Lange, C.M.; Westendorf, A.M.; Epple, M. Uptake of Functional Ultrasmall Gold Nanoparticles in 3D Gut Cell Models. Small 2022, 18, 2201167. [Google Scholar] [CrossRef]
- Tofani, L.B.; Luiz, M.T.; Paes Dutra, J.A.; Abriata, J.P.; Chorilli, M. Three-dimensional culture models: Emerging platforms for screening the antitumoral efficacy of nanomedicines. Nanomedicine 2023, 18, 633–647. [Google Scholar] [CrossRef]
- Tchoryk, A.; Taresco, V.; Argent, R.H.; Ashford, M.; Gellert, P.R.; Stolnik, S.; Grabowska, A.; Garnett, M.C. Penetration and uptake of nanoparticles in 3D tumor spheroids. Bioconjug. Chem. 2019, 30, 1371–1384. [Google Scholar] [CrossRef]
- Soeiro, J.F.; Sousa, F.L.; Monteiro, M.V.; Gaspar, V.M.; Silva, N.J.O.; Mano, J.F. Advances in screening hyperthermic nanomedicines in 3D tumor models. Nanoscale Horiz. 2024, 9, 334–364. [Google Scholar] [CrossRef]
- Foglietta, F.; Serpe, L.; Canaparo, R. The effective combination between 3d cancer models and stimuli-responsive nanoscale drug delivery systems. Cells 2021, 10, 3295. [Google Scholar] [CrossRef]
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning From 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 33. [Google Scholar] [CrossRef]
- Biju, T.S.; Priya, V.V.; Francis, A.P. Role of three-dimensional cell culture in therapeutics and diagnostics: An updated review. Drug Deliv. Transl. Res. 2023, 13, 2239–2253. [Google Scholar] [CrossRef]
- Toh, Y.-C.; Raja, A.; Yu, H.; van Noort, D. A 3D Microfluidic Model to Recapitulate Cancer Cell Migration and Invasion. Bioengineering 2018, 5, 29. [Google Scholar] [CrossRef]
- Lee, S.W.L.; Adriani, G.; Ceccarello, E.; Pavesi, A.; Tan, A.T.; Bertoletti, A.; Kamm, R.D.; Wong, S.C. Characterizing the Role of Monocytes in T Cell Cancer Immunotherapy Using a 3D Microfluidic Model. Front. Immunol. 2018, 9, 310560. [Google Scholar] [CrossRef]
- Sung, K.E.; Beebe, D.J. Microfluidic 3D models of cancer. Adv. Drug Deliv. Rev. 2014, 79–80, 68–78. [Google Scholar] [CrossRef]
- Guller, A.E.; Grebenyuk, P.N.; Shekhter, A.B.; Zvyagin, A.V.; Deyev, S.M. Bioreactor-Based Tumor Tissue Engineering. Acta Naturae 2016, 8, 44–58. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5081698/ (accessed on 12 March 2024). [CrossRef]
- Koh, M.A.Y.; Horst, E.; Neale, D.; Royzenblat, S.; Lahann, J.; Greineder, C.; Weivoda, M.; Mehta, G.; Keller, E.T. A Bioreactor for 3D In Vitro Modeling of the Mechanical Stimulation of Osteocytes. Front. Bioeng. Biotechnol. 2022, 10, 797542. [Google Scholar] [CrossRef]
- Bisgin, A.; Mujde, C. Bioreactor-Based Tissue Models as an Alternative Approach in Cancer Research BT-Handbook of Animal Models and its Uses in Cancer Research; Pathak, S., Banerjee, A., Bisgin, A., Eds.; Springer Nature: Singapore, 2022; pp. 1–16. [Google Scholar] [CrossRef]
- Altmann, B.; Grün, C.; Nies, C.; Gottwald, E. Advanced 3D cell culture techniques in micro-bioreactors, part II: Systems and applications. Processes 2020, 9, 21. [Google Scholar] [CrossRef]
- Bauer, M.; Su, G.; Beebe, D.J.; Friedl, A. 3D microchannel co-culture: Method and biological validation. Integr. Biol. 2010, 2, 371–378. [Google Scholar] [CrossRef][Green Version]
- Charelli, L.E.; Müller, N.; Silva, K.R.; Lima, L.M.T.R.; Sant’Anna, C.; Baptista, L.S. Biologically produced silver chloride nanoparticles from B. megaterium modulate interleukin secretion by human adipose stem cell spheroids. Cytotechnology 2018, 70, 1655–1669. [Google Scholar] [CrossRef]
- Sun, M.; Lee, J.; Chen, Y.; Hoshino, K. Studies of nanoparticle delivery with in vitro bio-engineered microtissues. Bioact. Mater. 2020, 5, 924–937. [Google Scholar] [CrossRef]
- Natarajan, P.; Tomich, J.M. Understanding the influence of experimental factors on bio-interactions of nanoparticles: Towards improving correlation between in vitro and in vivo studies. Arch. Biochem. Biophys. 2020, 694, 108592. [Google Scholar] [CrossRef]
- Lin, Q.; Fathi, P.; Chen, X. Nanoparticle delivery in vivo: A fresh look from intravital imaging. EBioMedicine 2020, 59, 102958. [Google Scholar] [CrossRef]
- Klapproth, A.P.; Shevtsov, M.; Stangl, S.; Li, W.B.; Multhoff, G. A New Pharmacokinetic Model Describing the Biodistribution of Intravenously and Intratumorally Administered Superparamagnetic Iron Oxide Nanoparticles (SPIONs) in a GL261 Xenograft Glioblastoma Model. Int. J. Nanomed. 2020, 15, 4677–4689. [Google Scholar] [CrossRef]
- Tate, J.A.; Petryk, A.A.; Giustini, A.J.; Hoopes, P.J. In vivo biodistribution of iron oxide nanoparticles: An overview. Proc. SPIE—Int. Soc. Opt. Eng. 2011, 7901, 790117. [Google Scholar] [CrossRef]
- Levy, M.; Luciani, N.; Alloyeau, D.; Elgrabli, D.; Deveaux, V.; Pechoux, C.; Chat, S.; Wang, G.; Vats, N.; Gendron, F.; et al. Long term in vivo biotransformation of iron oxide nanoparticles. Biomaterials 2011, 32, 3988–3999. [Google Scholar] [CrossRef]
- Ashby, J.; Pan, S.; Zhong, W. Size and surface functionalization of iron oxide nanoparticles influence the composition and dynamic nature of their protein corona. ACS Appl. Mater. Interfaces 2014, 6, 15412–15419. [Google Scholar] [CrossRef]
- Boyer, C.; Whittaker, M.R.; Bulmus, V.; Liu, J.; Davis, T.P. The design and utility of polymer-stabilized iron-oxide nanoparticles for nanomedicine applications. NPG Asia Mater. 2010, 2, 23–30. [Google Scholar] [CrossRef]
- Hyun Cho, I.K. Chapter 9—Microglia and Macrophages in Central Nervous Systems; Cho, M.D., Ed.; Academic Press: Cambridge, MA, USA, 2022; pp. 185–208. [Google Scholar] [CrossRef]
- Wani, T.U.; Raza, S.N.; Khan, N.A. Nanoparticle opsonization: Forces involved and protection by long chain polymers. Polym. Bull. 2020, 77, 3865–3889. [Google Scholar] [CrossRef]
- Mao, R.; Wang, J.; Pei, J.; Wu, S.; Feng, J.; Lin, Y.; Cai, X. Pharmacokinetics and Applications of Magnetic Nanoparticles. Curr. Drug Metab. 2013, 14, 872–878. [Google Scholar] [CrossRef][Green Version]
- Slavu, L.M.; Antonelli, A.; Scarpa, E.S.; Abdalla, P.; Wilhelm, C.; Silvestri, N.; Pellegrino, T.; Scheffler, K.; Magnani, M.; Rinaldi, R.; et al. Optimization of magnetic nanoparticles for engineering erythrocytes as theranostic agents. Biomater. Sci. 2023, 11, 3252–3268. [Google Scholar] [CrossRef]
- Yaremenko, A.V.; Zelepukin, I.V.; Ivanov, I.N.; Melikov, R.O.; Pechnikova, N.A.; Dzhalilova, D.S.; Mirkasymov, A.B.; Bragina, V.A.; Nikitin, M.P.; Deyev, S.M.; et al. Influence of magnetic nanoparticle biotransformation on contrasting efficiency and iron metabolism. J. Nanobiotechnol. 2022, 20, 535. [Google Scholar] [CrossRef]
- Poon, W.; Zhang, Y.-N.; Ouyang, B.; Kingston, B.R.; Wu, J.L.Y.; Wilhelm, S.; Chan, W.C.W. Elimination Pathways of Nanoparticles. ACS Nano 2019, 13, 5785–5798. [Google Scholar] [CrossRef]
- Liu, T.; Bai, R.; Zhou, H.; Wang, R.; Liu, J.; Zhao, Y.; Chen, C. The effect of size and surface ligands of iron oxide nanoparticles on blood compatibility. RSC Adv. 2020, 10, 7559–7569. [Google Scholar] [CrossRef]
- Sharma, A.; Cornejo, C.; Mihalic, J.; Geyh, A.; Bordelon, D.E.; Korangath, P.; Westphal, F.; Gruettner, C.; Ivkov, R. Physical characterization and in vivo organ distribution of coated iron oxide nanoparticles. Sci. Rep. 2018, 8, 4916. [Google Scholar] [CrossRef]
- Xie, M.; Li, Y.; Xu, Y.; Zhang, Z.; Ji, B.; Jones, J.B.; Wang, Z.; Mao, H. Brain Tumor Imaging and Delivery of Sub-5 nm Magnetic Iron Oxide Nanoparticles in an Orthotopic Murine Model of Glioblastoma. ACS Appl. Nano Mater. 2022, 5, 9706–9718. [Google Scholar] [CrossRef]
- Feng, X.; Xue, Y.; Gonca, S.; Ji, K.; Zhang, M.; García-García, F.R.; Li, Q.; Huang, Y.; Kamenev, K.V.; Chen, X. Ultrasmall superparamagnetic iron oxide nanoparticles for enhanced tumor penetration. J. Mater. Chem. B 2023, 11, 3422–3433. [Google Scholar] [CrossRef]
- Mao, Y.; Li, Y.; Zang, F.; Yu, H.; Yan, S.; Song, Q.; Qin, Z.; Sun, J.; Chen, B.; Huang, X.; et al. Continuous synthesis of extremely small-sized iron oxide nanoparticles used for T 1-weighted magnetic resonance imaging via a fluidic reactor. Sci. China Mater. 2022, 65, 1646–1654. [Google Scholar] [CrossRef]
- Radosinska, J.; Jasenovec, T.; Radosinska, D.; Balis, P.; Puzserova, A.; Skratek, M.; Manka, J.; Bernatova, I. Ultra-small superparamagnetic iron-oxide nanoparticles exert different effects on erythrocytes in normotensive and hypertensive rats. Biomedicines 2021, 9, 377. [Google Scholar] [CrossRef]
- Orlando, A.; Colombo, M.; Prosperi, D.; Gregori, M.; Panariti, A.; Rivolta, I.; Masserini, M.; Cazzaniga, E. Iron oxide nanoparticles surface coating and cell uptake affect biocompatibility and inflammatory responses of endothelial cells and macrophages. J. Nanopart. Res. 2015, 17, 351. [Google Scholar] [CrossRef]
- Laurent, S.; Henoumont, C.; Stanicki, D.; Boutry, S.; Lipani, E.; Belaid, S.; Muller, R.N.; Vander Elst, L. Superparamagnetic Iron Oxide Nanoparticles BT-MRI Contrast Agents: From Molecules to Particles; Springer: Singapore, 2017; pp. 55–109. [Google Scholar] [CrossRef]
- Gul, W.; Alrobei, H.; Shah, S.R.A.; Khan, A. Effect of iron oxide nanoparticles on the physical properties of medium density fiberboard. Polymers 2020, 12, 2911. [Google Scholar] [CrossRef]
- Abbasi, R.; Shineh, G.; Mobaraki, M.; Doughty, S.; Tayebi, L. Structural Parameters of Nanoparticles Affecting Their Toxicity for Biomedical Applications: A Review; Springer: Dordrecht, The Netherlands, 2023; Volume 25. [Google Scholar] [CrossRef]
- Arami, H.; Khandhar, A.; Liggitt, D.; Krishnan, K.M. In vivo delivery, pharmacokinetics, biodistribution and toxicity of iron oxide nanoparticles. Chem. Soc. Rev. 2015, 44, 8576–8607. [Google Scholar] [CrossRef]
- Popescu, R.C.; Vasile, B.Ş.; Savu, D.I.; Mogoşanu, G.D.; Bejenaru, L.E.; Andronescu, E.; Grumezescu, A.M.; Mogoantă, L. Influence of Polymer Shell Molecular Weight on Functionalized Iron Oxide Nanoparticles Morphology and In Vivo Biodistribution. Pharmaceutics 2022, 14, 1877. [Google Scholar] [CrossRef]
- Song, X.; Zheng, M.; Hu, H.; Chen, L.; Wang, S.; Ding, Z.; Fu, G.; Sun, L.; Zhao, L.; Zhang, L.; et al. Pharmacokinetic Study of Ultrasmall Superparamagnetic Iron Oxide Nanoparticles HY-088 in Rats. Eur. J. Drug Metab. Pharmacokinet. 2024, 49, 317–330. [Google Scholar] [CrossRef]
- Ali, A.; Zafar, H.; Zia, M.; Haq, I.U.; Phull, A.R.; Ali, J.S.; Hussain, A. Synthesis, characterization, applications, and challenges of iron oxide nanoparticles. Nanotechnol. Sci. Appl. 2016, 9, 49–67. [Google Scholar] [CrossRef]
- Perry, J.L.; Reuter, K.G.; Kai, M.P.; Herlihy, K.P.; Jones, S.W.; Luft, J.C.; Napier, M.; Bear, J.E.; DeSimone, J.M. PEGylated PRINT nanoparticles: The impact of PEG density on protein binding, macrophage association, biodistribution, and pharmacokinetics. Nano Lett. 2012, 12, 5304–5310. [Google Scholar] [CrossRef]
- Cole, A.J.; David, A.E.; Wang, J.; Galbán, C.J.; Yang, V.C. Magnetic brain tumor targeting and biodistribution of long-circulating PEG-modified, cross-linked starch-coated iron oxide nanoparticles. Biomaterials 2011, 32, 6291–6301. [Google Scholar] [CrossRef]
- Shubayev, V.I.; Pisanic, T.R., 2nd; Jin, S. Magnetic nanoparticles for theragnostics. Adv. Drug Deliv. Rev. 2009, 61, 467–477. [Google Scholar] [CrossRef]









| NPs Property | Parameter | System Description | Cell Type | Internalization Mechanism | Method | Observation on Internalization Efficiency | Reference |
|---|---|---|---|---|---|---|---|
| Size | 10 nm 70 nm 200 nm | protoporphyrin IX/IONPs | RAW264.7 cells | Endocytosis | FM | 70 nm NPs > 10 nm or 200 nm NPs ← more active in stimulating membrane receptors. | [62] |
| 60 nm, 110 nm, 142 nm | IONPs @ APTES, DMSA, AD | HeLa cells | Energy-dependent endocytosis | OM | Lower-hydrodynamic-diameter NPs > high-hydrodynamic-diameter NPs ← require less energy. | [63] | |
| Shape | Spheres, Bricks | IONPs | bEnd.3 cells | Caveolin-mediated endocytosis | [64] | Bricks > spheres ← interference with the caveolae. | |
| Spheres, Cubes, Plateles | IONPs | FaDu cells | Endocytosis | [65] | High-length IONP cubes > spheres and platelets ← they form aligned clusters. | ||
| Surface charge | Cationic, anionic | IONPs—CHIT, DEX, PAA, PEG, PC | A549 cells | Endocytosis | CM and TEM | Cationic IONPs > anionic IONPs. | [66] |
| Cationic, anionic | IONPs @ APTES, DMSA, AD | HeLa cells | Energy-dependent endocytosis | OM | Cationic IONPs > anionic IONPs in HeLa cells. | [63] | |
| Cationic, anionic | IONPs @ aminoPVA, OA | HT-29 and Caco-2 cells | Not studied | OM | Cationic IONPs > anionic IONPs in 2D cell models. Cationic NPs invade HT-29, Caco-2 3D cell spheroids. Anionic NPs invade only Caco-2 spheroids. None of the NPs cross the 3D membrane models. | [67] | |
| Cationic, anionic, neutral | Not described | RAW264.7 cells | Not studied | UV-VIS | Cationic and anionic IONPs > neutral IONPs ← non-specific electrostatic interactions with membrane proteins. | [68] | |
| Hydro-phobicity/Hydrophilicity | Hydro-philic | Hydrophobic Core- hydrophilic shell NPs of PLGA, PLGA@ CHI, PLGA@ PF68, PLGA@ GEL, PLA@ GEL, PCL@ GEL loaded with coumarin | In vivo biodistribution in mouse eye model | Passive transport | FM | Hydrophilic NPs → follow the conjunctival pathway in the eye → pass from clear to the iris–ciliary body through vessel uptake. | [69] |
| Hydro-phobic | IONPs@ MPS | HAoECs | Endocytosis | OM and FM | MPS-coated IONPs are internalized in HAoECs ← absorption through the plasma membrane is facilitated by the hydrophobic NPs. | [70] | |
| Rigidity | Stiffness | GM3- lipid- PLGA-PLA NPs | CD169, expressing macro-phage cells | Actin- dependent phagocytosis | FM | NPs with the stiffest cores are internalized in a higher manner in activated macrophages. | [71] |
| Stiffness | ALG@ lipidic bilayer | MDA-MB-231, MCF7, MCF10A cells | Not studied | FM | NPs with the highest stiffness are internalized in a lower manner in breast cancer cells. | [72] | |
| Functional groups | OH, NH2, COOH | IONPs @ BSA, PEG | A549 cells | Clathrin- mediated endocytosis and caveolin- mediated endocytosis | FM | BSA-coated IONPs are internalized via clathrin-mediated endocytosis ← (NH2) and (COOH). PEG-coated nanoparticles are taken up via caveolin-mediated endocytosis ← (OH). | [57] |
| OH | IONPs @ SiO2, DEX | HMDM, MDDC cells | Active actin cytoskeleton- dependent mechanism | TEM | IONPs@ SiO2 > DEX- coated IONPs ← the coating material can affect the protein interaction. | [73] | |
| OH | IONPs/PLGA/Cy5.5 | MSCs cells | Clathrin- mediated endocytosis | FM | SPION-clustered PLGA with average hydrodynamic size of 115.2 nm and negative charge are internalized in MSCs. | [74] | |
| COOH, NH2 | IONPs@ amphiphilic polymer terminated with (COOH) or (NH2) groups | HCAEC cells | Vesicle- mediated | TEM, ICP-MS | Both types of nanoparticles are internalized through vesicles. IONPs with (COOH) > IONPs with (NH2) groups. | [75] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petcov, T.E.; Straticiuc, M.; Iancu, D.; Mirea, D.A.; Trușcă, R.; Mereuță, P.E.; Savu, D.I.; Mogoșanu, G.D.; Mogoantă, L.; Popescu, R.C.; et al. Unveiling Nanoparticles: Recent Approaches in Studying the Internalization Pattern of Iron Oxide Nanoparticles in Mono- and Multicellular Biological Structures. J. Funct. Biomater. 2024, 15, 169. https://doi.org/10.3390/jfb15060169
Petcov TE, Straticiuc M, Iancu D, Mirea DA, Trușcă R, Mereuță PE, Savu DI, Mogoșanu GD, Mogoantă L, Popescu RC, et al. Unveiling Nanoparticles: Recent Approaches in Studying the Internalization Pattern of Iron Oxide Nanoparticles in Mono- and Multicellular Biological Structures. Journal of Functional Biomaterials. 2024; 15(6):169. https://doi.org/10.3390/jfb15060169
Chicago/Turabian StylePetcov, Teodora Eliana, Mihai Straticiuc, Decebal Iancu, Dragoș Alexandru Mirea, Roxana Trușcă, Paul Emil Mereuță, Diana Iulia Savu, George Dan Mogoșanu, Laurențiu Mogoantă, Roxana Cristina Popescu, and et al. 2024. "Unveiling Nanoparticles: Recent Approaches in Studying the Internalization Pattern of Iron Oxide Nanoparticles in Mono- and Multicellular Biological Structures" Journal of Functional Biomaterials 15, no. 6: 169. https://doi.org/10.3390/jfb15060169
APA StylePetcov, T. E., Straticiuc, M., Iancu, D., Mirea, D. A., Trușcă, R., Mereuță, P. E., Savu, D. I., Mogoșanu, G. D., Mogoantă, L., Popescu, R. C., Kopatz, V., & Jinga, S. I. (2024). Unveiling Nanoparticles: Recent Approaches in Studying the Internalization Pattern of Iron Oxide Nanoparticles in Mono- and Multicellular Biological Structures. Journal of Functional Biomaterials, 15(6), 169. https://doi.org/10.3390/jfb15060169








