Effects of Nickel Nanoparticles on Rhodococcus Cell Surface Morphology and Nanomechanical Properties
Abstract
:1. Introduction
2. Materials and Methods
2.1. Nickel Nanoparticles
2.2. Bacterial Strains and Culture Conditions
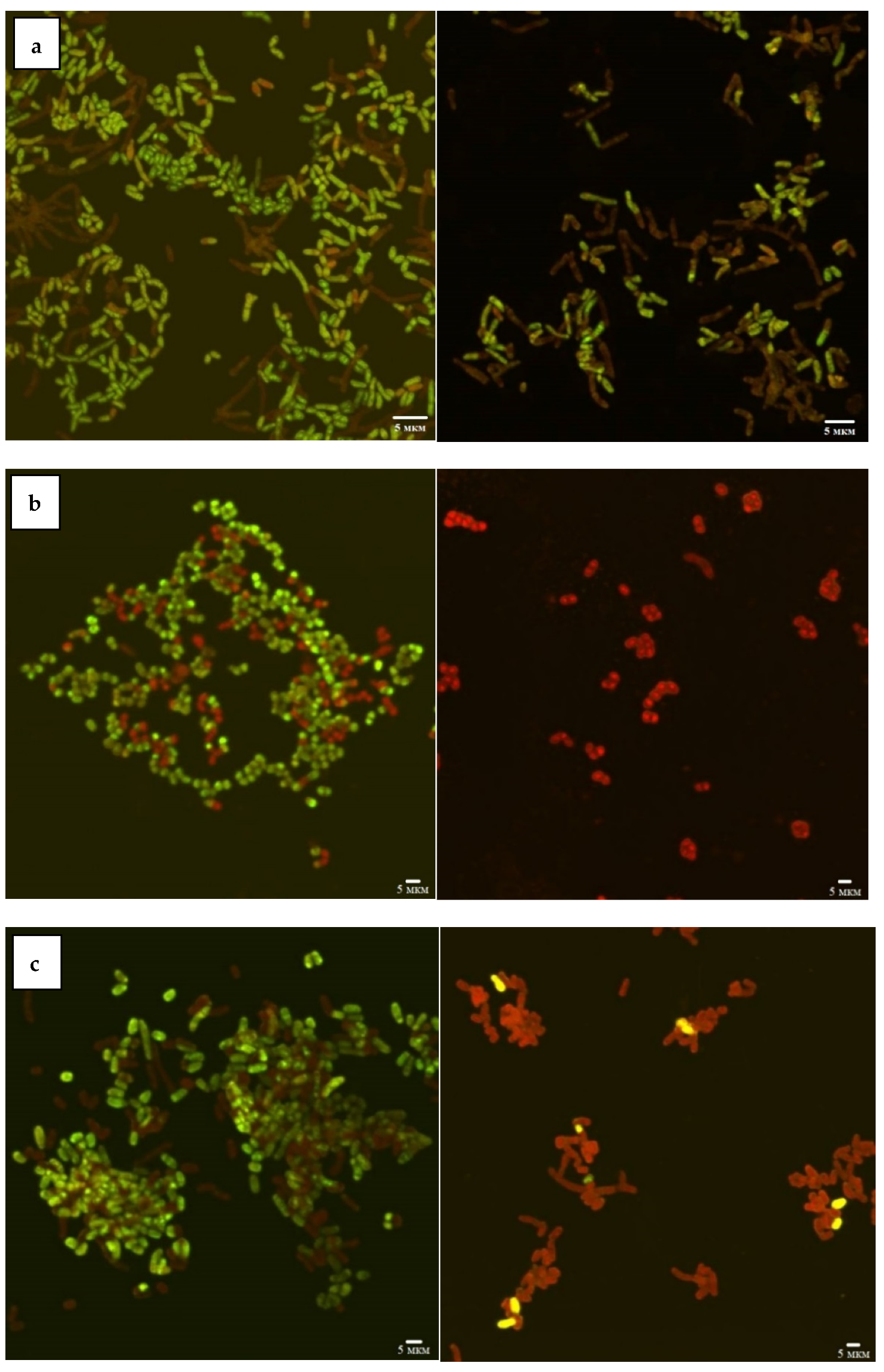
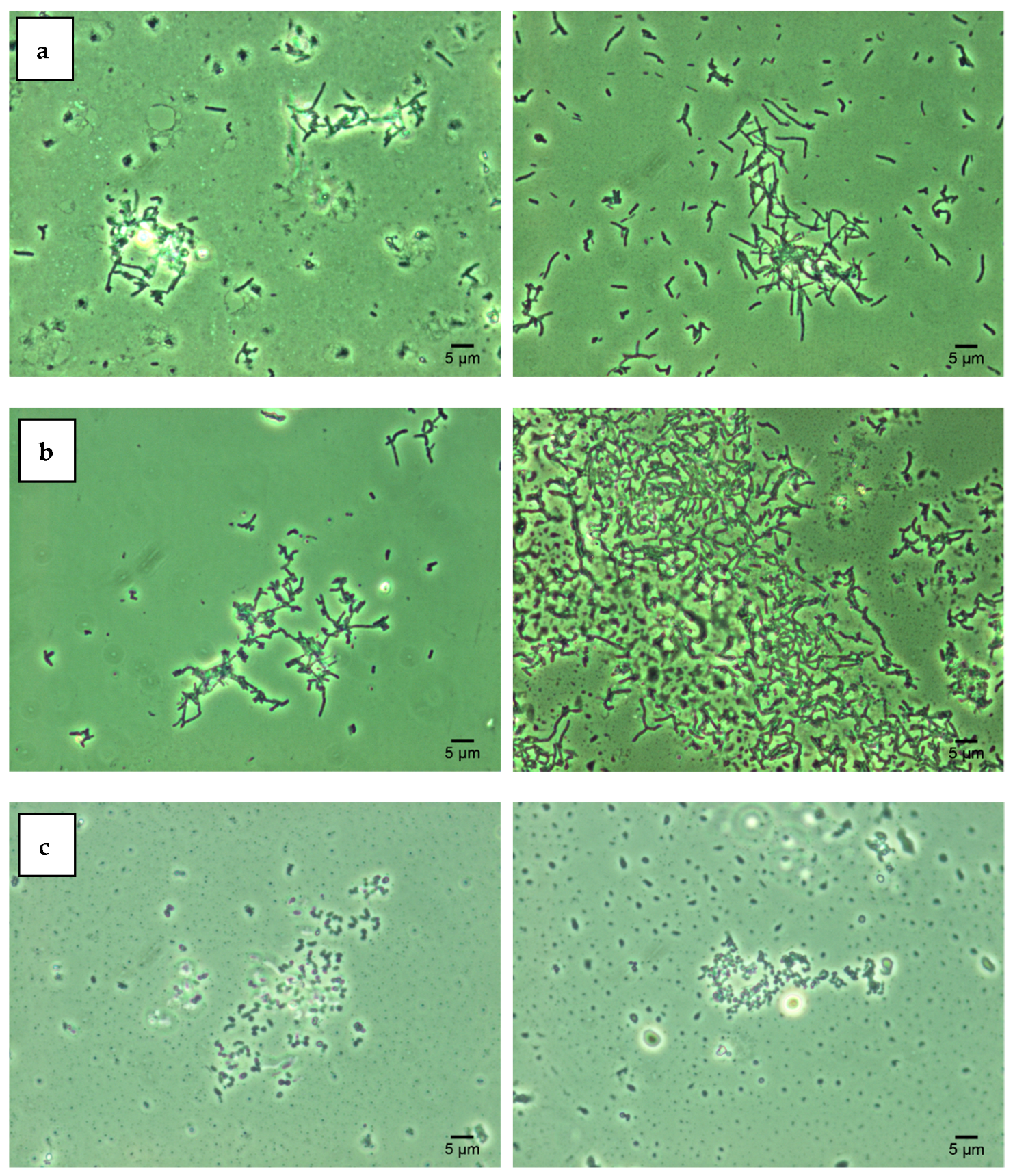
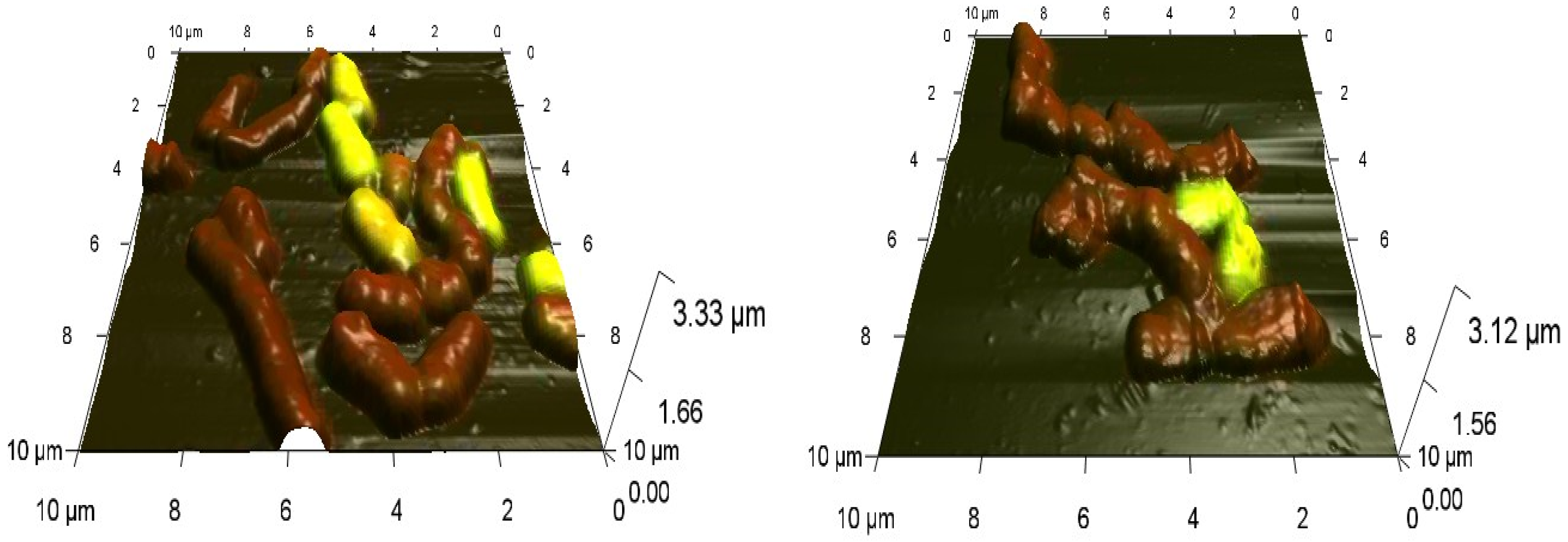
2.3. Combined Confocal Laser and Atomic Force Scanning
2.4. Nanomechanical Properties Using AFM Spectroscopy
2.5. Statistical Analysis
3. Results and Discussion
3.1. Effects of Nickel NPs on Viability and Hydrophobic Properties of Rhodococcus Cells
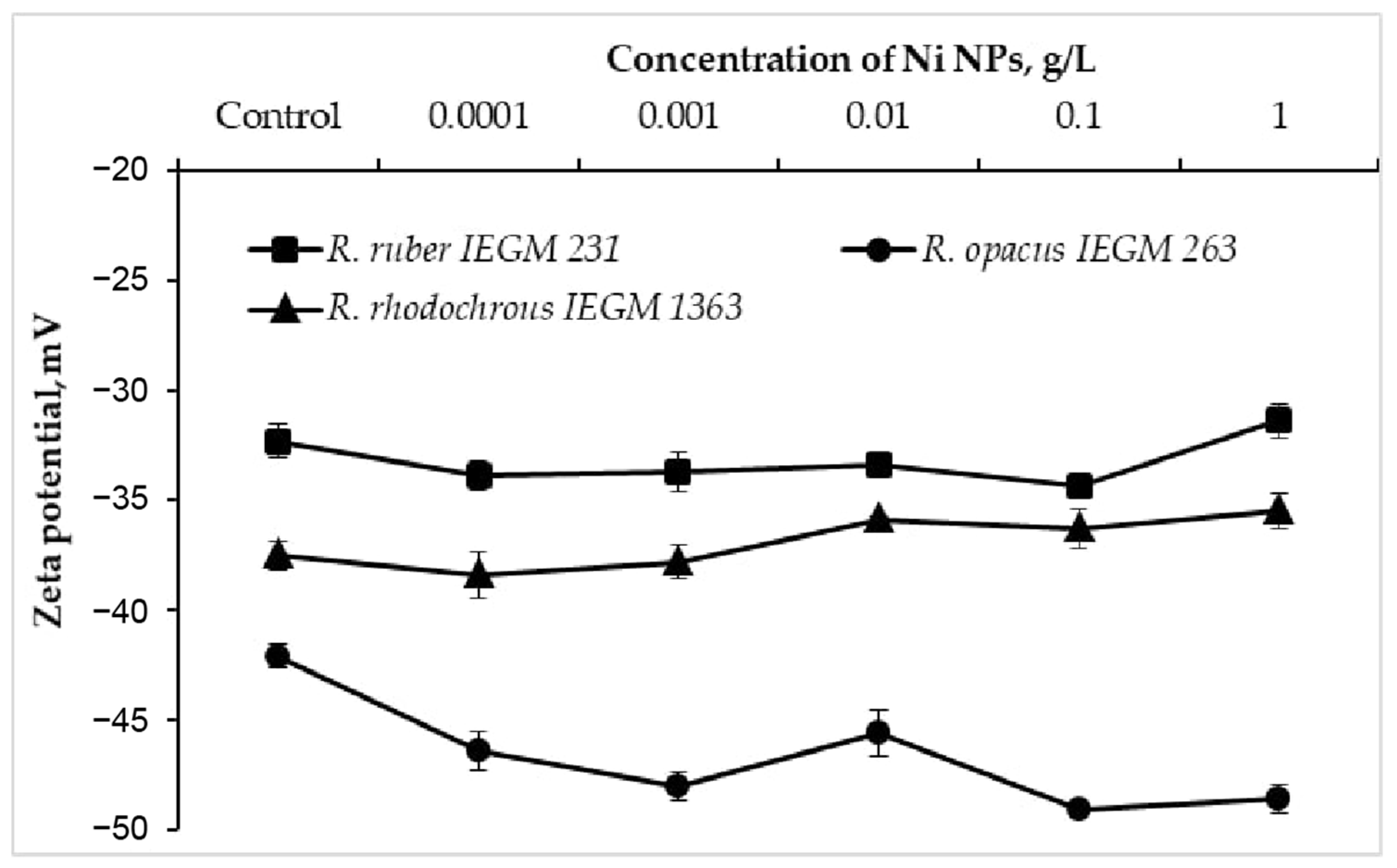
3.2. Effects of Nickel NPs on Zeta Potentials of Rhodococcus Cells
3.3. Effects of Nickel NPs on the Cell Size and Surface Roughness
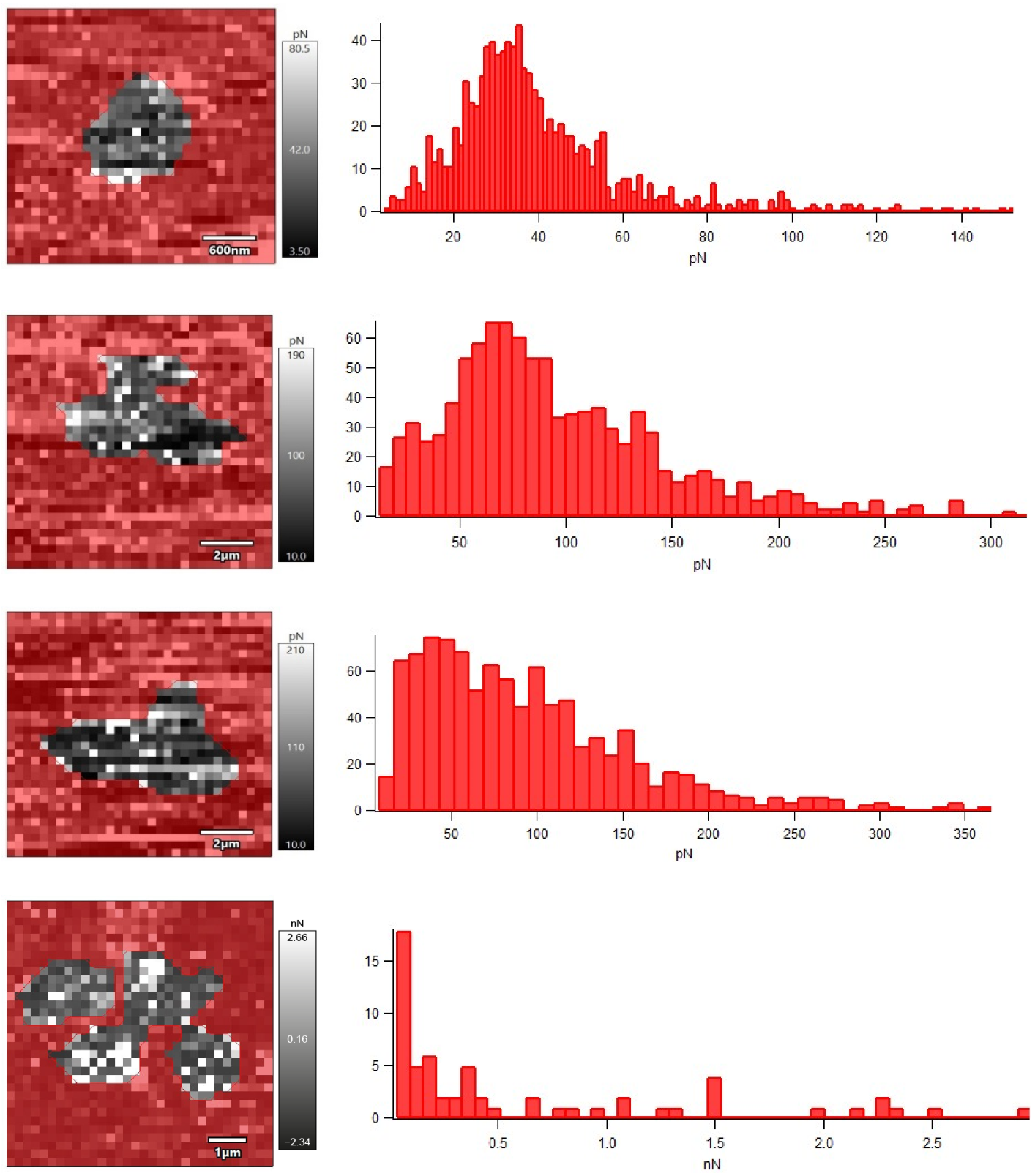
3.4. Effects of Nickel NPs on Nanomechanical Properties
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, G.; Li, J.; Zhang, G.; Zhao, L. Room-temperature synthesis of Ni nanoparticles as the absorbent used for sewage treatment. Adv. Mat. Sci. Engin. 2015, 2015, 973648. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Aker, W.G.; Fu, P.P.; Hwang, H.-M. Toxicity of engineered metal oxide nanomaterials mediated by nano–bio–eco–interactions: A review and perspective. Environ. Sci. Nano 2015, 2, 564–582. [Google Scholar] [CrossRef]
- Ahghari, M.R.; Soltaninejad, V.; Maleki, A. Synthesis of nickel nanoparticles by a green and convenient method as a magnetic mirror with antibacterial activities. Sci. Rep. 2020, 10, 12627. [Google Scholar] [CrossRef] [PubMed]
- Yin, I.X.; Zhang, J.; Zhao, I.S.; Mei, M.L.; Li, Q.; Chu, C.H. The antibacterial mechanism of silver nanoparticles and its application in dentistry. Int. J. Nanomed. 2020, 15, 2555–2562. [Google Scholar] [CrossRef] [Green Version]
- Raghupathi, K.R.; Koodali, R.T.; Manna, A.C. Size-dependent bacterial growth inhibition and mechanism of antibacterial activity of zinc oxide nanoparticles. Langmuir 2011, 27, 4020–4028. [Google Scholar] [CrossRef]
- Cui, Y.; Zhao, Y.; Tian, Y.; Zhang, W.; Lü, X.; Jiang, X. The molecular mechanism of action of bactericidal gold nanoparticles on Escherichia coli. Biomaterials 2012, 33, 2327–2333. [Google Scholar] [CrossRef]
- Zhang, W.; Hughes, J.; Chen, Y. Impacts of hematite nanoparticle exposure on biomechanical, adhesive, and surface electrical properties of Escherichia coli cells. Appl. Environ. Microbiol. 2012, 78, 3905–3915. [Google Scholar] [CrossRef] [Green Version]
- Ferreyra Maillard, A.; Gonçalves, S.; Santos, N.C.; López de Mishima, B.A.; Dalmasso, P.R.; Hollmann, A. Studies on interaction of green silver nanoparticles with whole bacteria by surface characterization techniques. Biochim. Biophys. Acta. Biomembr. 2019, 1861, 1086–1092. [Google Scholar] [CrossRef]
- Shi, X.; Shi, Z.; Wang, D.; Ullah, M.W.; Yang, G. Microbial cells with a Fe3O4 doped hydrogel extracellular matrix: Manipulation of living cells by magnetic stimulus. Macromol. Biosci. 2016, 16, 1506–1514. [Google Scholar] [CrossRef]
- Kuyukina, M.S.; Ivshina, I.B. Bioremediation of contaminated environments using Rhodococcus. In Biology of Rhodococcus, 2nd ed.; Microbiology Monographs; Alvarez, H.M., Ed.; Springer Nature: Cham, Switzerland, 2019; Volume 16, pp. 231–270. [Google Scholar] [CrossRef]
- Ivshina, I.B.; Kuyukina, M.S.; Krivoruchko, A.V.; Tyumina, E.A. Response mechanisms to ecopollutants and pathogenization risks of saprotrophic Rhodococcus. Pathogens 2021, 10, 974. [Google Scholar] [CrossRef]
- Ivshina, I.B.; Kuyukina, M.S.; Kostina, L.V. Adaptive mechanisms of nonspecific resistance to heavy metal ions in alkanotrophic actinobacteria. Russ. J. Ecol. 2013, 44, 123–130. [Google Scholar] [CrossRef]
- Wannoussa, W.; Hiligsmann, S.; Tasseroul, L.; Masy, T.; Lambert, S.D.; Heinrichs, B.; Al-Ahmad, A.E.; Weekers, F.; Thonart, P. Effect of metal ions and metal nanoparticles encapsulated in porous silica on biphenyl biodegradation by Rhodococcus erythropolis T902.1. J. Sol-Gel Sci. Technol. 2015, 75, 235–245. [Google Scholar] [CrossRef]
- Hou, J.; Liu, F.; Wu, N.; Ju, J.; Yu, B. Efficient biodegradation of chlorophenols in aqueous phase by magnetically immobilized aniline-degrading Rhodococcus rhodochrous strain. J. Nanobiotechnol. 2016, 14, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuyukina, M.S.; Ivshina, I.B.; Krivoruchko, A.V.; Peshkur, T.A.; Cunningham, C.J. Improvement of jet fuel contaminated water treatment in a fluidized-bed bioreactor by introducing nickel nanoparticles. Int. Biodeter. Biodegrad. 2021, 164, 105308. [Google Scholar] [CrossRef]
- Rubtsova, E.V.; Kuyukina, M.S.; Ivshina, I.B. Effect of cultivation conditions on the adhesive activity of Rhodococcus cells towards n-hexadecane. Appl. Biochem. Microbiol. 2012, 48, 452–459. [Google Scholar] [CrossRef]
- Kuyukina, M.S.; Ivshina, I.B.; Korshunova, I.O.; Rubtsova, E.V. Assessment of bacterial resistance to organic solvents using a combined confocal laser scanning and atomic force microscopy (CLSM/AFM). J. Microbiol. Methods 2014, 107, 23–29. [Google Scholar] [CrossRef]
- Krasowska, A.; Sigler, K. How microorganisms use hydrophobicity and what does this mean for human needs? Front. Cell. Infect. Microbiol. 2014, 4, 112. [Google Scholar] [CrossRef] [Green Version]
- Newmann, G.; Veeranagouda, Y.; Karegoudar, T.B.; Sahim, O.; Mausezahl, I.; Kabelitz, N.; Kappelmeyer, U.; Heipieper, H.J. Cells of Pseudomonas putida and Enterobacter sp. adapt to toxic organic compounds by increasing their size. Extremophiles 2005, 9, 163–168. [Google Scholar] [CrossRef]
- Korshunova, I.O.; Pistsova, O.N.; Kuyukina, M.S.; Ivshina, I.B. The effect of organic solvents on the viability and morphofunctional properties of Rhodococcus. Appl. Biochem. Microbiol. 2016, 52, 43–50. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, B.; Liu, Y.; Suo, X.; Li, H. Influence of surface topography on bacterial adhesion: A review (Review). Biointerphases 2018, 13, 060801. [Google Scholar] [CrossRef] [Green Version]
- Tamayo, L.; Melo, F.; Caballero, L.; Hamm, E.; Díaz, M.; Leal, M.S.; Guiliani, N.; Urzúa, M.D. Does bacterial elasticity affect adhesion to polymer fibers? ACS Appl. Mater. Interfaces 2020, 12, 14507–14517. [Google Scholar] [CrossRef] [PubMed]
- Viljoen, A.; Mathelié-Guinlet, M.; Ray, A.; Strohmeyer, N.; Oh, Y.J.; Hinterdorfer, P.; Müller, D.J.; Alsteens, D.; Dufrêne, Y.F. Force spectroscopy of single cells using atomic force microscopy. Nat. Rev. Methods Primers 2021, 1, 63. [Google Scholar] [CrossRef]
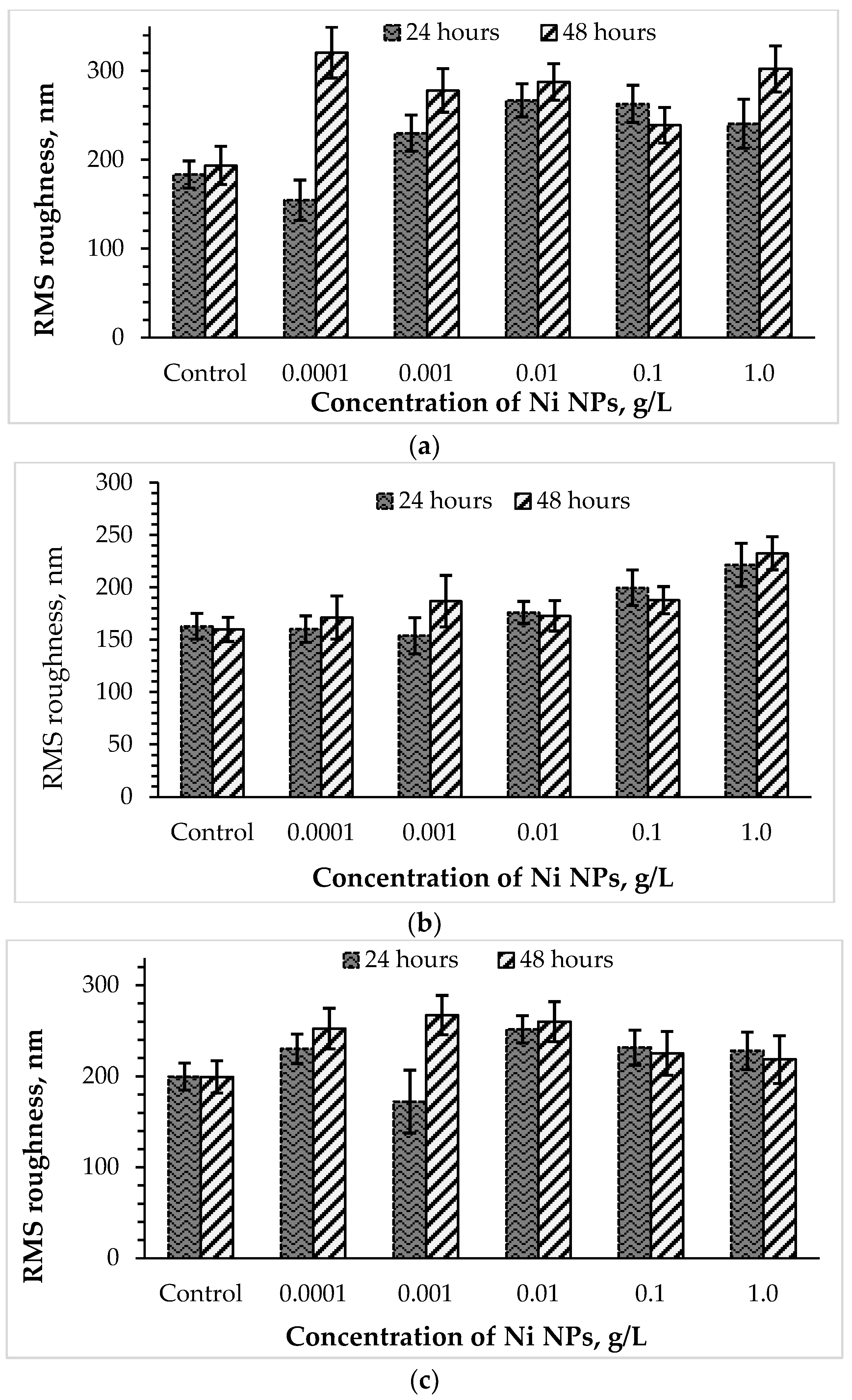
| Concentration of Ni NPs, g/L | Length, μm | Width, μm | Surface (S), μm2 | Volume (V), μm3 | Surface/Volume (S/V) |
|---|---|---|---|---|---|
| R. ruber IEGM 231 | |||||
| Control | 1.8 ± 0.2 a | 0.74 ± 0.08 | 5.0 ± 0.3 | 0.8 ± 0.08 | 6.5 ± 0.5 |
| 1.8 ± 0.1 b | 0.74 ± 0.09 | 4.9 ± 0.4 | 0.8 ± 0.07 | 6.6 ± 0.5 | |
| 0.0001 | 2.0 ± 0.3 | 0.57 ± 0.05 | 4.0 ± 0.4 | 0.5 ± 0.05 | 8.1 ± 0.7 |
| 1.4 ± 0.2 | 0.70 ± 0.06 | 3.9 ± 0.4 | 0.5 ± 0.06 | 7.1 ± 0.6 | |
| 0.001 | 2.0 ± 0.2 | 0.47 ± 0.02 | 3.3 ± 0.5 | 0.3 ± 0.05 | 9.6 ± 0.7 |
| 1.3 ± 0.3 | 0.85 ± 0.07 | 4.6 ± 0.4 | 0.7 ± 0.06 | 6.2 ± 0.8 | |
| 0.01 | 1.9 ± 0.3 | 0.73 ± 0.06 | 5.3 ± 0.5 | 0.8 ± 0.07 | 6.5 ± 0.5 |
| 1.8 ± 0.2 | 0.78 ± 0.09 | 5.3 ± 0.6 | 0.9 ± 0.07 | 6.2 ± 0.4 | |
| 0.1 | 2.2 ± 0.4 | 0.82 ± 0.07 | 6.6 ± 0.7 | 1.1 ± 0.09 | 5.8 ± 0.5 |
| 1.8 ± 0.2 | 0.85 ± 0.09 | 6.0 ± 0.8 | 1.0 ± 0.11 | 5.8 ± 0.7 | |
| 1.0 | 1.9 ± 0.3 | 0.83 ± 0.06 | 6.1 ± 0.6 | 1.0 ± 0.08 | 5.8 ± 0.4 |
| 1.2 ± 0.1 | 0.59 ± 0.04 | 2.8 ± 0.3 | 0.3 ± 0.04 | 8.4 ± 0.6 | |
| R. rhodochrous IEGM 1363 | |||||
| Control | 1.0 ± 0.1 | 0.58 ± 0.06 | 2.4 ± 0.2 | 0.3 ± 0.04 | 8.8 ± 0.9 |
| 1.0 ± 0.2 | 0.58 ± 0.03 | 2.4 ± 0.1 | 0.3 ± 0.03 | 8.9 ± 0.6 | |
| 0.0001 | 1.1 ± 0.2 | 0.50 ± 0.04 | 2.1 ± 0.1 | 0.2 ± 0.01 | 9.8 ± 0.7 |
| 1.2 ± 0.2 | 0.58 ± 0.07 | 2.7 ± 0.3 | 0.3 ± 0.02 | 8.5 ± 0.8 | |
| 0.001 | 1.1 ± 0.1 | 0.70 ± 0.05 | 3.3 ± 0.3 | 0.4 ± 0.05 | 7.5 ± 0.7 |
| 1.2 ± 0.2 | 0.88 ± 0.09 | 4.4 ± 0.4 | 0.7 ± 0.06 | 6.3 ± 0.7 | |
| 0.01 | 1.4 ± 0.1 | 0.73 ± 0.04 | 4.0 ± 0.5 | 0.6 ± 0.07 | 6.9 ± 0.5 |
| 1.3 ± 0.1 | 0.90 ± 0.07 | 4.8 ± 0.4 | 0.8 ± 0.06 | 6.0 ± 0.4 | |
| 0.1 | 1.1 ± 0.3 | 0.74 ± 0.02 | 3.4 ± 0.2 | 0.5 ± 0.04 | 7.2 ± 0.6 |
| 1.2 ± 0.1 | 0.95 ± 0.09 | 5.0 ± 0.3 | 0.9 ± 0.06 | 5.9 ± 0.7 | |
| 1.0 | 1.2 ± 0.1 | 1.23 ± 0.14 | 3.8 ± 0.3 | 0.6 ± 0.06 | 6.9 ± 0.8 |
| 1.4 ± 0.2 | 0.90 ± 0.11 | 7.8 ± 0.8 | 1.7 ± 0.12 | 4.7 ± 0.5 | |
| Concentration of Ni NPs, g/L | Adhesion Force, nN | Young’s Modulus, MPa |
|---|---|---|
| Control | 0.041 ± 0.018 | 48.8 ± 3.9 |
| 0.01 | 0.10 ± 0.04 | 20.8 ± 2.7 |
| 0.1 | 0.14 ± 0.05 | 3.2 ± 0.4 |
| 1.0 | 1.42 ± 0.13 | 6.0 ± 1.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuyukina, M.S.; Glebov, G.G.; Ivshina, I.B. Effects of Nickel Nanoparticles on Rhodococcus Cell Surface Morphology and Nanomechanical Properties. Nanomaterials 2022, 12, 951. https://doi.org/10.3390/nano12060951
Kuyukina MS, Glebov GG, Ivshina IB. Effects of Nickel Nanoparticles on Rhodococcus Cell Surface Morphology and Nanomechanical Properties. Nanomaterials. 2022; 12(6):951. https://doi.org/10.3390/nano12060951
Chicago/Turabian StyleKuyukina, Maria S., Grigorii G. Glebov, and Irena B. Ivshina. 2022. "Effects of Nickel Nanoparticles on Rhodococcus Cell Surface Morphology and Nanomechanical Properties" Nanomaterials 12, no. 6: 951. https://doi.org/10.3390/nano12060951






