Metal-Promoted Higher-Order Assembly of Disulfide-Stapled Helical Barrels
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. General Peptide Synthesis and Purification
2.3. Interhelical Disulfide Bond Formation
2.4. Circular Dichroism Spectroscopy
2.5. Molecular Modeling
2.6. Fluorescence Spectroscopy
2.7. Metal-Promoted Assembly
2.8. Scanning Electron Microscopy
2.9. Transmission Electron Microscopy
2.10. Cargo Inclusion before Assembly Formation
2.11. Cargo Inclusion after Assembly Formation
2.12. Confocal Microscopy
3. Results and Discussion
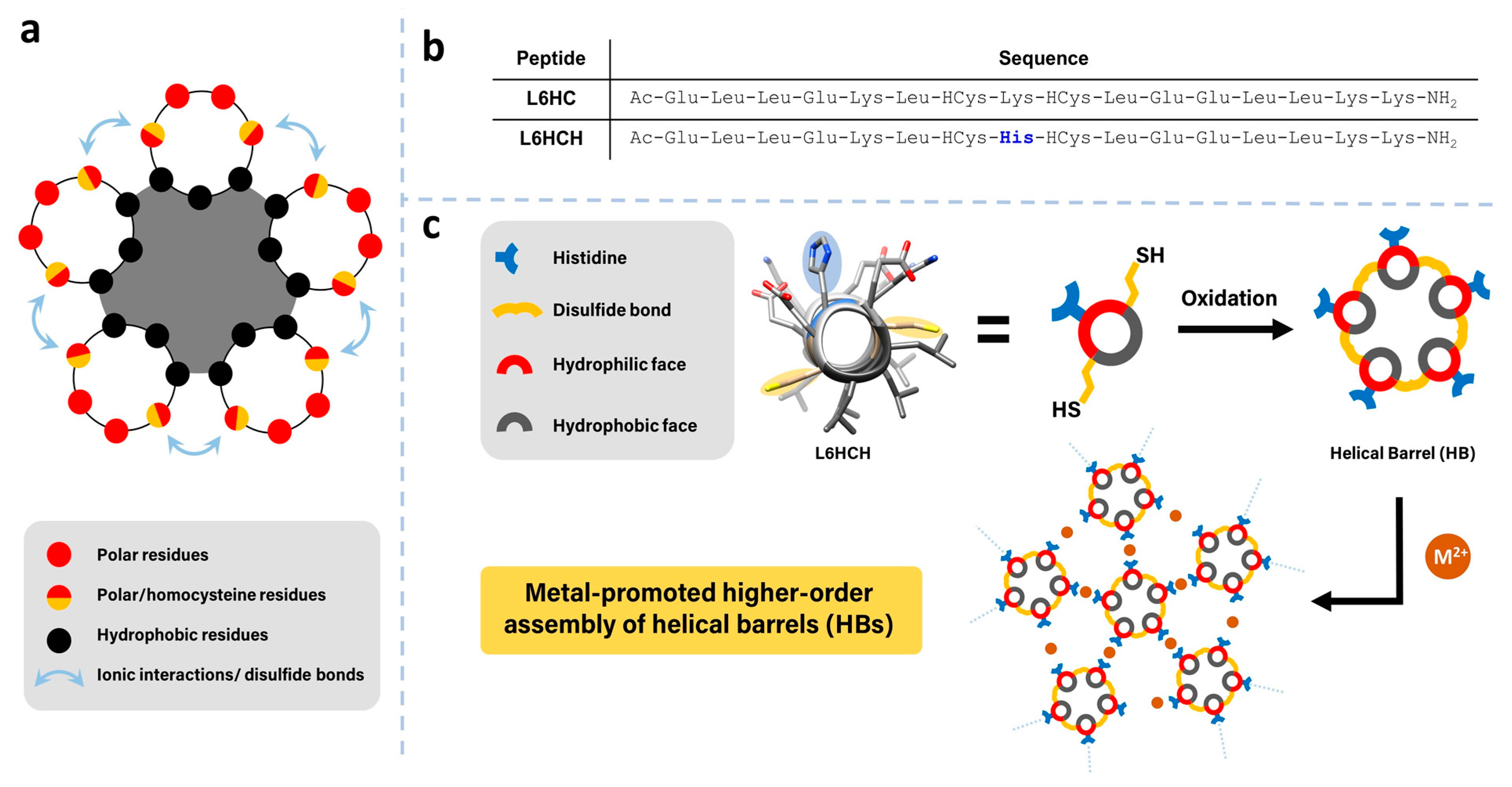
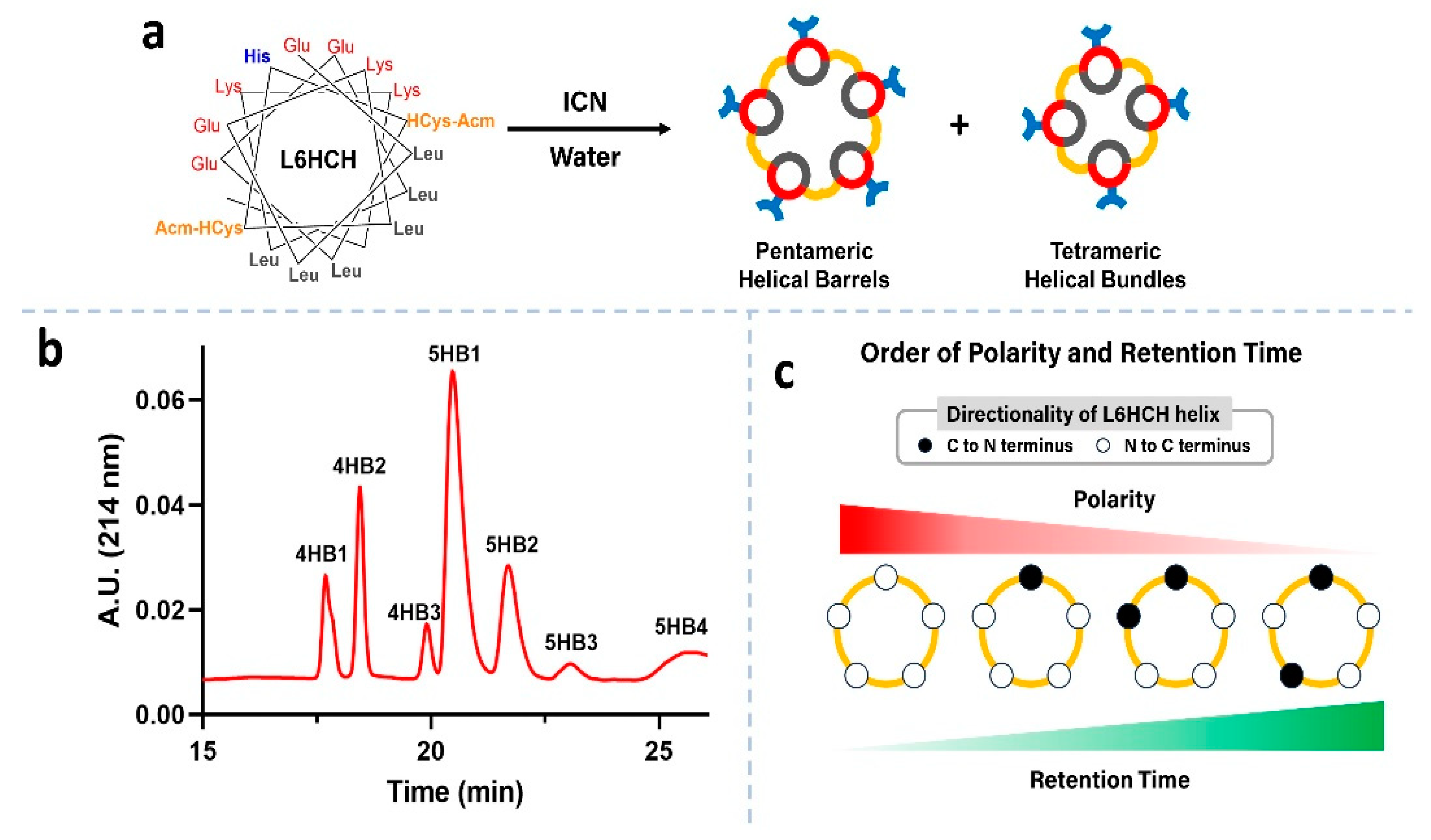
3.1. Peptide Design, Synthesis and Barrel Crosslinking
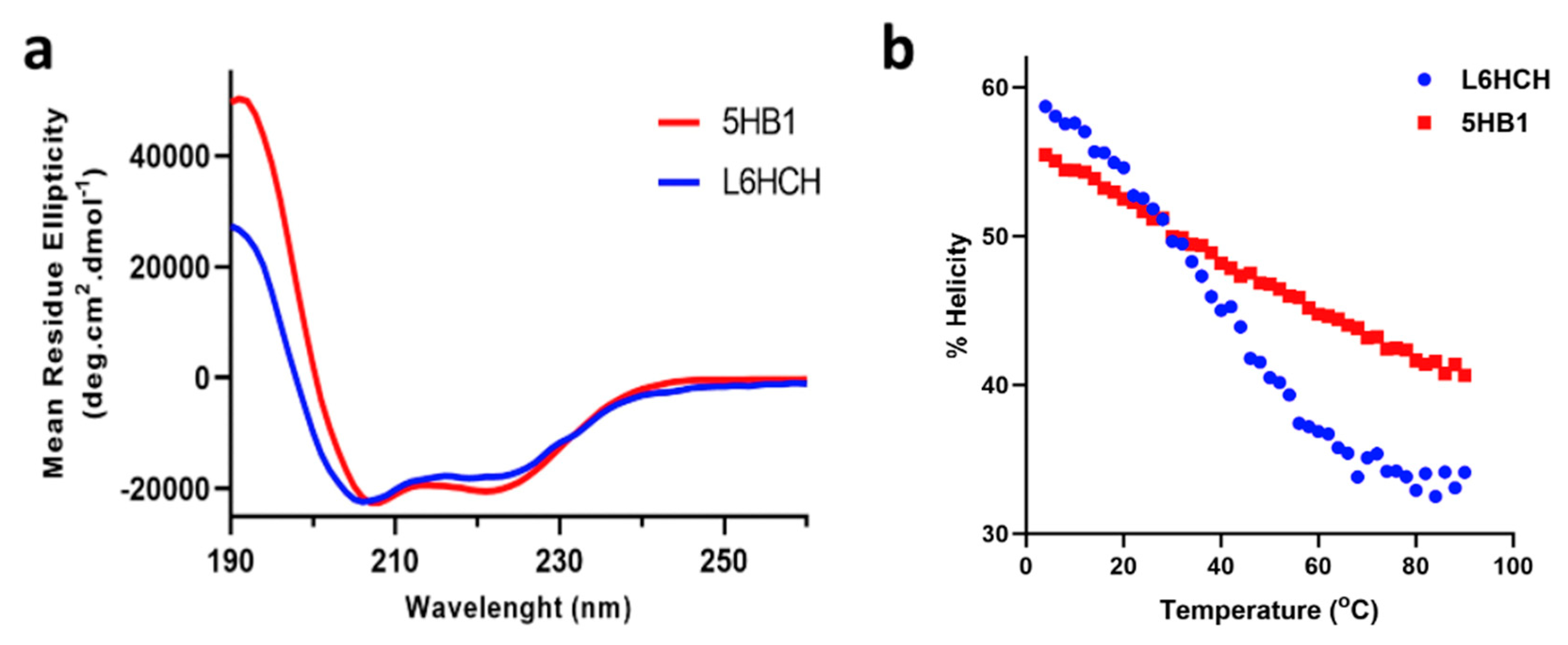
3.2. Secondary Structure Determination and Thermal Stability
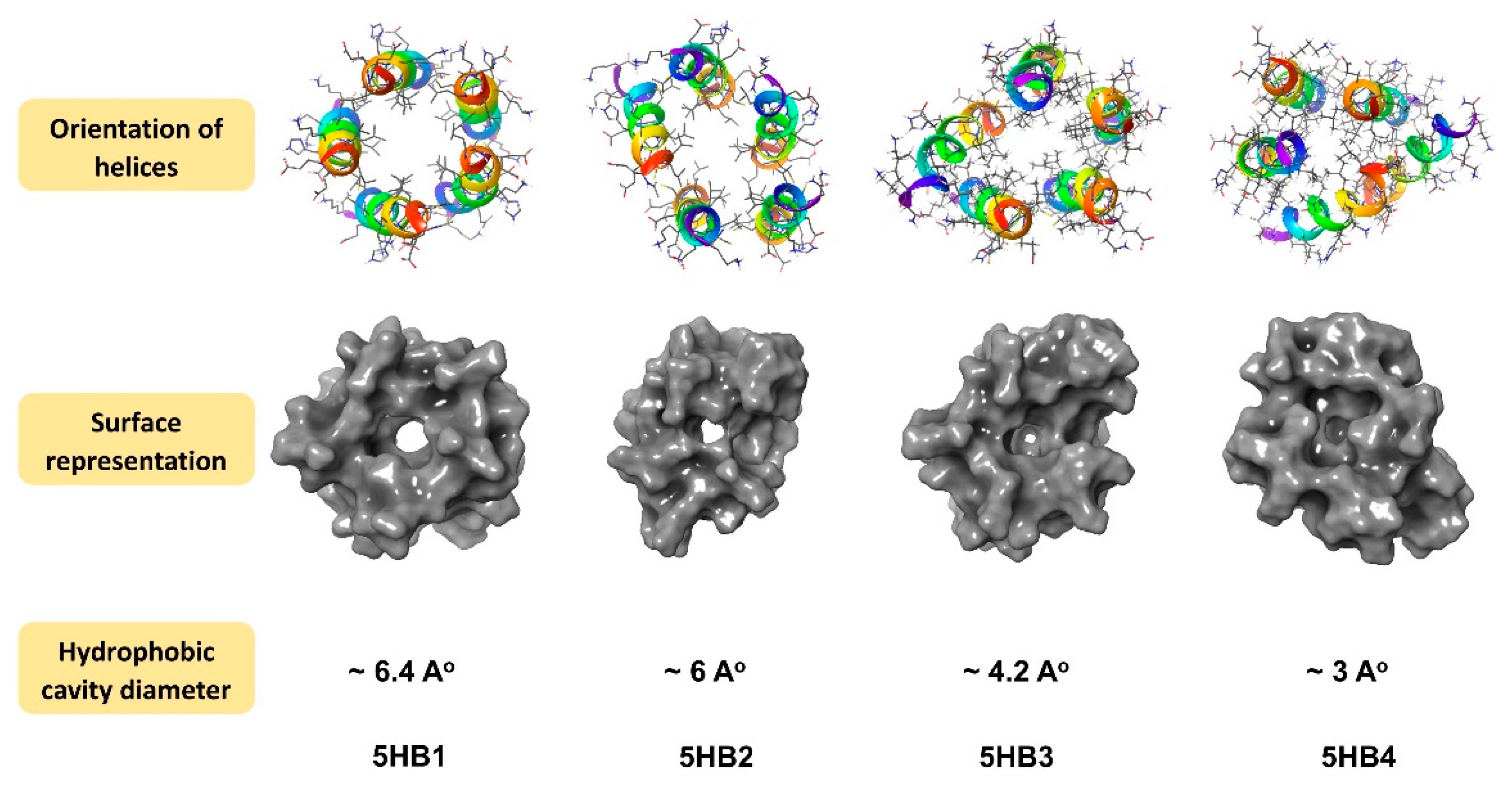
3.3. Molecular Modeling
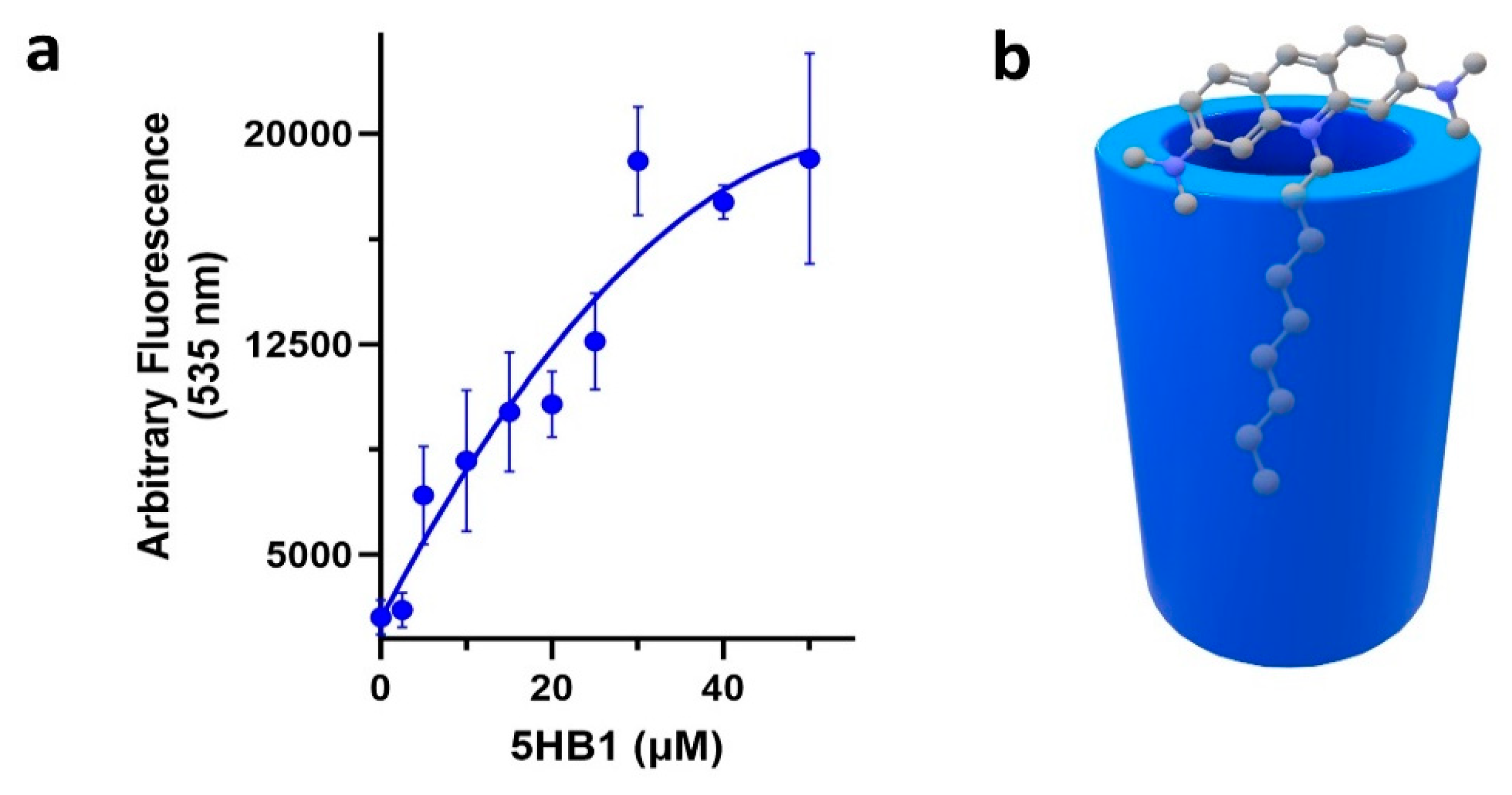
3.4. Inclusion of Hydrophobic Dyes
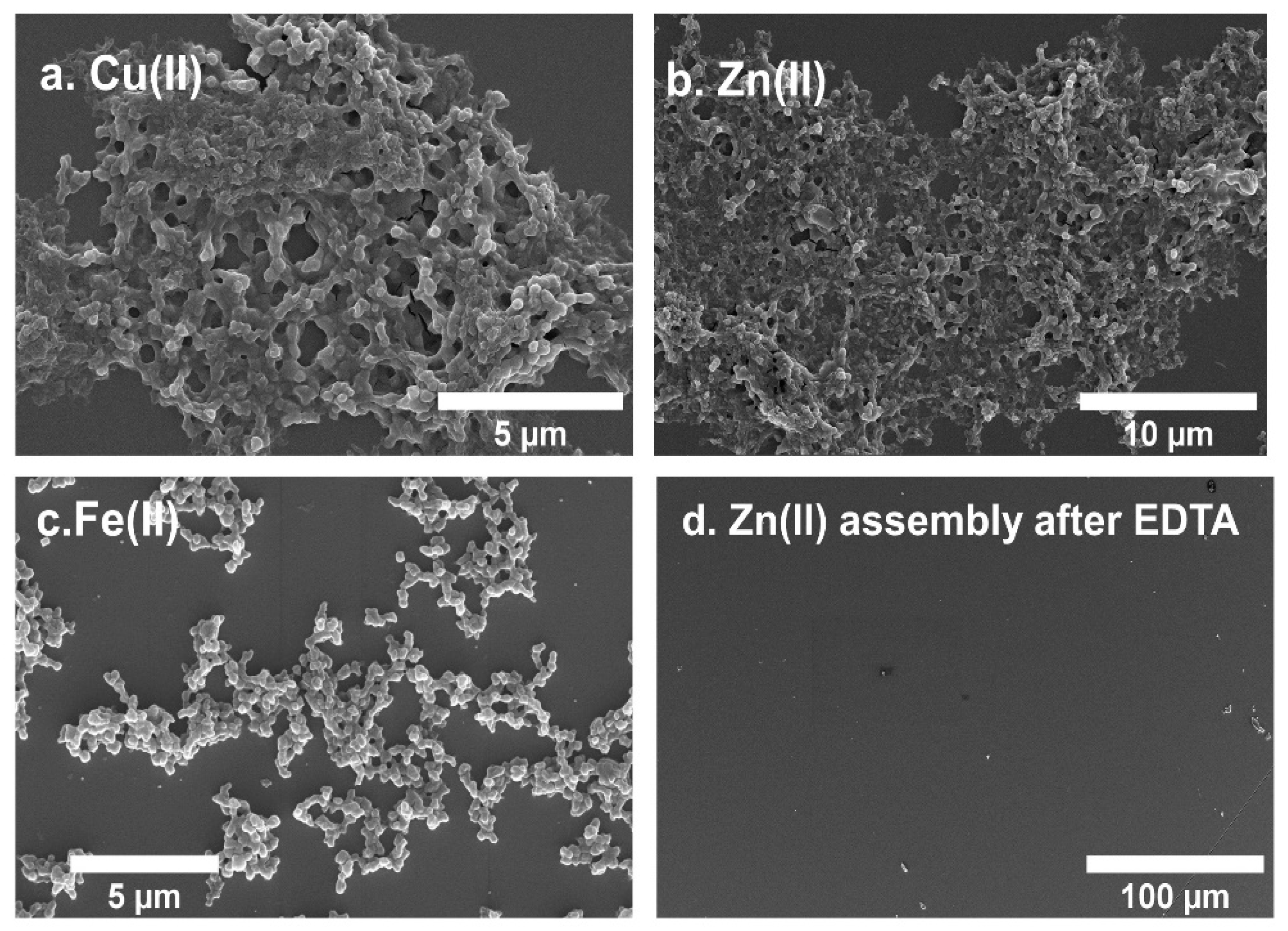
3.5. Metal-Promoted Higher-Order Assembly of 5HB1
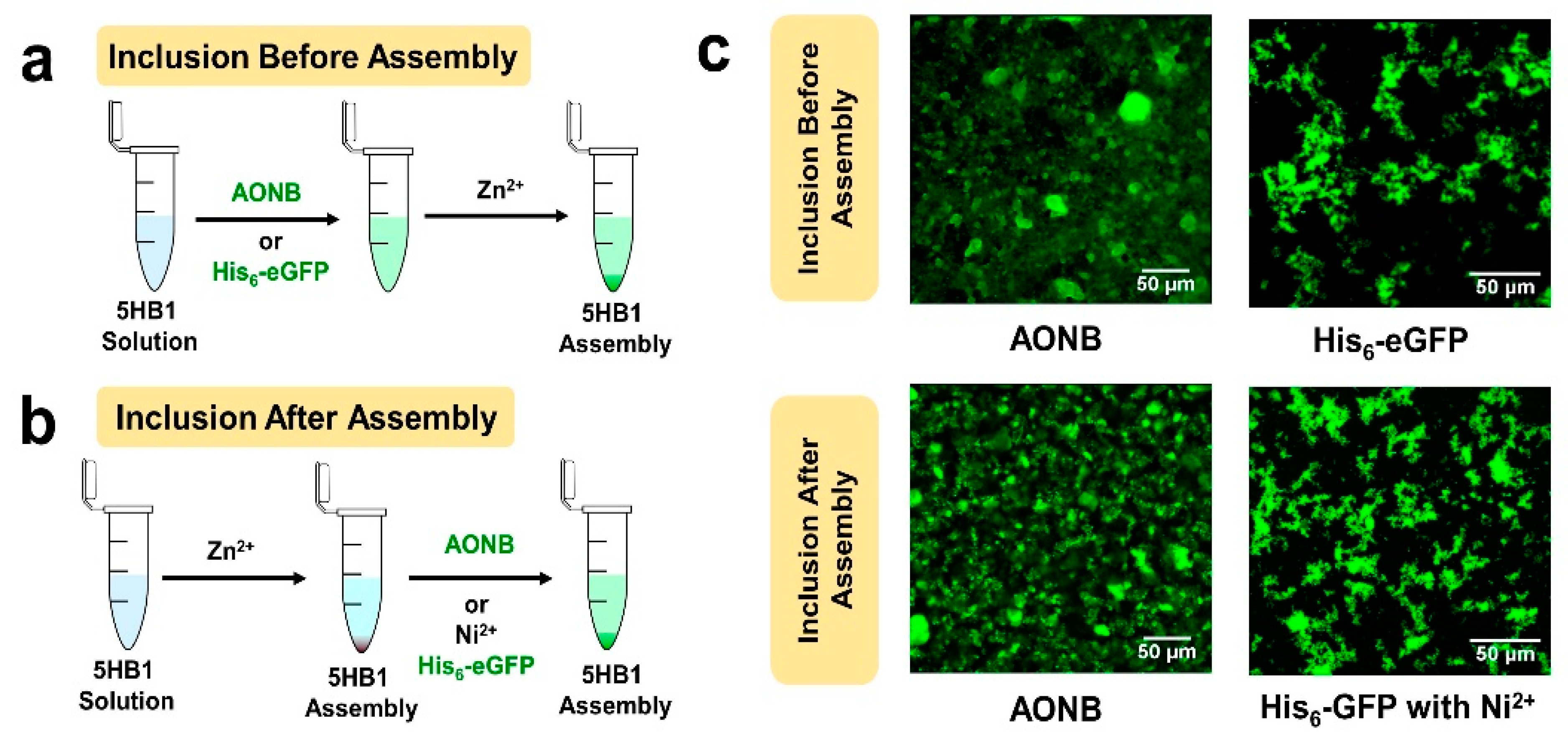
3.6. 3D-Matrix Cargo Loading
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Qiu, F.; Chen, Y.; Tang, C.; Zhao, X. Amphiphilic Peptides as Novel Nanomaterials: Design, Self-Assembly and Application. Int. J. Nanomed. 2018, 13, 5003–5022. [Google Scholar] [CrossRef]
- Hendricks, M.P.; Sato, K.; Palmer, L.C.; Stupp, S.I. Supramolecular Assembly of Peptide Amphiphiles. Acc. Chem. Res. 2017, 50, 2440–2448. [Google Scholar] [CrossRef] [PubMed]
- Curtis, R.W.; Chmielewski, J. A Comparison of the Collagen Triple Helix and Coiled-Coil Peptide Building Blocks on Metal Ion-Mediated Supramolecular Assembly. Pept. Sci. 2021, 113, e24190. [Google Scholar] [CrossRef]
- Jorgensen, M.D.; Chmielewski, J. Recent Advances in Coiled-Coil Peptide Materials and Their Biomedical Applications. Chem. Commun. 2022, 58, 11625–11636. [Google Scholar] [CrossRef]
- Thomas, F.; Dawson, W.M.; Lang, E.J.M.; Burton, A.J.; Bartlett, G.J.; Rhys, G.G.; Mulholland, A.J.; Woolfson, D.N. De Novo-Designed α-Helical Barrels as Receptors for Small Molecules. ACS Synth. Biol. 2018, 7, 1808–1816. [Google Scholar] [CrossRef] [PubMed]
- Dawson, W.M.; Martin, F.J.O.; Rhys, G.G.; Shelley, K.L.; Brady, R.L.; Woolfson, D.N. Coiled Coils 9-to-5: Rational: De Novo Design of α-Helical Barrels with Tunable Oligomeric States. Chem. Sci. 2021, 12, 6923–6928. [Google Scholar] [CrossRef] [PubMed]
- Dawson, W.M.; Shelley, K.L.; Fletcher, J.M.; Scott, D.A.; Lombardi, L.; Rhys, G.G.; LaGambina, T.J.; Obst, U.; Burton, A.J.; Cross, J.A.; et al. Differential Sensing with Arrays of de Novo Designed Peptide Assemblies. Nat. Commun. 2023, 14, 383. [Google Scholar] [CrossRef]
- Dawson, W.M.; Lang, E.J.M.; Rhys, G.G.; Shelley, K.L.; Williams, C.; Brady, R.L.; Crump, M.P.; Mulholland, A.J.; Woolfson, D.N. Structural Resolution of Switchable States of a de Novo Peptide Assembly. Nat. Commun. 2021, 12, 1530. [Google Scholar] [CrossRef]
- Rhys, G.G.; Wood, C.W.; Lang, E.J.M.; Mulholland, A.J.; Brady, R.L.; Thomson, A.R.; Woolfson, D.N. Maintaining and Breaking Symmetry in Homomeric Coiled-Coil Assemblies. Nat. Commun. 2018, 9, 4132. [Google Scholar] [CrossRef]
- Beesley, J.L.; Woolfson, D.N. The de Novo Design of α-Helical Peptides for Supramolecular Self-Assembly. Curr. Opin. Biotechnol. 2019, 58, 175–182. [Google Scholar] [CrossRef]
- Xu, C.; Liu, R.; Mehta, A.K.; Guerrero-Ferreira, R.C.; Wright, E.R.; Dunin-Horkawicz, S.; Morris, K.; Serpell, L.C.; Zuo, X.; Wall, J.S.; et al. Rational Design of Helical Nanotubes from Self-Assembly of Coiled-Coil Lock Washers. J. Am. Chem. Soc. 2013, 135, 15565–15578. [Google Scholar] [CrossRef] [PubMed]
- More, H.T.; Zhang, K.S.; Srivastava, N.; Frezzo, J.A.; Montclare, J.K. Influence of Fluorination on Protein-Engineered Coiled-Coil Fibers. Biomacromolecules 2015, 16, 1210–1217. [Google Scholar] [CrossRef] [PubMed]
- Britton, D.; Monkovic, J.; Jia, S.; Liu, C.; Mahmoudinobar, F.; Meleties, M.; Renfrew, P.D.; Bonneau, R.; Montclare, J.K. Supramolecular Assembly and Small-Molecule Binding by Protein-Engineered Coiled-Coil Fibers. Biomacromolecules 2022, 23, 4851–4859. [Google Scholar] [CrossRef] [PubMed]
- Hume, J.; Sun, J.; Jacquet, R.; Renfrew, P.D.; Martin, J.A.; Bonneau, R.; Gilchrist, M.L.; Montclare, J.K. Engineered Coiled-Coil Protein Microfibers. Biomacromolecules 2014, 15, 3503–3510. [Google Scholar] [CrossRef]
- Burgess, N.C.; Sharp, T.H.; Thomas, F.; Wood, C.W.; Thomson, A.R.; Zaccai, N.R.; Brady, R.L.; Serpell, L.C.; Woolfson, D.N. Modular Design of Self-Assembling Peptide-Based Nanotubes. J. Am. Chem. Soc. 2015, 137, 10554–10562. [Google Scholar] [CrossRef]
- Lutgring, R.; Chmielewski, J. General Strategy for Covalently Stabilizing Helical Bundles: A Novel Five-Helix Bundle Protein. J. Am. Chem. Soc. 1994, 116, 6451–6452. [Google Scholar] [CrossRef]
- Woolfson, D.N.; Bartlett, G.J.; Burton, A.J.; Heal, J.W.; Niitsu, A.; Thomson, A.R.; Wood, C.W. De Novo Protein Design: How Do We Expand into the Universe of Possible Protein Structures? Curr. Opin. Struct. Biol. 2015, 33, 16–26. [Google Scholar] [CrossRef]
- Morii, H.; Ichimura, K.; Uedaira, H. Asymmetric Inclusion by de Novo Designed Proteins: Fluorescence Probe Studies on Amphiphilic A-helix Bundles. Proteins Struct. Funct. Bioinform. 1991, 11, 133–141. [Google Scholar] [CrossRef]
- Britton, D.; Meleties, M.; Liu, C.; Jia, S.; Mahmoudinobar, F.; Renfrew, P.D.; Bonneau, R.; Montclare, J.K. Tuning a Coiled-Coil Hydrogel via Computational Design of Supramolecular Fiber Assembly. Mol. Syst. Des. Eng. 2023, 8, 217–226. [Google Scholar] [CrossRef]
- Thomas, F.; Burgess, N.C.; Thomson, A.R.; Woolfson, D.N. Controlling the Assembly of Coiled-Coil Peptide Nanotubes. Angew. Chem. 2016, 128, 999–1003. [Google Scholar] [CrossRef]
- Bishop, P.; Chmielewski, J. Cyanogen Iodide: A New Reagent for Disulfide Bond Formation in Peptides. Tetrahedron Lett. 1992, 33, 6263–6266. [Google Scholar] [CrossRef]
- Sommese, R.F.; Sivaramakrishnan, S.; Baldwin, R.L.; Spudich, J.A. Helicity of Short E-R/K Peptides. Protein Sci. 2010, 19, 2001–2005. [Google Scholar] [CrossRef] [PubMed]
- Mohamadi, F.; Richards, N.G.J.; Guida, W.C.; Liskamp, R.; Lipton, M.; Caufield, C.; Chang, G.; Hendrickson, T.; Still, W.C. Macromodel—An Integrated Software System for Modeling Organic and Bioorganic Molecules Using Molecular Mechanics. J. Comput. Chem. 1990, 11, 440–467. [Google Scholar] [CrossRef]
- Cornell, W.D.; Cieplak, P.; Bayly, I.; Gould, I.R.; Merz, K.M.; Ferguson, D.M.; Spellmeyer, D.C.; Fox, T.; Caldwell, J.W.; Kollman, P.A. A Second. Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. J. Am. Chem. Soc. 1995, 117, 5179–5197. [Google Scholar] [CrossRef]
- Qiu, D.; Shenkin, P.S.; Hollinger, F.P.; Still, W.C. The GB/SA Continuum Model for Solvation. A Fast Analytical Method. for the Calculation of Approximate Born Radii. J. Phys. Chem. A 1996, 101, 3005–3014. [Google Scholar] [CrossRef]
- Ponder, J.W.; Richards, F.M. An Efficient Newton-like Method for Molecular Mechanics Energy Minimization of Large Molecules. J. Comput. Chem. 1987, 8, 1016–1024. [Google Scholar] [CrossRef]
- Van Gunsteren, W.F.; Berendsen, H.J.C. A Leap-Frog Algorithm for Stochastic Dynamics. Mol. Simul. 1988, 1, 173–185. [Google Scholar] [CrossRef]
- Ho, S.P.; DeGrado, W.F. Design of a 4-Helix Bundle Protein: Synthesis of Peptides Which Self-Associate into a Helical Protein. J. Am. Chem. Soc. 1987, 109, 6751–6758. [Google Scholar] [CrossRef]
- Marqusee, S.; Baldwin, R.L. Helix Stabilization by Glu-...Lys+ Salt Bridges in Short Peptides of de Novo Design. Proc. Natl. Acad. Sci. USA 1987, 84, 8898–8902. [Google Scholar] [CrossRef]
- Lyu, P.C.; Marky, L.A.; Kallenbach, N.R. The Role of Ion Pairs in alpha-Helix Stability: Two New Designed Helical Peptides. J. Am. Chem. Soc. 1989, 111, 2733–2734. [Google Scholar] [CrossRef]
- Shoemaker, K.R.; Kim, P.S.; York, E.J.; Stewart, J.M.; Baldwin, R.L. Tests of the Helix Dipole Model for Stabilization of α-Helices. Nature 1987, 326, 563–567. [Google Scholar] [CrossRef] [PubMed]
- Fairman, R.; Shoemaker, K.R.; York, E.J.; Stewart, J.M.; Baldwin, R.L. Further Studies of the Helix Dipole Model: Effects of a Free α-NH3+ or α-COO− Group on Helix Stability. Proteins Struct. Funct. Genet. 1989, 5, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Kim, E.-H.; Hibino, T.; Ooi, T. Comparison of alpha-Helix Stability in Peptides Having a Negatively or Positively Charged Residue Block Attached Either to the N- or C-Terminus of an alpha-Helix: The Electrostatic Contribution and Anisotropic Stability of the alpha-Helix. Biopolymers 1989, 28, 995–1009. [Google Scholar] [CrossRef] [PubMed]
- Tidor, B.; Karplus, M. Simulation Analysis of the Stability Mutant R96H of T4 Lysozyme. Biochemistry 1991, 30, 3217–3228. [Google Scholar] [CrossRef]
- Kakkis, A.; Gagnon, D.; Esselborn, J.; Britt, R.D.; Tezcan, F.A. Metal-Templated Design of Chemically Switchable Protein Assemblies with High-Affinity Coordination Sites. Angew. Chem. Int. Ed. 2020, 59, 21940–21944. [Google Scholar] [CrossRef]
- Thomas, F.; Boyle, A.L.; Burton, A.J.; Woolfson, D.N. A Set of de Novo Designed Parallel Heterodimeric Coiled Coils with Quantified Dissociation Constants in the Micromolar to Sub-Nanomolar Regime. J. Am. Chem. Soc. 2013, 135, 5161–5166. [Google Scholar] [CrossRef]
- Tavenor, N.A.; Murnin, M.J.; Horne, W.S. Supramolecular Metal-Coordination Polymers, Nets, and Frameworks from Synthetic Coiled-Coil Peptides. J. Am. Chem. Soc. 2017, 139, 2212–2215. [Google Scholar] [CrossRef]
- Hernandez-Gordillo, V.; Chmielewski, J. Mimicking the Extracellular Matrix with Functionalized, Metal-Assembled Collagen Peptide Scaffolds. Biomaterials 2014, 35, 7363–7373. [Google Scholar] [CrossRef]
- Pires, M.M.; Ernenwein, D.; Chmielewski, J. Selective Decoration and Release of His-Tagged Proteins from Metal-Assembled Collagen Peptide Microflorettes. Biomacromolecules 2011, 12, 2429–2433. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agrahari, A.; Lipton, M.; Chmielewski, J. Metal-Promoted Higher-Order Assembly of Disulfide-Stapled Helical Barrels. Nanomaterials 2023, 13, 2645. https://doi.org/10.3390/nano13192645
Agrahari A, Lipton M, Chmielewski J. Metal-Promoted Higher-Order Assembly of Disulfide-Stapled Helical Barrels. Nanomaterials. 2023; 13(19):2645. https://doi.org/10.3390/nano13192645
Chicago/Turabian StyleAgrahari, Ashutosh, Mark Lipton, and Jean Chmielewski. 2023. "Metal-Promoted Higher-Order Assembly of Disulfide-Stapled Helical Barrels" Nanomaterials 13, no. 19: 2645. https://doi.org/10.3390/nano13192645
APA StyleAgrahari, A., Lipton, M., & Chmielewski, J. (2023). Metal-Promoted Higher-Order Assembly of Disulfide-Stapled Helical Barrels. Nanomaterials, 13(19), 2645. https://doi.org/10.3390/nano13192645





