A Comparison of Common Quantum Dot Alternatives to Cadmium-Based Quantum Dots on the Basis of Liver Cytotoxicity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Quantum Dots
2.2. Cell Lines
2.3. Cell Culture
2.4. QD Characterization
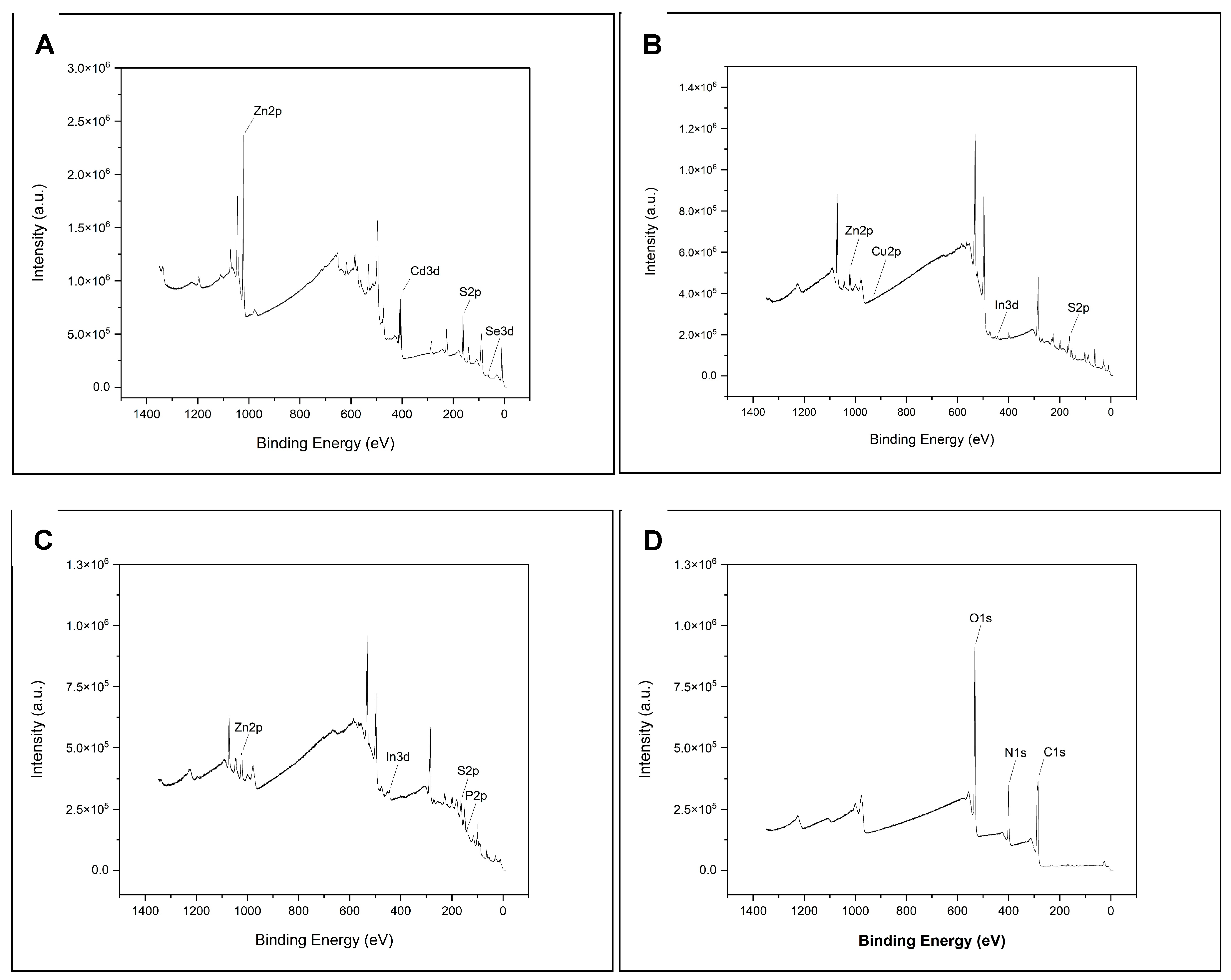
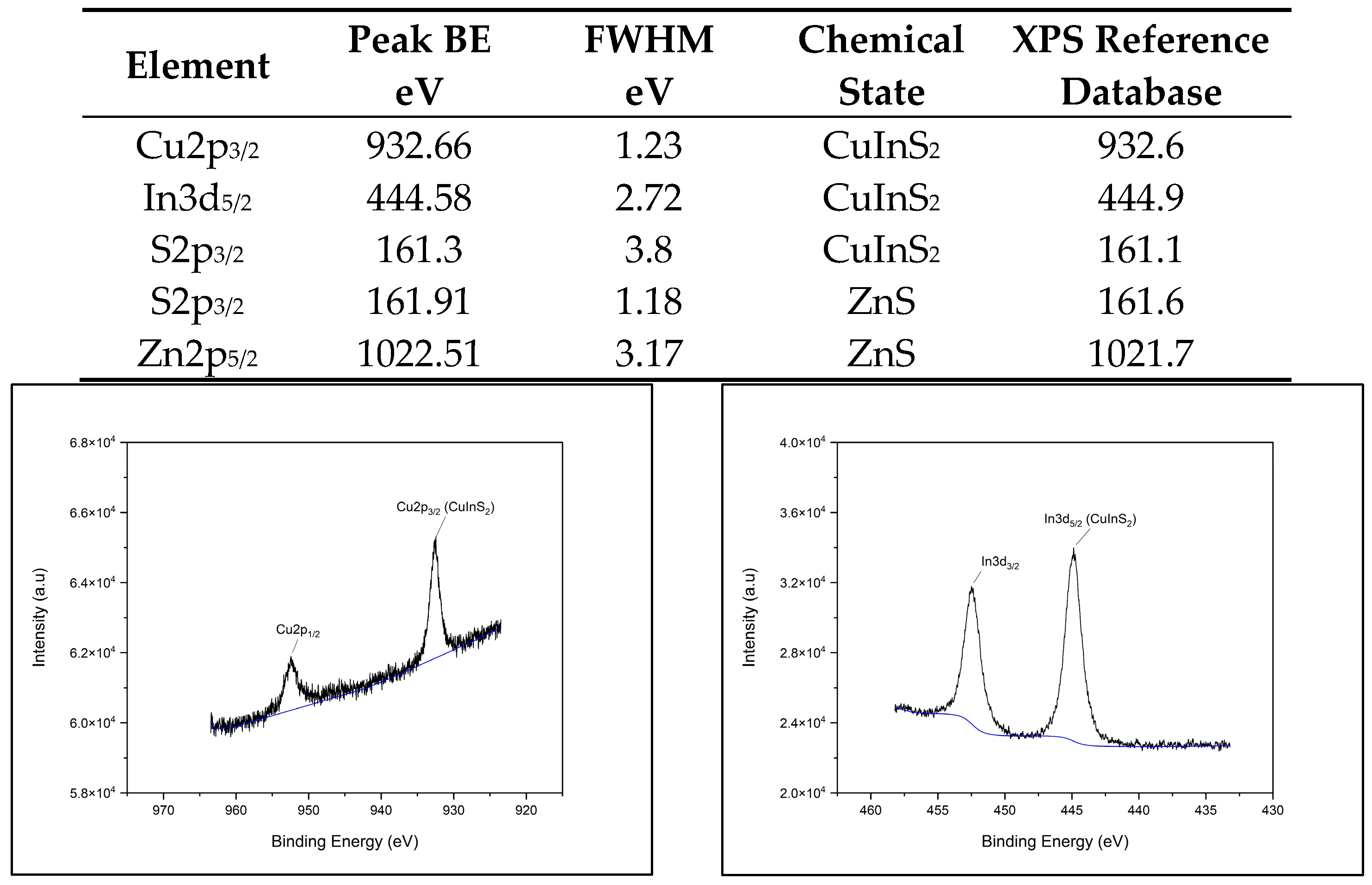
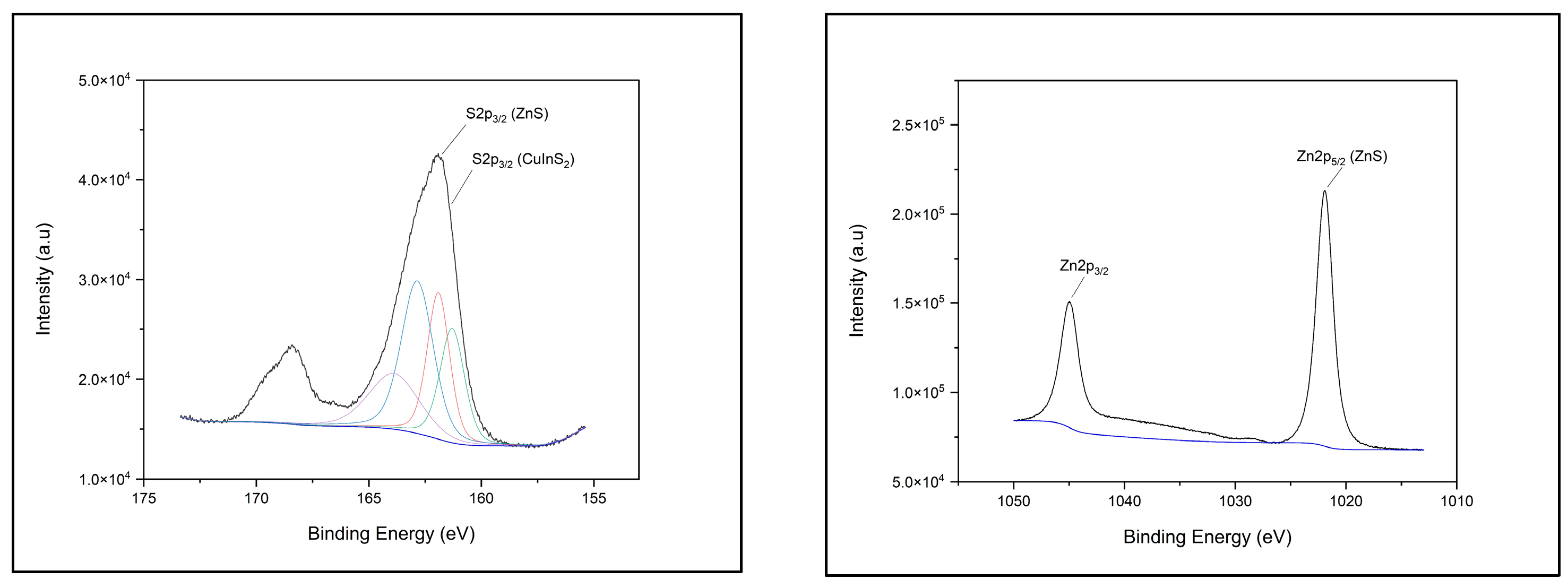
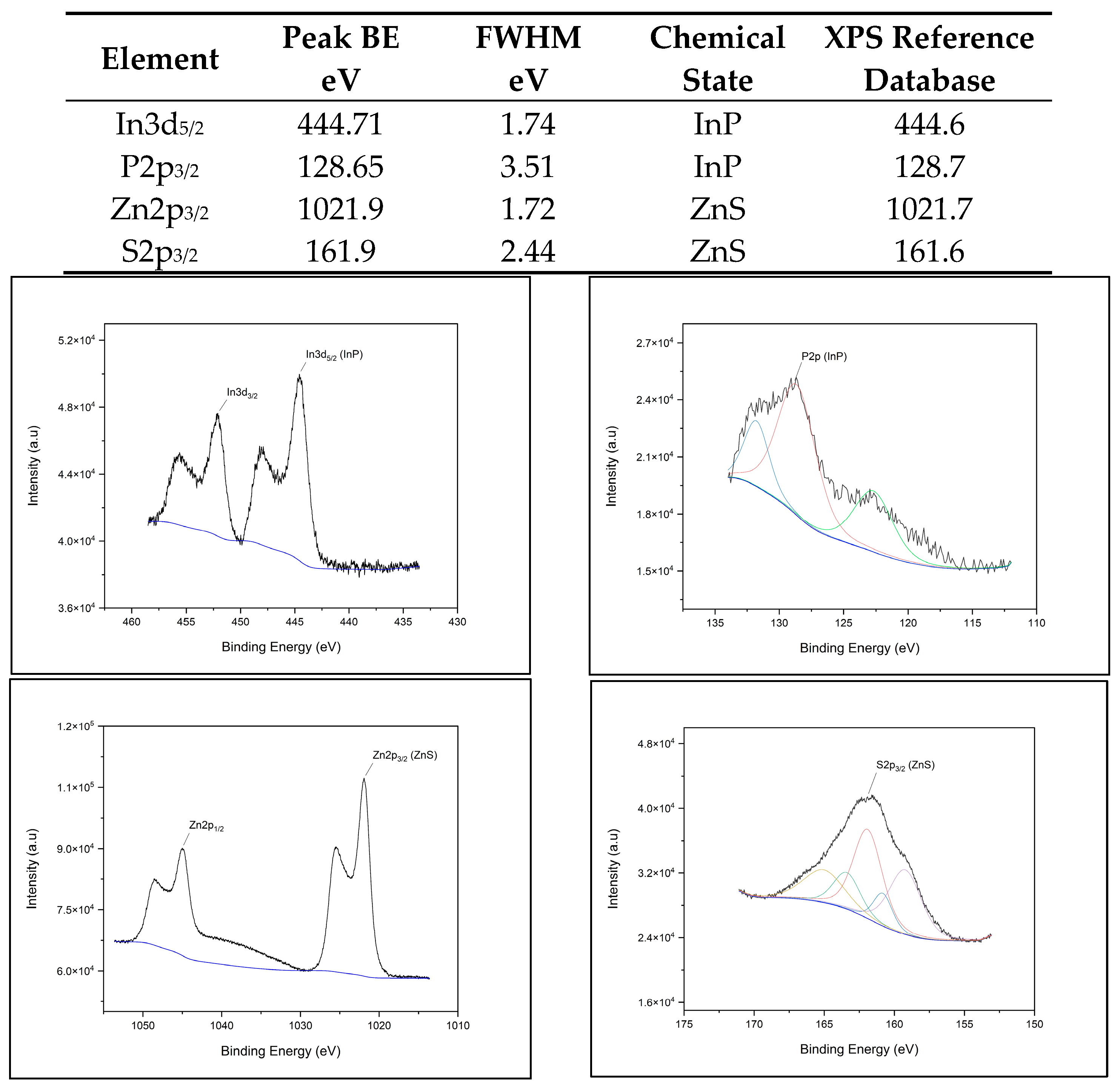
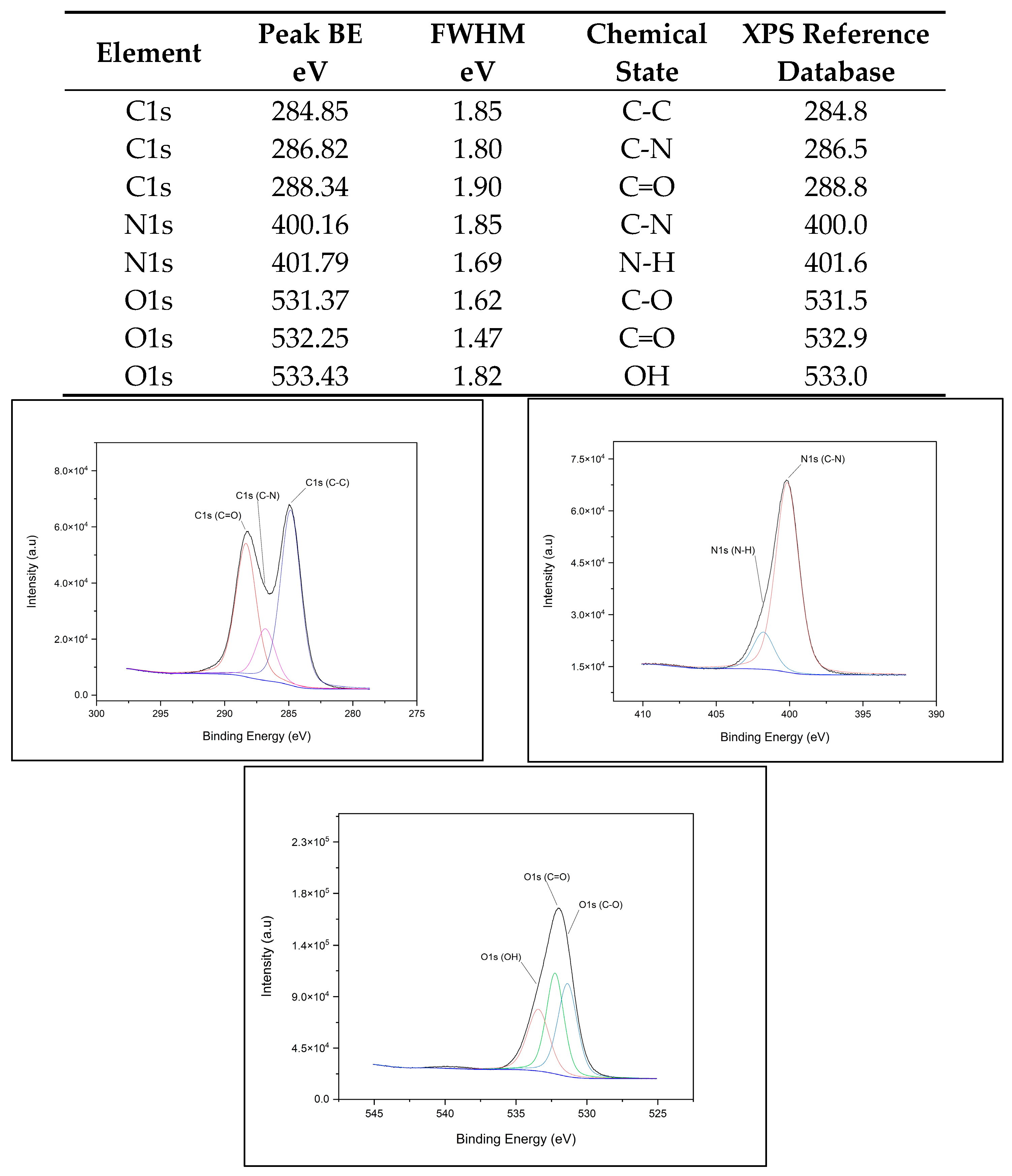
2.4.1. XPS Chemical Composition
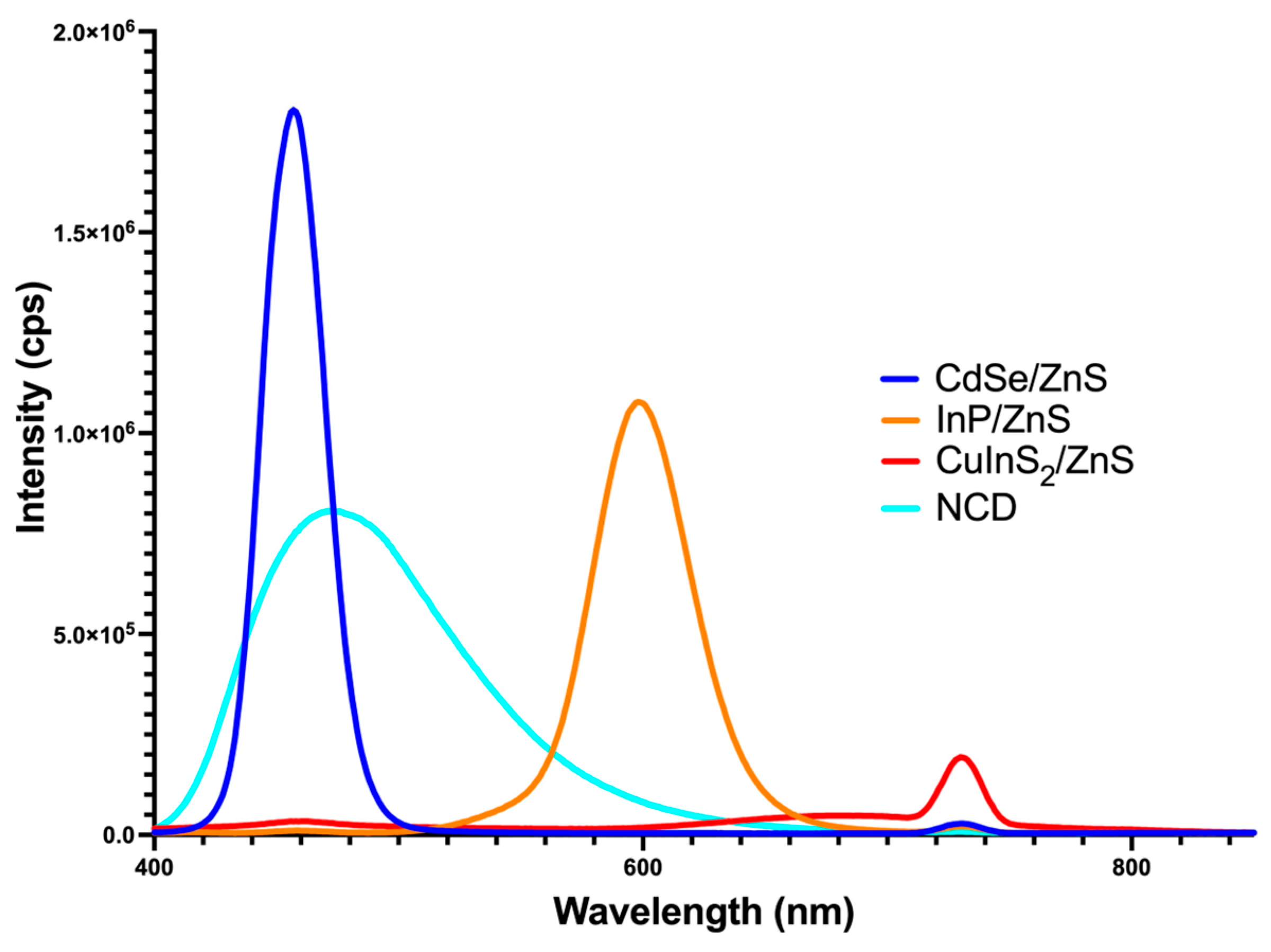
2.4.2. Fluorescent Spectra Analysis
2.5. XTT Viability Assay
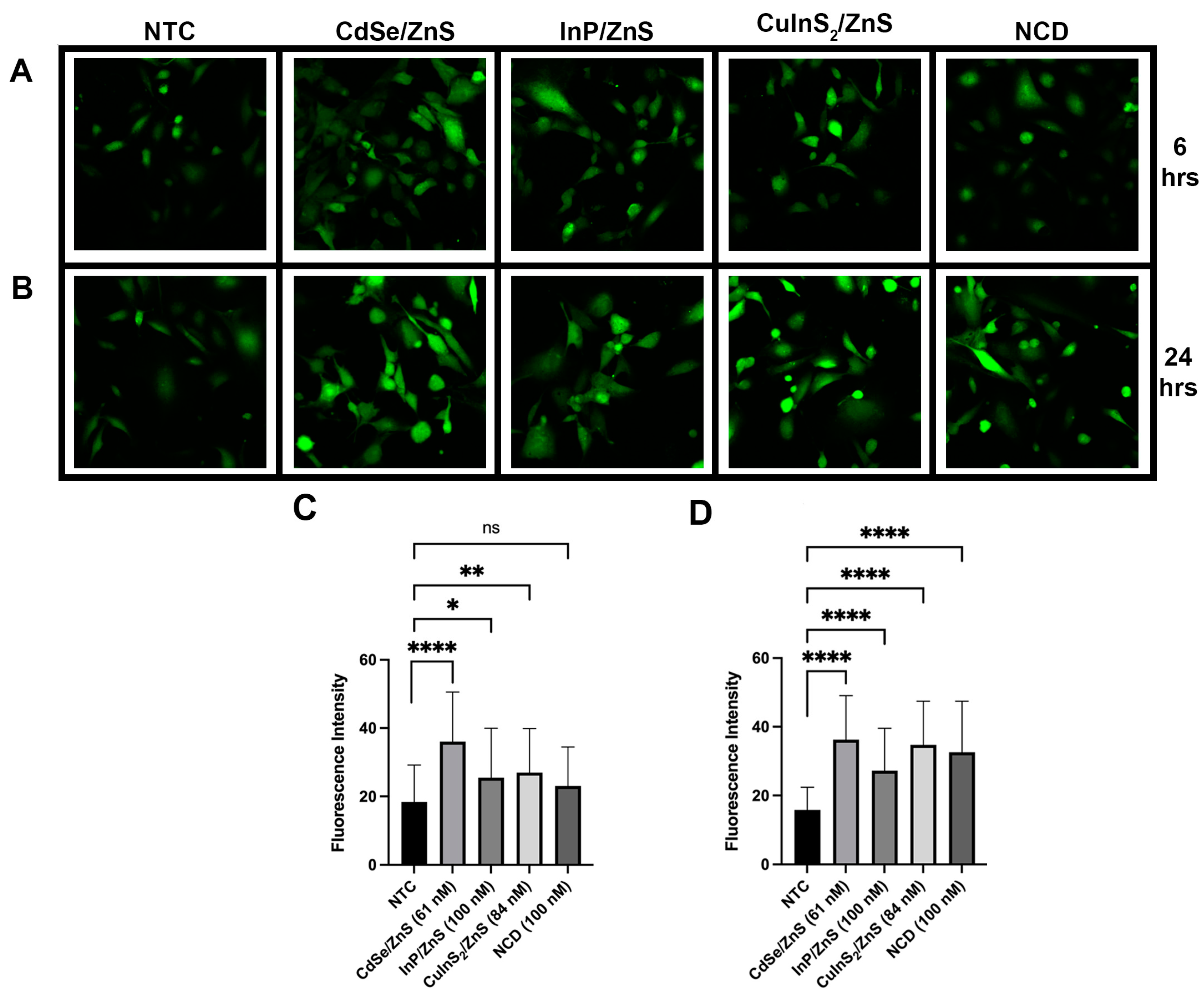
2.6. ROS Assay
Confocal Microscopy
2.7. Apoptosis Assay
Flow Cytometry Gating
2.8. RNA Transcriptomics
2.8.1. RNA Isolation
2.8.2. RNA Seq Analysis
2.9. Statistical Analysis
3. Results and Discussion
3.1. XPS Component Analysis
3.2. QD Peak Fluorescence
3.3. CdSe/ZnS and CuInS2/ZnS Show Reduction in THLE-2 Cell Viability
3.4. CdSe/ZnS- and CuInS2/ZnS-Treated THLE-2 Cells Exhibit Early ROS Generation
3.5. CdSe/ZnS and CuInS2/ZnS Treatment Causes THLE-2 Cells to Enter Early Stages of Apoptosis
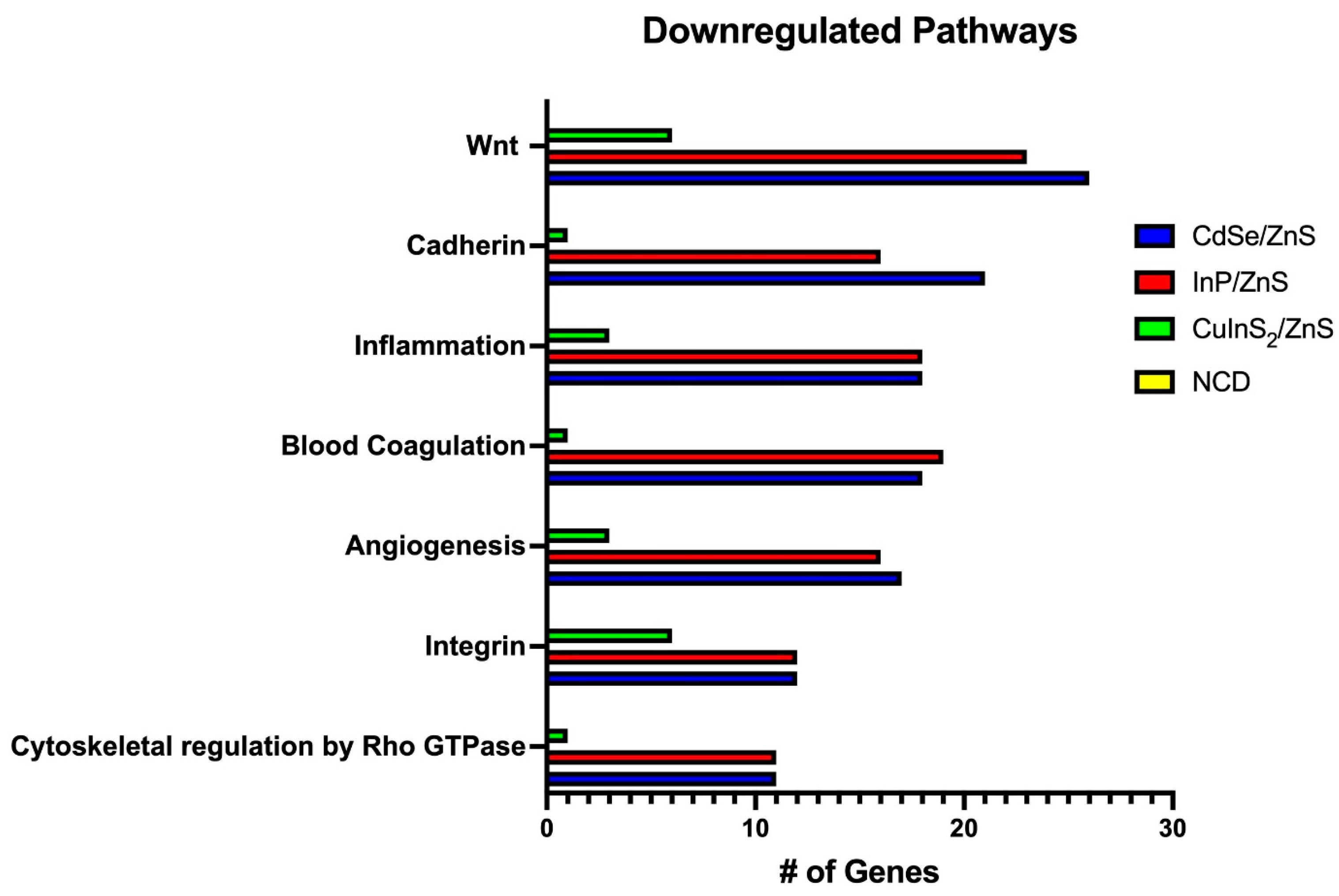
3.6. Transcriptomics Reveals Heavy Downregulation of Adherence Pathways
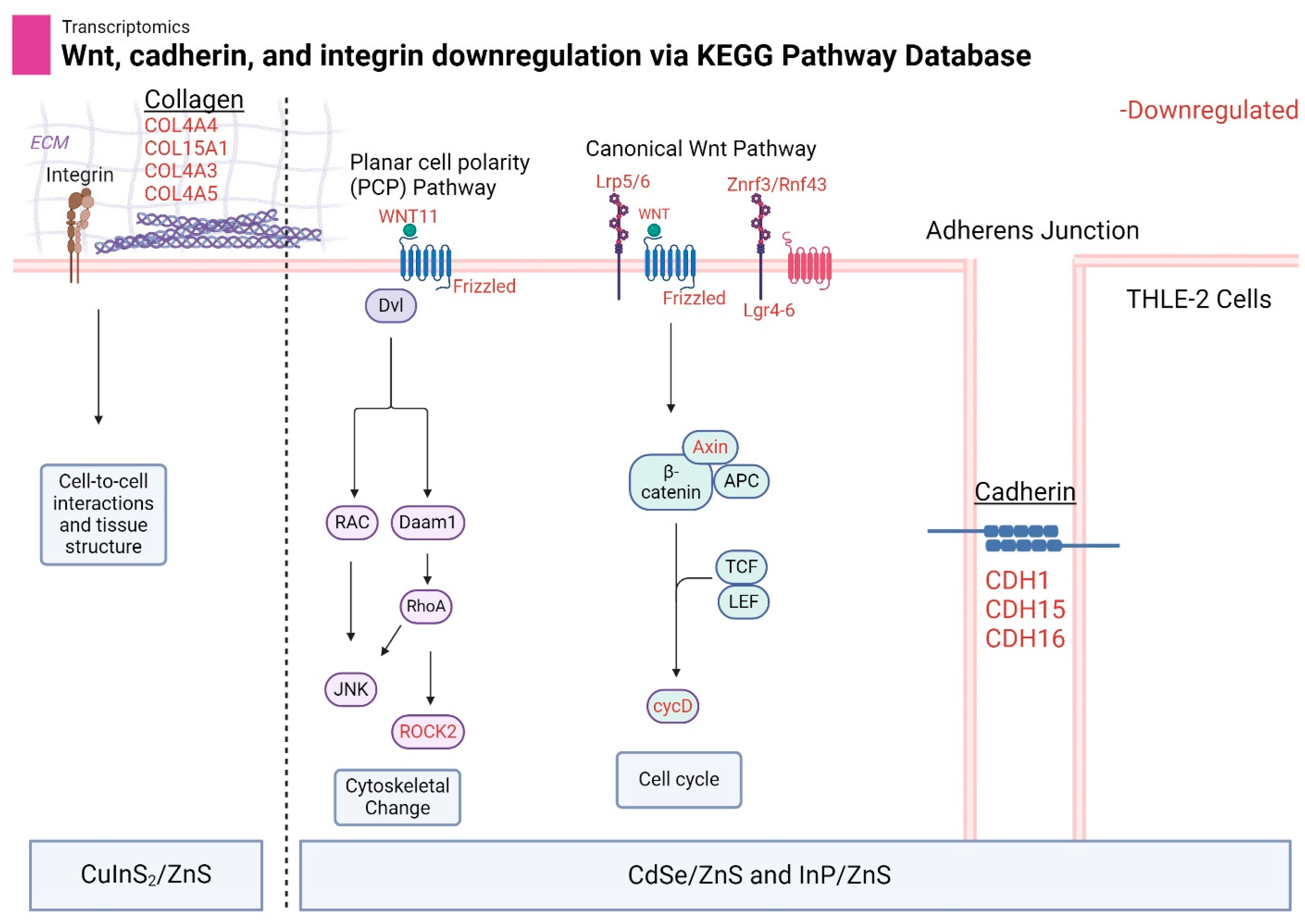
3.7. Wnt, Cadherin, and Integrin Pathways Are Downregulated by Metallic QDs
3.8. CD82, SYT12, and DUSP4 Are Upregulated by Metallic QDs
4. Future Directions
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, Z.; Lin, C.-H.; Hyun, B.-R.; Sher, C.-W.; Lv, Z.; Luo, B.; Jiang, F.; Wu, T.; Ho, C.-H.; Kuo, H.-C.; et al. Micro-light-emitting Diodes with Quantum Dots in Display Technology. Light Sci. Appl. 2020, 9, 83. [Google Scholar] [CrossRef] [PubMed]
- Baig, N.; Kammakakam, I.; Falath, W. Nanomaterials: A Review of Synthesis Methods, Properties, Recent Progress, and Challenges. Mater. Adv. 2021, 2, 1821–1871. [Google Scholar] [CrossRef]
- Abdellatif, A.A.H.; Younis, M.A.; Alsharidah, M.; Al Rugaie, O.; Tawfeek, H.M. Biomedical Applications of Quantum Dots: Overview, Challenges, and Clinical Potential. Int. J. Nanomed. 2022, 17, 1951–1970. [Google Scholar] [CrossRef] [PubMed]
- Le, N.; Zhang, M.; Kim, K. Quantum Dots and Their Interaction with Biological Systems. Int. J. Mol. Sci. 2022, 23, 763. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Tian, J.; Wang, X.; Xu, G.; Jiang, W.; Yang, Z.; Liu, D.; Lin, G. Cardiotoxicity of Intravenously Administered CdSe/ZnS Quantum Dots in BALB/c Mice. Front. Pharmacol. 2019, 10, 1179. [Google Scholar] [CrossRef] [PubMed]
- Hens, B.; Smothers, J.; Rizvanovic, H.; Patel, R.; Wu, Q.; Kim, K. The Future of Anticancer Drugs: A Cytotoxicity Assessment Study of CdSe/ZnS Quantum Dots. J. Nanotheranostics 2020, 1, 19–38. [Google Scholar] [CrossRef]
- Zhang, M.; Kim, D.S.; Patel, R.; Wu, Q.; Kim, K. Intracellular Trafficking and Distribution of Cd and InP Quantum Dots in HeLa and ML-1 Thyroid Cancer Cells. Nanomaterials 2022, 12, 1517. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; He, M.; Chen, B.; Wu, Q.; Zhang, Z.; Pang, D.; Zhu, Y.; Hu, B. Cellular uptake, elimination and toxicity of CdSe/ZnS quantum dots in HepG2 cells. Biomaterials 2013, 34, 9545–9558. [Google Scholar] [CrossRef]
- Chung, C.-Y.; Chen, Y.-J.; Kang, C.-H.; Lin, H.-Y.; Huang, C.-C.; Hsu, P.-H.; Lin, H.-J. Toxic or Not Toxic, That Is the Carbon Quantum Dot’s Question: A Comprehensive Evaluation with Zebrafish Embryo, Eleutheroembryo, and Adult Models. Polymers 2021, 13, 1598. [Google Scholar] [CrossRef]
- Derfus, A.M.; Chan, W.C.W.; Bhatia, S.N. Probing the Cytotoxicity of Semiconductor Quantum Dots. Nano Lett. 2004, 4, 11–18. [Google Scholar] [CrossRef]
- Manshian, B.B.; Martens, T.F.; Kantner, K.; Braeckmans, K.; De Smedt, S.C.; Demeester, J.; Jenkins, G.J.S.; Parak, W.J.; Pelaz, B.; Doak, S.H.; et al. The role of intracellular trafficking of CdSe/ZnS QDs on their consequent toxicity profile. J. Nanobiotechnol. 2017, 15, 45. [Google Scholar] [CrossRef]
- Abdal Dayem, A.; Hossain, M.K.; Lee, S.B.; Kim, K.; Saha, S.K.; Yang, G.-M.; Choi, H.Y.; Cho, S.-G. The Role of Reactive Oxygen Species (ROS) in the Biological Activities of Metallic Nanoparticles. Int. J. Mol. Sci. 2017, 18, 120. [Google Scholar] [CrossRef]
- Le, N.; Routh, J.; Kirk, C.; Wu, Q.; Patel, R.; Keyes, C.; Kim, K. Red CdSe/ZnS QDs’ Intracellular Trafficking and Its Impact on Yeast Polarization and Actin Filament. Cells 2023, 12, 484. [Google Scholar] [CrossRef]
- Zhang, Y.N.; Poon, W.; Tavares, A.J.; McGilvray, I.D.; Chan, W.C.W. Nanoparticle–liver Interactions: Cellular Uptake and Hepatobiliary Elimination. J. Control. Release 2016, 240, 332–348. [Google Scholar] [CrossRef] [PubMed]
- Kermanizadeh, A.; Powell, L.G.; Stone, V. A Review of Hepatic Nanotoxicology—Summation of Recent Findings and Considerations for the Next Generation of Study Designs. J. Toxicol. Environ. Health B Crit. Rev. 2020, 23, 137–176. [Google Scholar] [CrossRef] [PubMed]
- Tsoi, K.M.; MacParland, S.A.; Ma, X.Z.; Spetzler, V.N.; Echeverri, J.; Ouyang, B.; Fadel, S.M.; Sykes, E.A.; Goldaracena, N.; Kaths, J.M.; et al. Mechanism of Hard Nanomaterial Clearance by the Liver. Nat. Mater. 2016, 15, 1212–1221. [Google Scholar] [CrossRef]
- Arzumanian, V.A.; Kiseleva, O.I.; Poverennaya, E.V. The Curious Case of the HepG2 Cell Line: 40 Years of Expertise. Int. J. Mol. Sci. 2021, 22, 3135. [Google Scholar] [CrossRef]
- Pfeifer, A.M.; Cole, K.E.; Smoot, D.T.; Weston, A.; Groopman, J.D.; Shields, P.G.; Vignaud, J.M.; Juillerat, M.; Lipsky, M.M.; Trump, B.F. Simian Virus 40 Large Tumor Antigen-Immortalized Normal Human Liver Epithelial Cells Express Hepatocyte Characteristics and Metabolize Chemical Carcinogens. Proc. Natl. Acad. Sci. USA 1993, 90, 5123–5127. [Google Scholar] [CrossRef] [PubMed]
- Heng, W.S.; Cheah, S.C. Chelerythrine Chloride Downregulates β-Catenin and Inhibits Stem Cell Properties of Non-Small Cell Lung Carcinoma. Molecules 2020, 25, 224. [Google Scholar] [CrossRef]
- Prado, M.B.; Truong, N.T.; Wanekaya, A.K. Improving the quantum yield of nitrogen-doped carbon dots by varying dopant ratios and pH. Sens. Actuators Rep. 2023, 6, 100165. [Google Scholar] [CrossRef]
- Zou, W.; Li, L.; Chen, Y.; Chen, T.; Yang, Z.; Wang, J.; Liu, D.; Lin, G.; Wang, X. In Vivo Toxicity Evaluation of PEGylated CuInS2/ZnS Quantum Dots in BALB/c Mice. Front. Pharmacol. 2019, 10, 437. [Google Scholar] [CrossRef]
- Liu, L.; Xiao, Y.Y.; Ji, Y.H.; Liu, M.Z.; Chen, Y.; Zeng, Y.L.; Zhang, Y.; Jin, L. CuInS2/ZnS QD Exposure Induces Developmental Toxicity, Oxidative Stress and DNA Damage in Rare Minnow (Gobiocypris rarus) Embryos and Larvae. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2017, 198, 19–27. [Google Scholar] [CrossRef]
- Singh, V.; Kashyap, S.; Yadav, U.; Srivastava, A.; Singh, A.V.; Singh, R.K.; Singh, S.K.; Saxena, P.S. Nitrogen Doped Carbon Quantum Dots Demonstrate No Toxicity Under in vitro Conditions in a Cervical Cell Line and in vivo in Swiss Albino Mice. Toxicol. Res. 2019, 8, 395–406. [Google Scholar] [CrossRef]
- Lu, Y.; Xu, S.; Chen, H.; He, M.; Deng, Y.; Cao, Z.; Pi, H.; Chen, C.; Li, M.; Ma, Q.; et al. CdSe/ZnS Quantum Dots Induce Hepatocyte Pyroptosis and Liver Inflammation via NLRP3 Inflammasome Activation. Biomaterials 2016, 90, 27–39. [Google Scholar] [CrossRef]
- Yang, M.Y.; Hong, J.; Zhang, Y.; Gao, Z.; Jiang, T.-T.; Song, J.-L.; Xu, X.-L.; Zhu, L.-X. Bio-compatibility and Cytotoxicity Studies of Water-Soluble CuInS2-ZnS-AFP Fluorescence Probe in Liver Cancer Cells. Hepatobiliary Pancreat. Dis. Int. 2016, 15, 406–411. [Google Scholar] [CrossRef]
- Shlomovitz, I.; Speir, M.; Gerlic, M. Flipping the Dogma—Phosphatidylserine in Non-Apoptotic Cell Death. Cell Commun. Signal. 2019, 17, 139. [Google Scholar] [CrossRef]
- Sharma, M.; Pruitt, K. Wnt Pathway: An Integral Hub for Developmental and Oncogenic Signaling Networks. Int. J. Mol. Sci. 2020, 21, 8018. [Google Scholar] [CrossRef]
- Qin, K.; Yu, M.; Fan, J.; Wang, H.; Zhao, P.; Zhao, G.; Zeng, W.; Chen, C.; Wang, Y.; Wang, A.; et al. Canonical and Noncanonical Wnt Signaling: Multilayered Mediators, Signaling Mechanisms and Major Signaling Crosstalk. Genes. Dis. 2024, 11, 103–134. [Google Scholar] [CrossRef]
- Le, N.; Chand, A.; Braun, E.; Keyes, C.; Wu, Q.; Kim, K. Interactions between Quantum Dots and G-Actin. Int. J. Mol. Sci. 2023, 24, 14760. [Google Scholar] [CrossRef]
- Maître, J.L.; Heisenberg, C.P. Three Functions of Cadherins in Cell Adhesion. Curr. Biol. 2013, 23, R626–R633. [Google Scholar] [CrossRef]
- Zismanov, V.; Lishner, M.; Tartakover-Matalon, S.; Radnay, J.; Shapiro, H.; Drucker, L. Tetraspanin-induced Death of Myeloma Cell Lines is Autophagic and Involves Increased UPR Signalling. Br. J. Cancer 2009, 101, 1402–1409. [Google Scholar] [CrossRef] [PubMed]
- Ono, M.; Handa, K.; Withers, D.A.; Hakomori, S. Motility Inhibition and Apoptosis are Induced by Metastasis-suppressing Gene Product CD82 and its Analogue CD9, with Concurrent Glycosylation. Cancer Res. 1999, 59, 2335–2339. [Google Scholar] [PubMed]
- Schoenfeld, N.; Bauer, M.K.; Grimm, S. The Metastasis Suppressor Gene C33/CD82/KAI1 Induces Apoptosis Through Reactive Oxygen Intermediates. FASEB J. 2004, 18, 158–160. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.C.; Weissman, A.M. Dissecting the Diverse Functions of the Metastasis Suppressor CD82/KAI1. FEBS Lett. 2011, 585, 3166–3173. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, J.A.; Kilbane Myers, J.; Khan, M.; Angelos, M.G.; Chen, C.A. Dual-Specificity Phosphatase 4 Overexpression in Cells Prevents Hypoxia/Reoxygenation-Induced Apoptosis via the Upregulation of eNOS. Front. Cardiovasc. Med. 2017, 4, 22. [Google Scholar] [CrossRef]
- Kaeser-Woo, Y.J.; Younts, T.J.; Yang, X.; Zhou, P.; Wu, D.; Castillo, P.E.; Südhof, T.C. Synaptotagmin-12 Phosphorylation by cAMP-dependent Protein Kinase is Essential for Hippocampal Mossy Fiber LTP. J. Neurosci. 2013, 33, 9769–9780. [Google Scholar] [CrossRef]



| QD Type | Number of Genes | |
|---|---|---|
| Upregulated | Downregulated | |
| CdSe/ZnS | 53 | 1105 |
| InP/ZnS | 53 | 1030 |
| CuInS2/ZnS | 191 | 392 |
| NCD | 38 | 49 |
| Gene | CdSe/ZnS | InP/ZnS | CuInS2/ZnS | NCD |
|---|---|---|---|---|
| CD82 | 1.706 | 1.713 | 2.089 | n/a |
| SYT12 | 1.835 | 1.745 | 1.944 | n/a |
| DUSP4 | 1.644 | 1.508 | 1.577 | n/a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harris, S.; Kim, K. A Comparison of Common Quantum Dot Alternatives to Cadmium-Based Quantum Dots on the Basis of Liver Cytotoxicity. Nanomaterials 2024, 14, 1086. https://doi.org/10.3390/nano14131086
Harris S, Kim K. A Comparison of Common Quantum Dot Alternatives to Cadmium-Based Quantum Dots on the Basis of Liver Cytotoxicity. Nanomaterials. 2024; 14(13):1086. https://doi.org/10.3390/nano14131086
Chicago/Turabian StyleHarris, Seth, and Kyoungtae Kim. 2024. "A Comparison of Common Quantum Dot Alternatives to Cadmium-Based Quantum Dots on the Basis of Liver Cytotoxicity" Nanomaterials 14, no. 13: 1086. https://doi.org/10.3390/nano14131086





