Instrumentation-Free Semiquantitative Immunoanalysis Using a Specially Patterned Lateral Flow Assay Device
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Instruments
2.2. Preparation of Optical Probes, Including AuNPs and Ab-AuNPs
2.3. Preparation of SQ-LFI Strips
2.4. Operation of SQ-LFI Strips
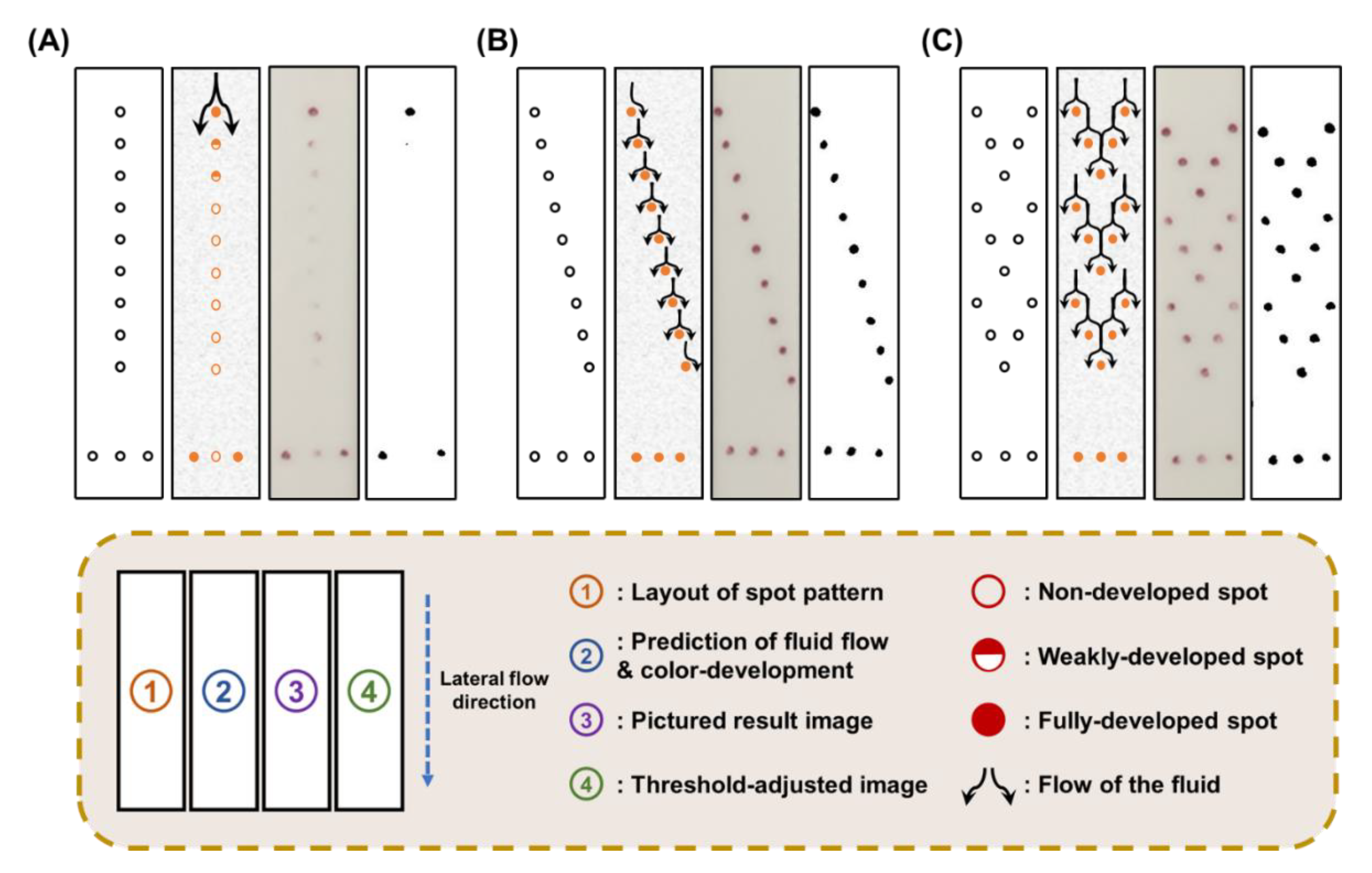
2.5. Examination of the Effect of Spot-Pattern Alterations on Color Development
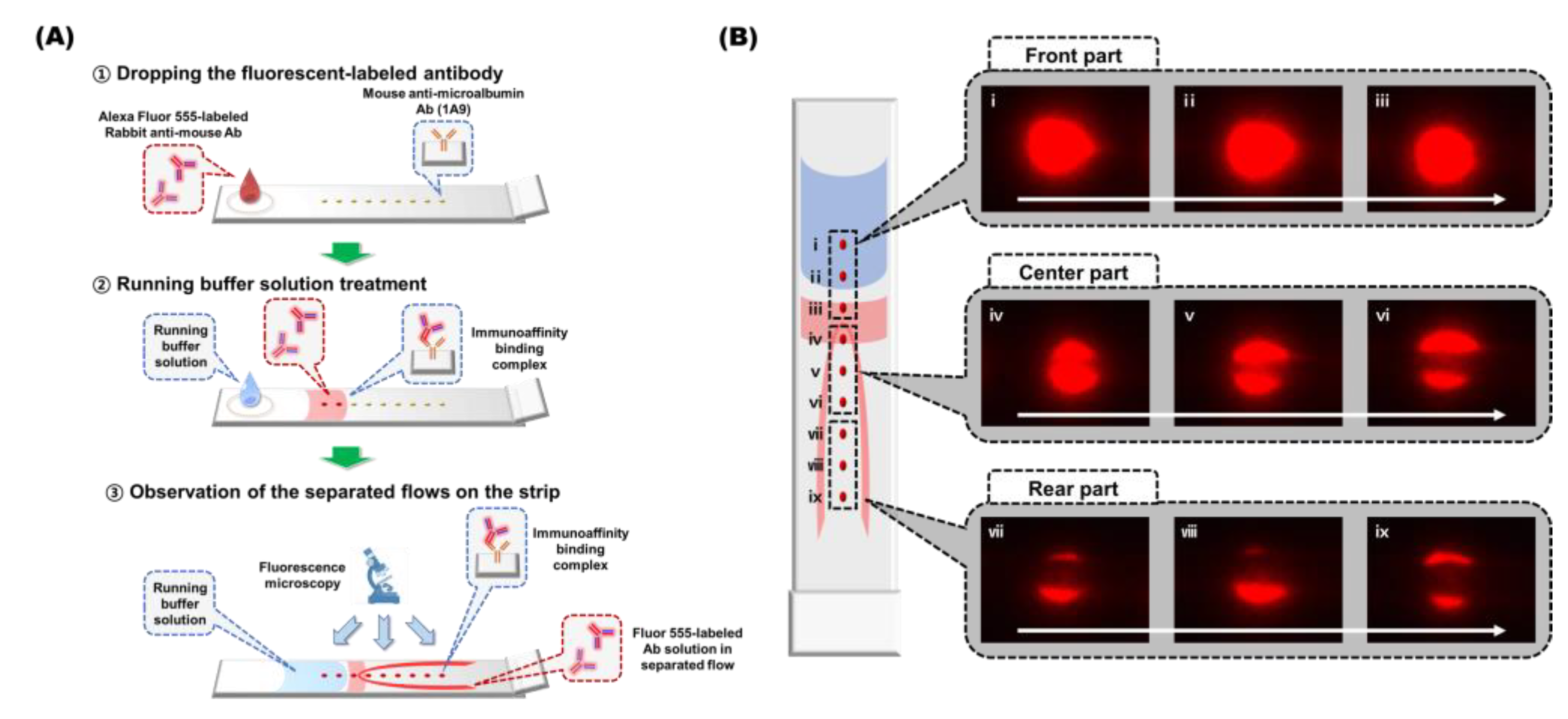
2.6. Observation of Flows in the SQ-LFI
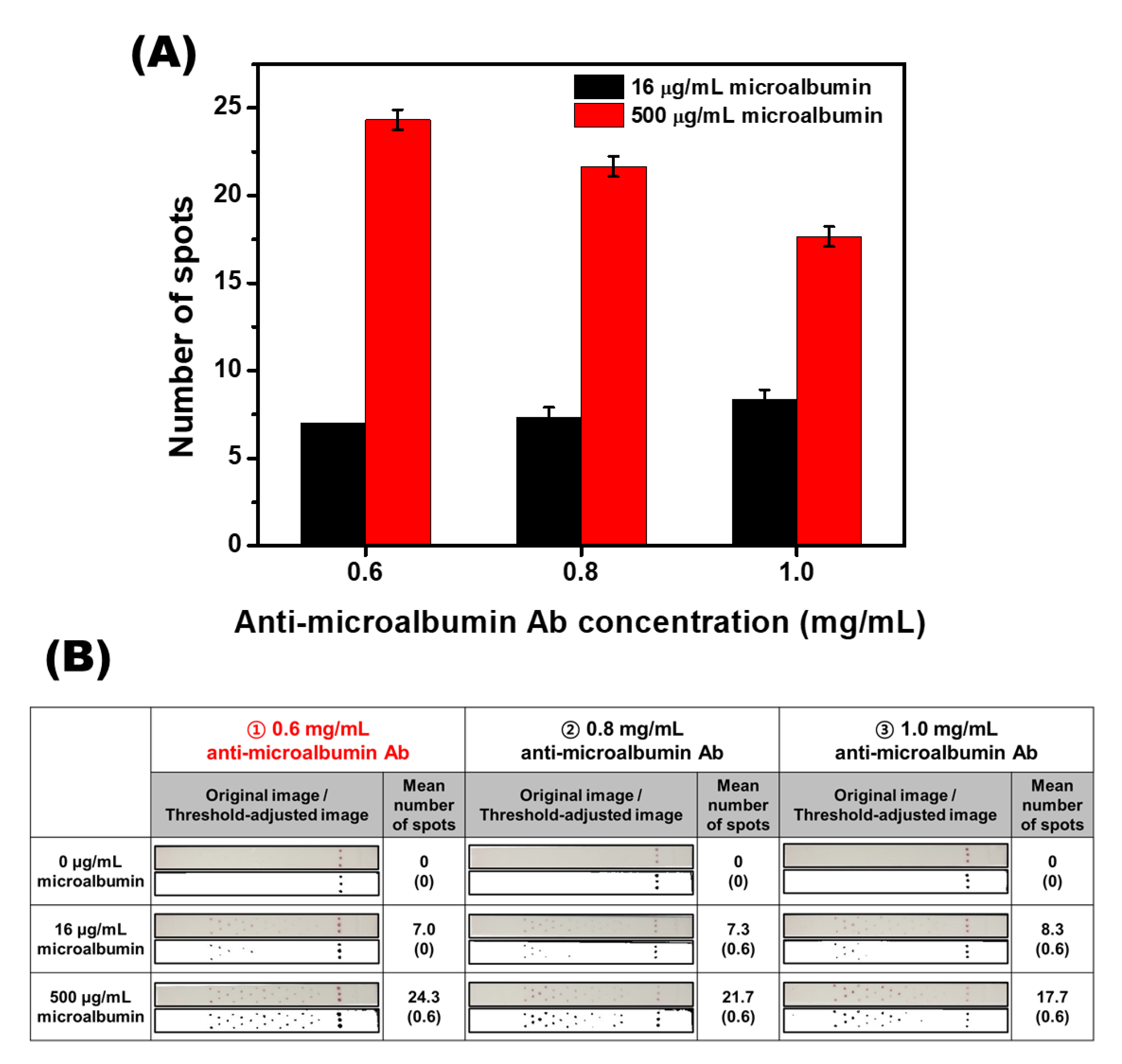
2.7. Signal Optimization of the SQ-LFI
2.8. Quantification of Microalbumin in Buffer Samples and Artificial Urine Samples
3. Results and Discussion
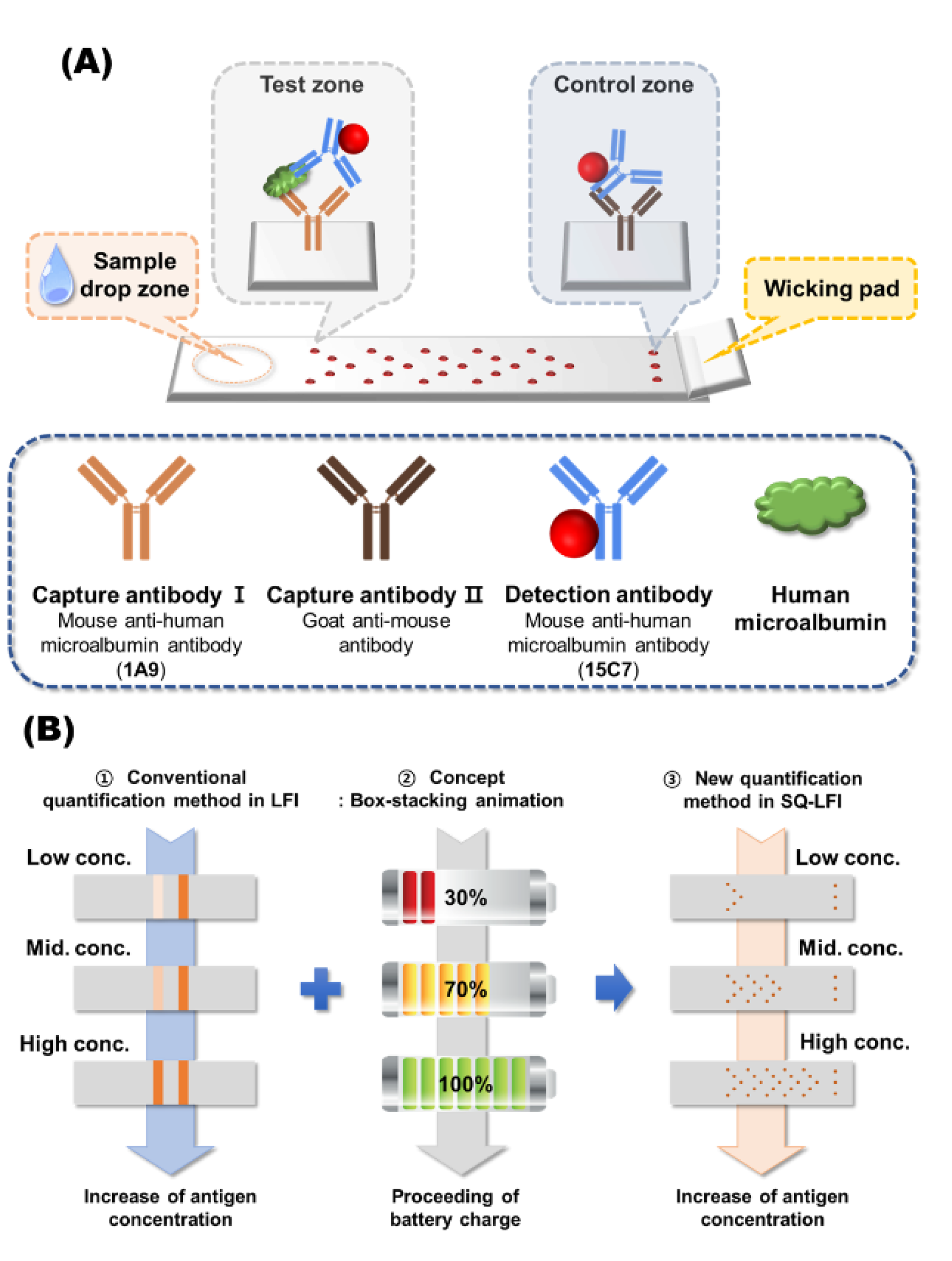
3.1. Signaling Principle of the Developed SQ-LFI System
3.2. Effect of Spot-Pattern Alterations on the Signal Development of LFI
3.3. Flow Behavior Observation of the SQ-LFI Strip
3.4. Signal Optimization of the Herringbone Pattern SQ-LFI
3.5. Calibration Study with Microalbumin-Spiked PBS Samples
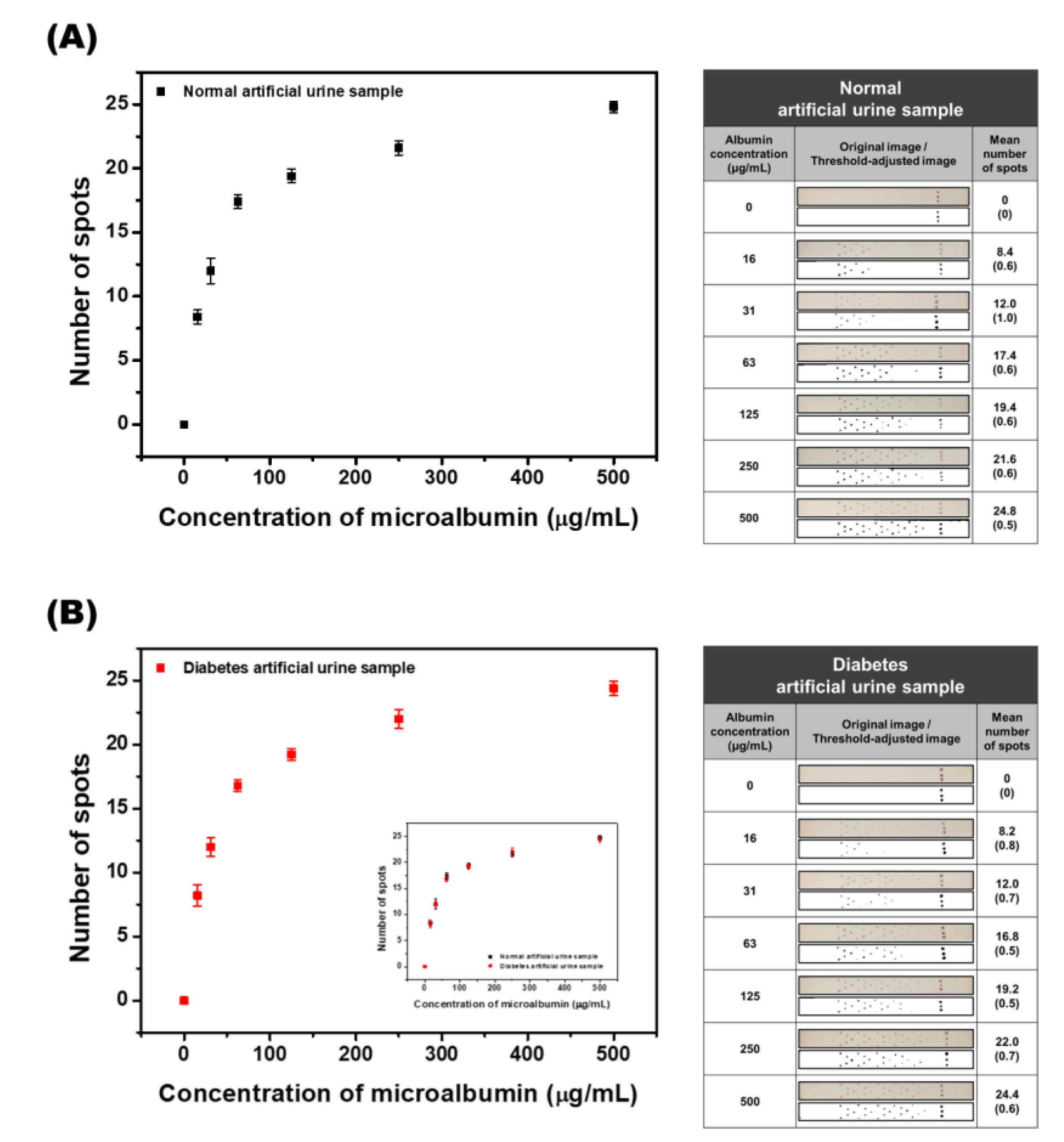
3.6. Calibration Study with Standard Artificial Urine and Diabetes Artificial Urine Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Dhiman, A.; Kalra, P.; Bansal, V.; Bruno, J.G.; Sharma, T.K. Aptamer-based point-of-care diagnostic platforms. Sens. Actuators B Chem. 2017, 246, 535–553. [Google Scholar] [CrossRef]
- Darwish, N.T.; Sekaran, S.D.; Khor, S.M. Point-of-care tests: A review of advances in the emerging diagnostic tools for dengue virus infection. Sens. Actuators B Chem. 2018, 255, 3316–3331. [Google Scholar] [CrossRef]
- Wang, P.; Kricka, L.J. Current and emerging trends in point-of-care technology and strategies for clinical validation and implementation. Clin. Chem. 2018, 64, 1439–1452. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Wang, K.; Xu, H.; Yan, W.; Jin, Q.; Cui, D. Detection platforms for point-of-care testing based on colorimetric, luminescent and magnetic assays: A review. Talanta 2019, 202, 96–110. [Google Scholar] [CrossRef] [PubMed]
- Syedmoradi, L.; Daneshpour, M.; Alvandipour, M.; Gomez, F.A.; Hajghassem, H.; Omidfar, K. Point of care testing: The impact of nanotechnology. Biosens. Bioelectron. 2017, 87, 373–387. [Google Scholar] [CrossRef]
- Nasseri, B.; Soleimani, N.; Rabiee, N.; Kalbasi, A.; Karimi, M.; Hamblin, M.R. Point-of-care microfluidic devices for pathogen detection. Biosens. Bioelectron. 2018, 117, 112–128. [Google Scholar] [CrossRef]
- Han, Y.D.; Chun, H.J.; Yoon, H.C. Low-cost point-of-care biosensors using common electronic components as transducers. Biochip J. 2020, 14, 32–47. [Google Scholar] [CrossRef] [Green Version]
- Mansfield, M.A. Nitrocellulose membranes for lateral flow immunoassays: A technical treatise. In Lateral Flow Immunoassay; Wong, R.C., Tse, H.Y., Eds.; Springer Science Business Media: New York, NY, USA, 2009; pp. 1–19. [Google Scholar]
- Huang, X.; Aguilar, Z.P.; Xu, H.; Lai, W.; Xiong, Y. Membrane-based lateral flow immunochromatographic strip with nanoparticles as reporters for detection: A review. Biosens. Bioelectron. 2016, 75, 166–180. [Google Scholar] [CrossRef]
- Bahadır, E.B.; Sezgintürk, M.K. Lateral flow assays: Principles, designs and labels. Trends Anal. Chem. 2016, 82, 286–306. [Google Scholar] [CrossRef]
- Wu, Y.; Zhou, Y.; Leng, Y.; Lai, W.; Huang, X.; Xiong, Y. Emerging design strategies for constructing multiplex lateral flow test strip sensors. Biosens. Bioelectron. 2020, 157, 112168. [Google Scholar] [CrossRef]
- Rodríguez, M.O.; Covián, L.B.; García, A.C.; Blanco-López, M.C. Silver and gold enhancement methods for lateral flow immunoassays. Talanta 2016, 148, 272–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, X.; Liu, Z.; Yang, Y.; Li, L.; Wang, L.; Li, A.; Qu, Z.; Xu, F. Sensitivity enhancement of nucleic acid lateral flow assays through a physical−chemical coupling method: Dissoluble saline barriers. ACS Sens. 2019, 4, 1691–1700. [Google Scholar] [CrossRef] [PubMed]
- Han, S.-M.; Kim, Y.-W.; Kim, Y.-K.; Chun, J.-H.; Oh, H.-B.; Paek, S.-H. Performance characterization of two-dimensional paper chromatography-based biosensors for biodefense, exemplified by detection of Bacillus anthracis spores. BioChip J. 2018, 12, 59–68. [Google Scholar] [CrossRef]
- Yuzon, M.K.; Kim, J.-H.; Kim, S. A novel paper-plastic microfluidic hybrid chip integrated with a lateral flow immunoassay for Dengue nonstructural protein 1 antigen detection. BioChip J. 2019, 13, 277–287. [Google Scholar] [CrossRef]
- Zhu, H.; Isikman, S.O.; Mudanyali, O.; Greenbaum, A.; Ozcan, A. Optical imaging techniques for point-of-care diagnostics. Lab Chip 2013, 13, 51–67. [Google Scholar] [CrossRef]
- Martinez, A.W.; Phillips, S.T.; Carrilho, E.; Thomas, S.W., III; Sindi, H.; Whitesides, G.M. Simple telemedicine for developing regions: Camera phones and paper-based microfluidic devices for real-time, off-site diagnosis. Anal. Chem. 2008, 80, 3699–3707. [Google Scholar] [CrossRef] [Green Version]
- Pilavaki, E.; Demosthenous, A. Optimized lateral flow immunoassay reader for the detection of infectious diseases in developing countries. Sensors 2017, 17, 2673. [Google Scholar] [CrossRef] [Green Version]
- O’Farrell, B. Lateral flow technology for field-based applications—Basics and advanced developments. Top. Companion Anim. Med. 2015, 30, 139–147. [Google Scholar] [CrossRef]
- Saisin, L.; Amarit, R.; Somboonkaew, A.; Gajanandana, O.; Himananto, O.; Sutapun, B. Significant sensitivity improvement for camera-based lateral flow immunoassay readers. Sensors 2018, 18, 4026. [Google Scholar] [CrossRef] [Green Version]
- Urusov, A.E.; Zherdev, A.V.; Dzantiev, B.B. Towards lateral flow quantitative assays: Detection approaches. Biosensors 2019, 9, 89. [Google Scholar] [CrossRef] [Green Version]
- Leung, W.; Chan, C.P.; Rainer, T.H.; Ip, M.; Cautherley, G.W.H.; Renneberg, R. InfectCheck CRP barcode-style lateral flow assay for semi-quantitative detection of C-reactive protein in distinguishing between bacterial and viral infections. J. Immunol. Methods 2008, 336, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Chen, Z.; Li, L.; Xia, J. Barcode lateral flow immunochromatographic strip for prostate acid phosphatase determination. J. Pharm. Biomed. 2011, 56, 1035–1040. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.-Y.; Chu, P.-T.; Tsai, W.-C.; Wen, H.-W. Development of a barcode-style lateral flow immunoassay for the rapid semi-quantification of gliadin in foods. Food Chem. 2016, 192, 934–942. [Google Scholar] [CrossRef] [PubMed]
- Toto, R.D. Microalbuminuria: Definition, detection, and clinical significance. J. Clin. Hypertens. 2004, 6, 2–7. [Google Scholar] [CrossRef] [Green Version]
- Rabelink, T.J.; Heerspink, H.J.L.; de Zeeuw, D. The pathophysiology of proteinuria. In Chronic Renal Disease, 1st ed.; Kimmel, P.L., Rosenberg, M.E., Eds.; Elsevier Inc.: London, UK, 2015; pp. 92–105. [Google Scholar]
- Futrakul, N.; Sridama, V.; Futrakul, P. Microalbuminuria—A biomarker of renal microvascular disease. Ren. Fail. 2009, 31, 140–143. [Google Scholar] [CrossRef]
- Jawa, A.; Kcomt, J.; Fonseca, V.A. Diabetic nephropathy and retinopathy. Med. Clin. N. Am. 2004, 88, 1001–1036. [Google Scholar] [CrossRef]
- Satchell, S.C.; Tooke, J.E. What is the mechanism of microalbuminuria in diabetes: A role for the glomerular endothelium? Diabetologia 2008, 51, 714–725. [Google Scholar] [CrossRef] [Green Version]
- Parving, H.-H.; Persson, F.; Rossing, P. Microalbuminuria: A parameter that has changed diabetes care. Diabetes Res. Clin. Pract. 2015, 107, 1–8. [Google Scholar] [CrossRef]
- Roett, M.A.; Liegl, S.; Jabbarpour, Y. Diabetic nephropathy—The family physician’s role. Am. Fam. Physician 2012, 85, 883–889. [Google Scholar]
- Frens, G. Controlled nucleation for the regulation of the particle size in monodisperse gold suspensions. Nat. Phys. Sci. 1973, 241, 20–22. [Google Scholar] [CrossRef]
- Hong, S.Y.; Park, Y.M.; Jang, Y.H.; Min, B.-H.; Yoon, H.C. Quantitative lateral-flow immunoassay for the assessment of the cartilage oligomeric matrix protein as a marker of osteoarthritis. Biochip J. 2012, 6, 213–220. [Google Scholar] [CrossRef]
- Thanh, N.T.K.; Aveyard, J.; Fernig, D.G. Determination of size and concentration of gold nanoparticles from UV-Vis spectra. Anal. Chem. 2007, 79, 4215–4221. [Google Scholar]
- Mendez, S.; Fenton, E.M.; Gallegos, G.R.; Petsev, D.N.; Sibbett, S.S.; Stone, H.A.; Zhang, Y.; Lopez, G.P. Imbibition in porous membranes of complex shape: Quasi-stationary flow in thin rectangular segments. Langmuir 2010, 26, 1380–1385. [Google Scholar] [CrossRef] [PubMed]
- Osborn, J.L.; Lutz, B.; Fu, E.; Kauffman, P.; Stevens, D.Y.; Yager, P. Microfluidics without pumps: Reinventing the T-sensor and H-filter in paper networks. Lab Chip 2010, 10, 2659–2665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fridley, D.E.; Le, H.Q.; Fu, E.; Yager, P. Controlled release of dry reagents in porous media for tunable temporal and spatial distribution upon rehydration. Lab Chip 2012, 12, 4321–4327. [Google Scholar] [CrossRef] [Green Version]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, K.W.; Yu, Y.C.; Chun, H.J.; Jang, Y.H.; Han, Y.D.; Yoon, H.C. Instrumentation-Free Semiquantitative Immunoanalysis Using a Specially Patterned Lateral Flow Assay Device. Biosensors 2020, 10, 87. https://doi.org/10.3390/bios10080087
Lee KW, Yu YC, Chun HJ, Jang YH, Han YD, Yoon HC. Instrumentation-Free Semiquantitative Immunoanalysis Using a Specially Patterned Lateral Flow Assay Device. Biosensors. 2020; 10(8):87. https://doi.org/10.3390/bios10080087
Chicago/Turabian StyleLee, Kyung Won, Ye Chan Yu, Hyeong Jin Chun, Yo Han Jang, Yong Duk Han, and Hyun C. Yoon. 2020. "Instrumentation-Free Semiquantitative Immunoanalysis Using a Specially Patterned Lateral Flow Assay Device" Biosensors 10, no. 8: 87. https://doi.org/10.3390/bios10080087





