Bioengineering of Genetically Encoded Gene Promoter Repressed by the Flavonoid Apigenin for Constructing Intracellular Sensor for Molecular Events
Abstract
:1. Introduction
2. Materials and Methods
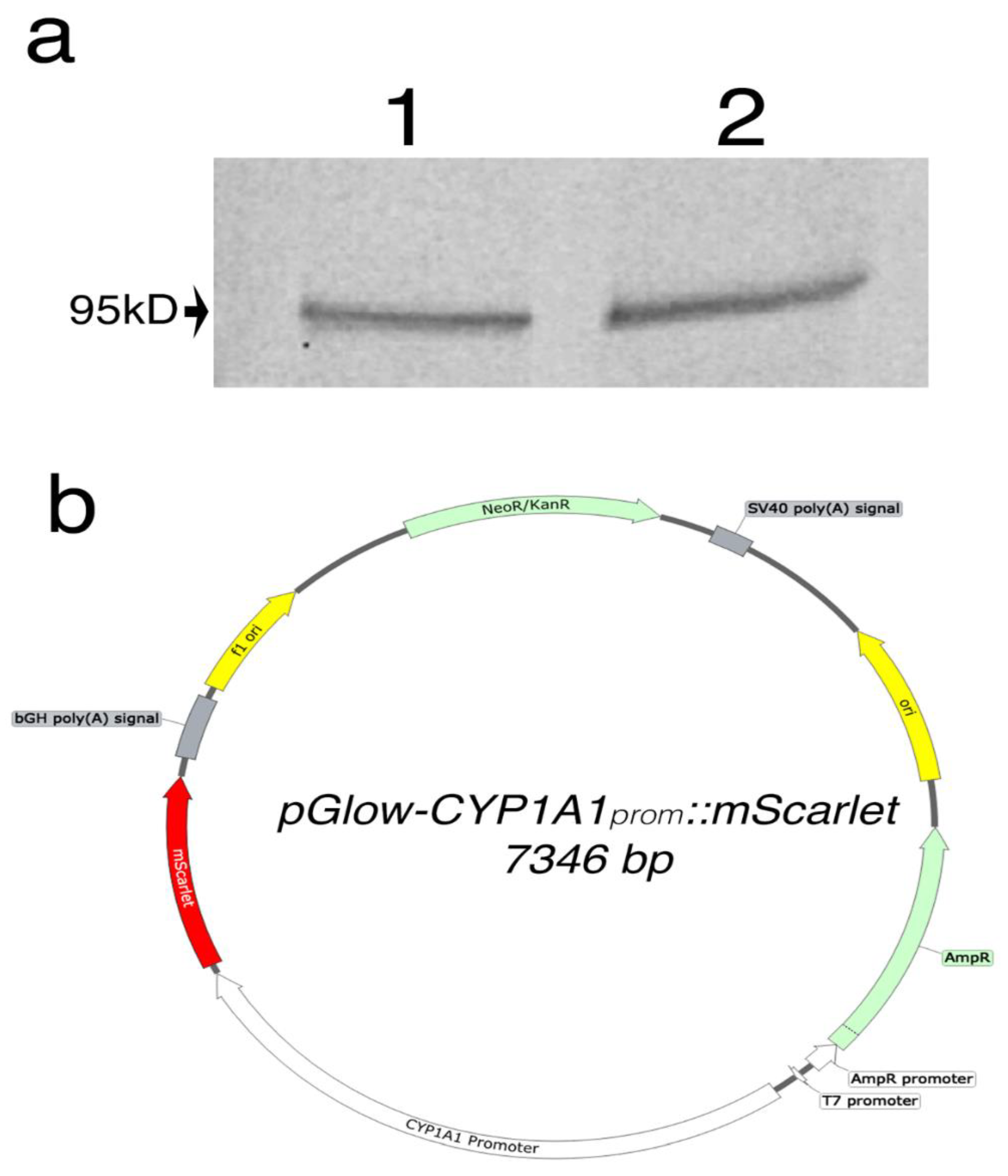
2.1. Plasmid Synthesis and Construction
2.2. Cell Culture and Western Blot
2.3. Transfection
2.4. Flavonoid Treatment
2.5. Imaging
2.6. Quantitative Analysis
2.7. Structure Prediction, Docking and Validation
3. Results and Discussion
3.1. Identifying the Optimal Promoter Sequence
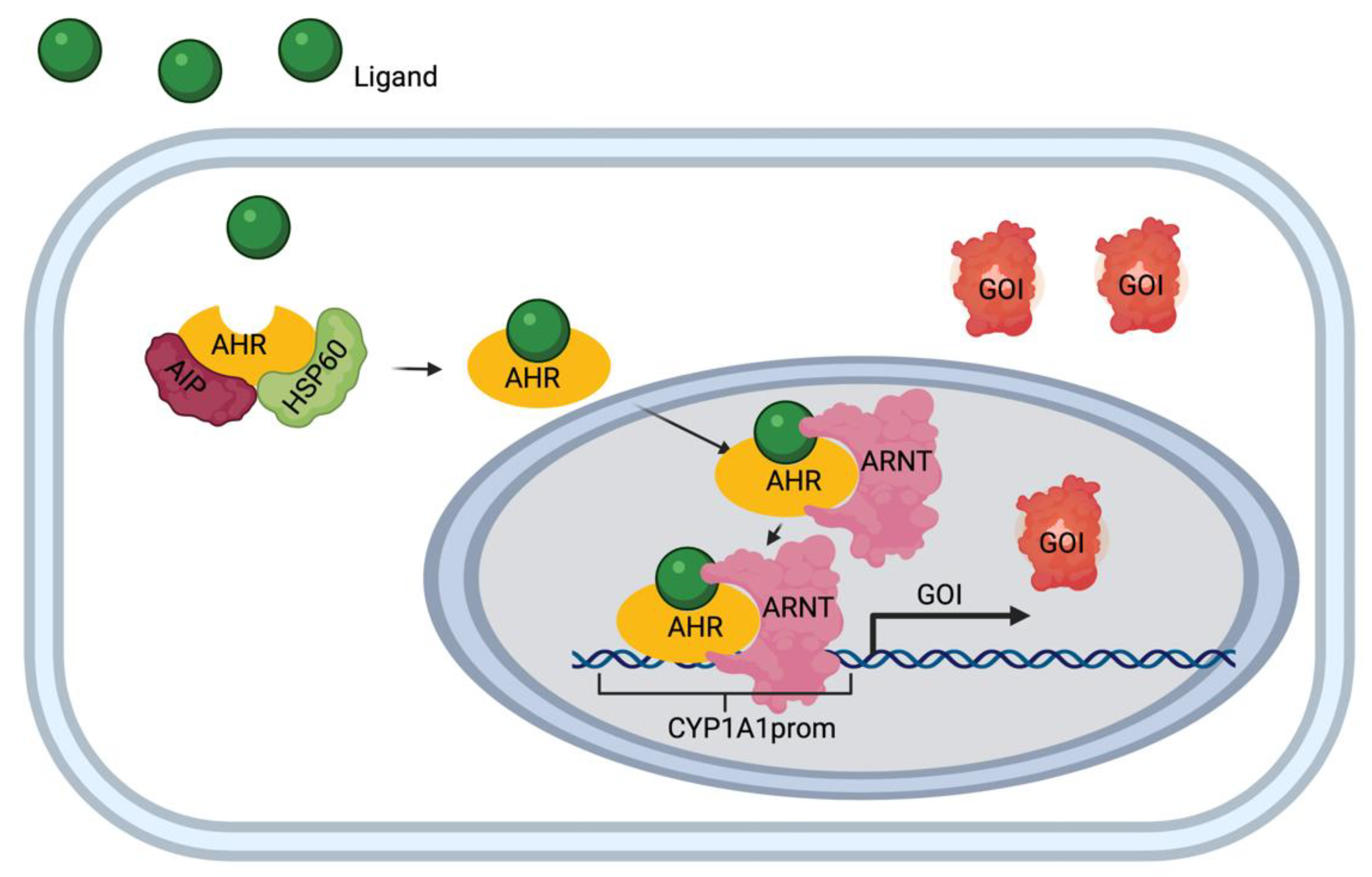
3.2. Bioengineering an Optical Reporter System
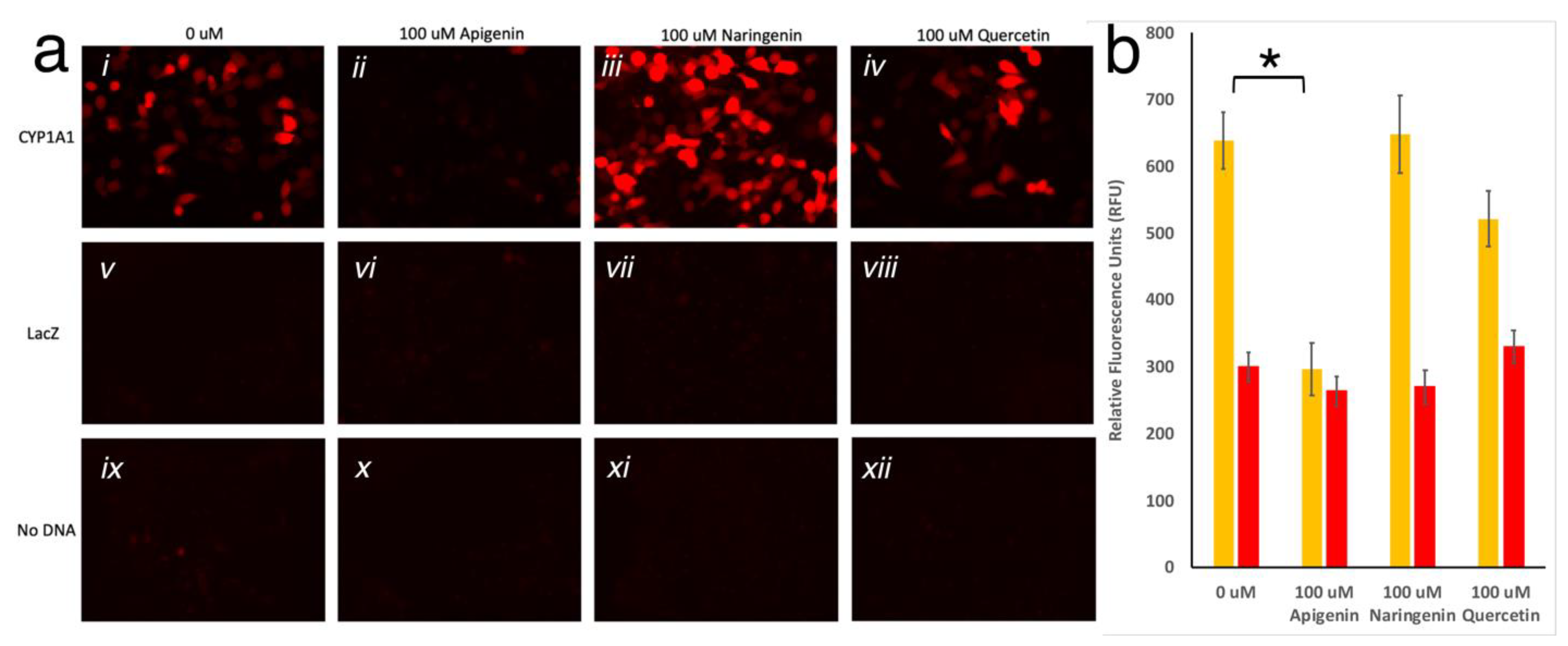
3.3. Testing the Gene Switch Reporter System In Vivo
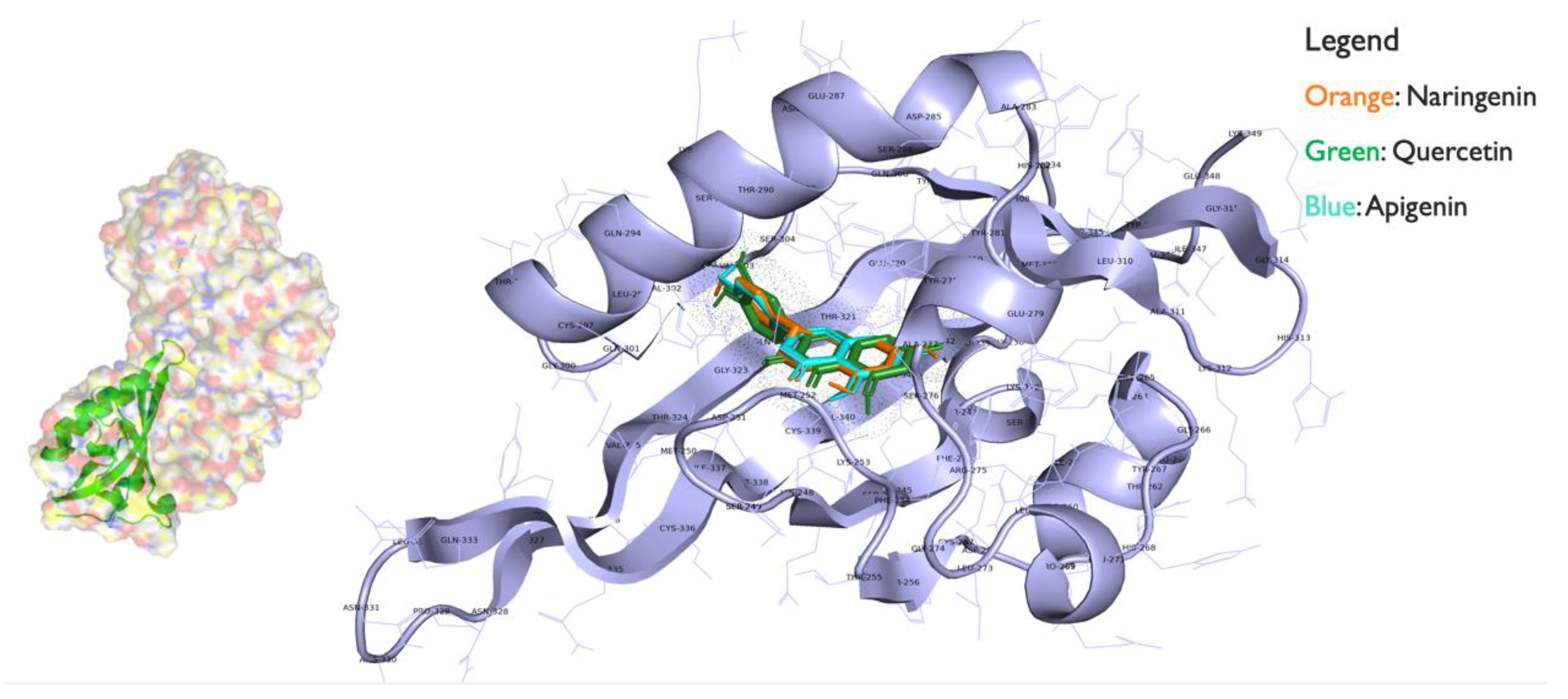
3.4. Ligand/Receptor Binding Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ruder, W.C.; Lu, T.; Collins, J.J. Synthetic biology moving into the clinic. Science 2011, 333, 1248–1252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cameron, D.E.; Bashor, C.J.; Collins, J.J. A brief history of synthetic biology. Nat. Rev. Microbiol. 2014, nrmicro3239. [Google Scholar] [CrossRef]
- Nussinovitch, U.; Gepstein, L. Optogenetics for suppression of cardiac electrical activity in human and rat cardiomyocyte cultures. NEUROW 2015, 2, 031204. [Google Scholar] [CrossRef] [PubMed]
- Airan, R.D.; Li, N.; Gilad, A.A.; Pelled, G. Genetic tools to manipulate MRI contrast. NMR Biomed. 2013, 26, 803–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, F.; Gradinaru, V.; Adamantidis, A.R.; Durand, R.; Airan, R.D.; de Lecea, L.; Deisseroth, K. Optogenetic interrogation of neural circuits: Technology for probing mammalian brain structures. Nat. Protoc. 2010, 5, 439–456. [Google Scholar] [CrossRef] [Green Version]
- Motta-Mena, L.B.; Reade, A.; Mallory, M.J.; Glantz, S.; Weiner, O.D.; Lynch, K.W.; Gardner, K.H. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat. Chem. Biol. 2014, 10, 196–202. [Google Scholar] [CrossRef]
- St-Pierre, F.; Marshall, J.D.; Yang, Y.; Gong, Y.; Schnitzer, M.J.; Lin, M.Z. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat. Neurosci. 2014, 17, 884–889. [Google Scholar] [CrossRef] [Green Version]
- Gomez, J.L.; Bonaventura, J.; Lesniak, W.; Mathews, W.B.; Sysa-Shah, P.; Rodriguez, L.A.; Ellis, R.J.; Richie, C.T.; Harvey, B.K.; Dannals, R.F.; et al. Chemogenetics revealed: DREADD occupancy and activation via converted clozapine. Science 2017, 357, 503–507. [Google Scholar] [CrossRef] [Green Version]
- Wess, J.; Nakajima, K.; Jain, S. Novel designer receptors to probe GPCR signaling and physiology. Trends Pharmacol. Sci. 2013, 34, 385–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cywiak, C.; Ashbaugh, R.C.; Metto, A.C.; Udpa, L.; Qian, C.; Gilad, A.A.; Reimers, M.; Zhong, M.; Pelled, G. Non-invasive neuromodulation using rTMS and the electromagnetic-perceptive gene (EPG) facilitates plasticity after nerve injury. Brain Stimul. 2020, 13, 1774–1783. [Google Scholar] [CrossRef]
- Hwang, J.; Choi, Y.; Lee, K.; Krishnan, V.; Pelled, G.; Gilad, A.A.; Choi, J. Regulation of Electromagnetic Perceptive Gene Using Ferromagnetic Particles for the External Control of Calcium Ion Transport. Biomolecules 2020, 10, 308. [Google Scholar] [CrossRef] [Green Version]
- Krishnan, V.; Park, S.A.; Shin, S.S.; Alon, L.; Tressler, C.M.; Stokes, W.; Banerjee, J.; Sorrell, M.E.; Tian, Y.; Fridman, G.Y.; et al. Wireless control of cellular function by activation of a novel protein responsive to electromagnetic fields. Sci. Rep. 2018, 8, 8764. [Google Scholar] [CrossRef]
- Gilad, A.A.; Shapiro, M.G. Molecular Imaging in Synthetic Biology, and Synthetic Biology in Molecular Imaging. Mol. Imaging Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Gilad, A.A.; McMahon, M.T.; Walczak, P.; Winnard, P.T., Jr.; Raman, V.; van Laarhoven, H.W.; Skoglund, C.M.; Bulte, J.W.; van Zijl, P.C. Artificial reporter gene providing MRI contrast based on proton exchange. Nat. Biotechnol. 2007, 25, 217–219. [Google Scholar] [CrossRef] [PubMed]
- Myklatun, A.; Lauri, A.; Eder, S.H.K.; Cappetta, M.; Shcherbakov, D.; Wurst, W.; Winklhofer, M.; Westmeyer, G.G. Author Correction: Zebrafish and medaka offer insights into the neurobehavioral correlates of vertebrate magnetoreception. Nat. Commun. 2018, 9, 2859. [Google Scholar] [CrossRef] [Green Version]
- Xie, M.; Fussenegger, M. Designing cell function: Assembly of synthetic gene circuits for cell biology applications. Nat. Rev. Mol. Cell Biol. 2018, 19, 507–525. [Google Scholar] [CrossRef] [PubMed]
- Kemmer, C.; Gitzinger, M.; Daoud-El Baba, M.; Djonov, V.; Stelling, J.; Fussenegger, M. Self-sufficient control of urate homeostasis in mice by a synthetic circuit. Nat. Biotech. 2010, 28, 355–360. [Google Scholar] [CrossRef]
- Mesko, M.; Lebar, T.; Dekleva, P.; Jerala, R.; Bencina, M. Engineering and Rewiring of a Calcium-Dependent Signaling Pathway. ACS Synth. Biol. 2020, 9, 2055–2065. [Google Scholar] [CrossRef]
- Scheller, L.; Strittmatter, T.; Fuchs, D.; Bojar, D.; Fussenegger, M. Generalized extracellular molecule sensor platform for programming cellular behavior. Nat. Chem. Biol. 2018. [Google Scholar] [CrossRef]
- Bloom, R.J.; Winkler, S.M.; Smolke, C.D. A quantitative framework for the forward design of synthetic miRNA circuits. Nat. Methods 2014, 11, 1147–1153. [Google Scholar] [CrossRef] [PubMed]
- Gossen, M.; Freundlieb, S.; Bender, G.; Muller, G.; Hillen, W.; Bujard, H. Transcriptional Activation by Tetracyclines in Mammalian-Cells. Science 1995, 268, 1766–1769. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.Z.; Hogenesch, J.B.; Bradfield, C.A. The PAS superfamily: Sensors of environmental and developmental signals. Annu. Rev. Pharmacol. Toxicol. 2000, 40, 519–561. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.P.; Bradfield, C.A. The search for endogenous activators of the aryl hydrocarbon receptor. Chem. Res. Toxicol. 2008, 21, 102–116. [Google Scholar] [CrossRef] [Green Version]
- Beischlag, T.V.; Morales , J.L.; Hollingshead, B.D.; Perdew, G.H. The aryl hydrocarbon receptor complex and the control of gene expression. Crit. Rev. Eukaryot. Gene Expr. 2008, 18, 207–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puga, A.; Ma, C.; Marlowe, J.L. The aryl hydrocarbon receptor cross-talks with multiple signal transduction pathways. Biochem. Pharmacol. 2009, 77, 713–722. [Google Scholar] [CrossRef] [Green Version]
- Jin, U.H.; Park, H.; Li, X.; Davidson, L.A.; Allred, C.; Patil, B.; Jayaprakasha, G.; Orr, A.A.; Mao, L.; Chapkin, R.S.; et al. Structure-Dependent Modulation of Aryl Hydrocarbon Receptor-Mediated Activities by Flavonoids. Toxicol. Sci. 2018, 164, 205–217. [Google Scholar] [CrossRef] [Green Version]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 31–17 August 2016. [Google Scholar] [CrossRef] [Green Version]
- Scheuermann, T.H.; Sleet, C.E.; Bayeh, L.; Shokri, C.; Wang, H.; Caldwell, C.G.; Longgood, J.; MacMillan, J.B.; Bruick, R.K.; Gardner, K.H.; et al. Isoform-Selective and Stereoselective Inhibition of Hypoxia Inducible Factor-2. J. Med. Chem. 2015, 58, 5930–5941. [Google Scholar] [CrossRef]
- Tian, W.; Chen , C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef] [Green Version]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Y.; Ahmed, S.; Satheesh, S.V.; Matthews, J. Genome-wide mapping and analysis of aryl hydrocarbon receptor (AHR)- and aryl hydrocarbon receptor repressor (AHRR)-binding sites in human breast cancer cells. Arch. Toxicol. 2018, 92, 225–240. [Google Scholar] [CrossRef] [Green Version]
- Arango, D.; Morohashi, K.; Yilmaz, A.; Kuramochi, K.; Parihar, A.; Brahimaj, B.; Grotewold, E.; Doseff, A.I. Molecular basis for the action of a dietary flavonoid revealed by the comprehensive identification of apigenin human targets. Proc. Natl. Acad. Sci. USA 2013, 110, E2153–E2162. [Google Scholar] [CrossRef] [Green Version]
- Subhasitanont, P.; Chokchaichamnankit, D.; Chiablaem, K.; Keeratichamroen, S.; Ngiwsara, L.; Paricharttanakul, N.M.; Lirdprapamongkol, K.; Weeraphan, C.; Svasti, J.; Srisomsap, C. Apigenin inhibits growth and induces apoptosis in human cholangiocarcinoma cells. Oncol. Lett. 2017, 14, 4361–4371. [Google Scholar] [CrossRef] [Green Version]
- Woo, J.S.; Choo, G.S.; Yoo, E.S.; Kim, S.H.; Lee, J.H.; Han, S.H.; Kim, H.J.; Jung, S.H.; Park, Y.S.; Kim, B.S.; et al. Apigenin induces apoptosis by regulating Akt and MAPK pathways in human melanoma cell A375SM. Mol. Med. Rep. 2020, 22, 4877–4889. [Google Scholar] [CrossRef] [PubMed]
- Vargo, M.A.; Voss, O.H.; Poustka, F.; Cardounel, A.J.; Grotewold, E.; Doseff, A.I. Apigenin-induced-apoptosis is mediated by the activation of PKCdelta and caspases in leukemia cells. Biochem. Pharmacol. 2006, 72, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Masuelli, L.; Benvenuto, M.; Mattera, R.; Di Stefano, E.; Zago, E.; Taffera, G.; Tresoldi, I.; Giganti, M.G.; Frajese, G.V.; Berardi, G.; et al. In Vitro and In Vivo Anti-tumoral Effects of the Flavonoid Apigenin in Malignant Mesothelioma. Front. Pharmacol. 2017, 8, 373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alon, A.; Kraitchman, D.L.; Schär, M.; Cortez, A.; Yadav, N.; Krimins, R.; Johnston, P.V.; McMahon, M.T.; van Zijl, P.C.M.; Nimmagadda, S.; et al. Molecular imaging of CXCL12 promoter-driven HSV1-TK reporter gene expression. Biotechnol. Bioprocess Eng. 2018, 23, 208–217. [Google Scholar] [CrossRef]
- Jouroukhin, Y.; Nonyane, B.A.; Gilad, A.A.; Pelled, G. Molecular Neuroimaging of Post-Injury Plasticity. J. Mol. Neurosci. 2014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Motif | Binding Probability |
|---|---|
| ACGCTGGGCGTGCAGATGC | 0.17791858 |
| CCGGCTCGCGTGCGCCGGC | 0.6637833 |
| CTAGCTTGCGTGCGCCGGC | 0.5893966 |
| AGGCGTTGCGTGAGAAGGA | 0.82652086 |
| GCGCGCGGCGTGGGGTTGG | 0.15912299 |
| TAGGTCTGCGTGTGGCTTC | 0.6745489 |
| TGTATTTGCGTGCCTAGCT | 0.87979823 |
| CCCCCTCGCGTGACTGCGA | 0.4820609 |
| GCCACAGGCGTGGACCGAA | 0.15255722 |
| ATTACAGGCGTGGGCCACC | 0.2313509 |
| Ligands | Binding Energies (Kcal/mol) |
|---|---|
| Apigenin 5280443 1 | −5.4 |
| Naringenin 932 1 | −5.6 |
| Quercetin 5280343 1 | −5.5 |
| 43L 2 | −5.8 |
| TCDD | −5.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Desmet, N.M.; Dhusia, K.; Qi, W.; Doseff, A.I.; Bhattacharya, S.; Gilad, A.A. Bioengineering of Genetically Encoded Gene Promoter Repressed by the Flavonoid Apigenin for Constructing Intracellular Sensor for Molecular Events. Biosensors 2021, 11, 137. https://doi.org/10.3390/bios11050137
Desmet NM, Dhusia K, Qi W, Doseff AI, Bhattacharya S, Gilad AA. Bioengineering of Genetically Encoded Gene Promoter Repressed by the Flavonoid Apigenin for Constructing Intracellular Sensor for Molecular Events. Biosensors. 2021; 11(5):137. https://doi.org/10.3390/bios11050137
Chicago/Turabian StyleDesmet, Nicole M., Kalyani Dhusia, Wenjie Qi, Andrea I. Doseff, Sudin Bhattacharya, and Assaf A. Gilad. 2021. "Bioengineering of Genetically Encoded Gene Promoter Repressed by the Flavonoid Apigenin for Constructing Intracellular Sensor for Molecular Events" Biosensors 11, no. 5: 137. https://doi.org/10.3390/bios11050137
APA StyleDesmet, N. M., Dhusia, K., Qi, W., Doseff, A. I., Bhattacharya, S., & Gilad, A. A. (2021). Bioengineering of Genetically Encoded Gene Promoter Repressed by the Flavonoid Apigenin for Constructing Intracellular Sensor for Molecular Events. Biosensors, 11(5), 137. https://doi.org/10.3390/bios11050137






