CBM: An IoT Enabled LiDAR Sensor for In-Field Crop Height and Biomass Measurements
Abstract
:1. Introduction
2. Materials and Methods
2.1. System Architecture
2.2. System 3D Design
2.3. Software Architecture
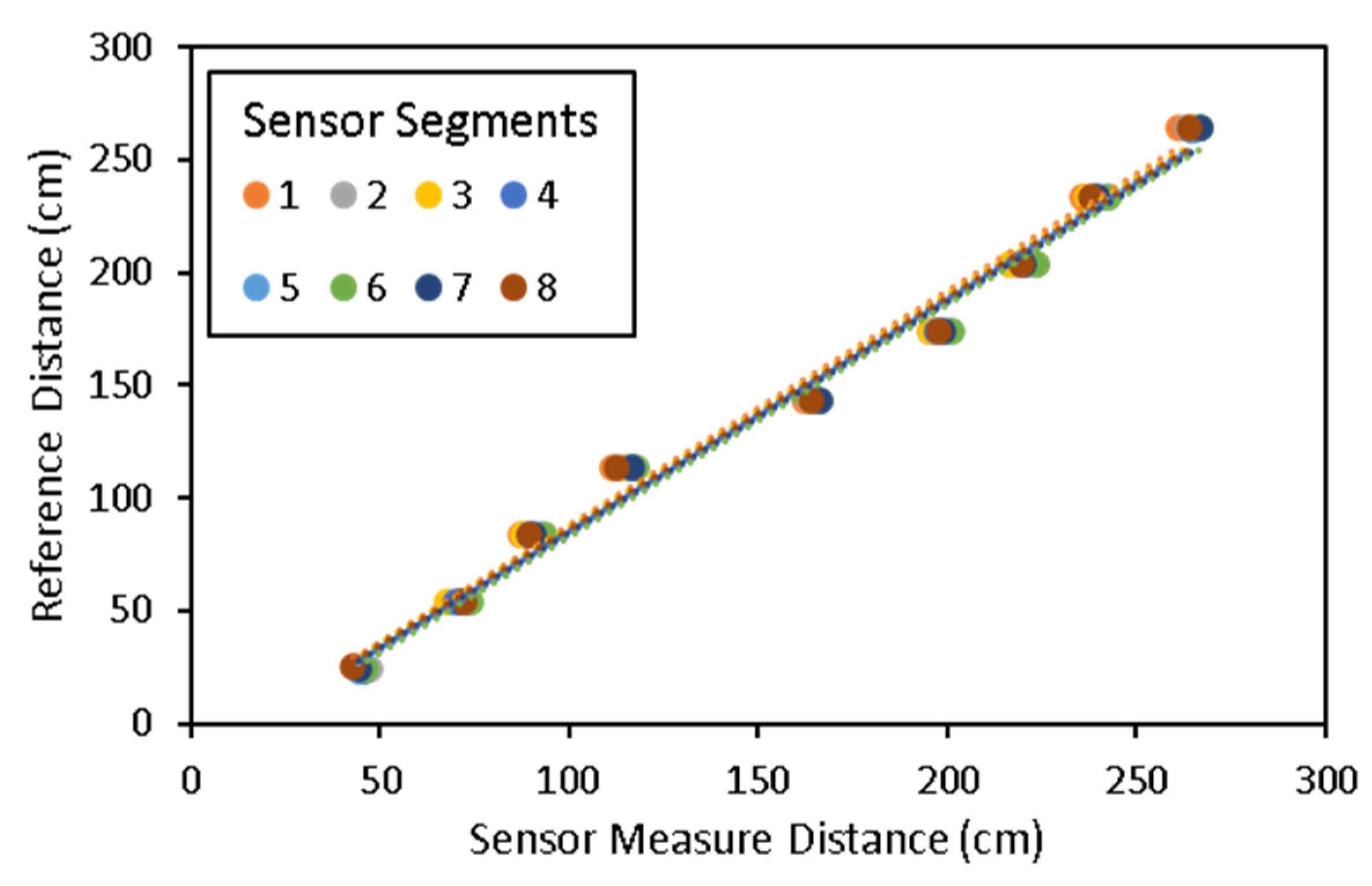
2.4. Laboratory Sensor Calibration
2.5. Field Experiment and Data Collection
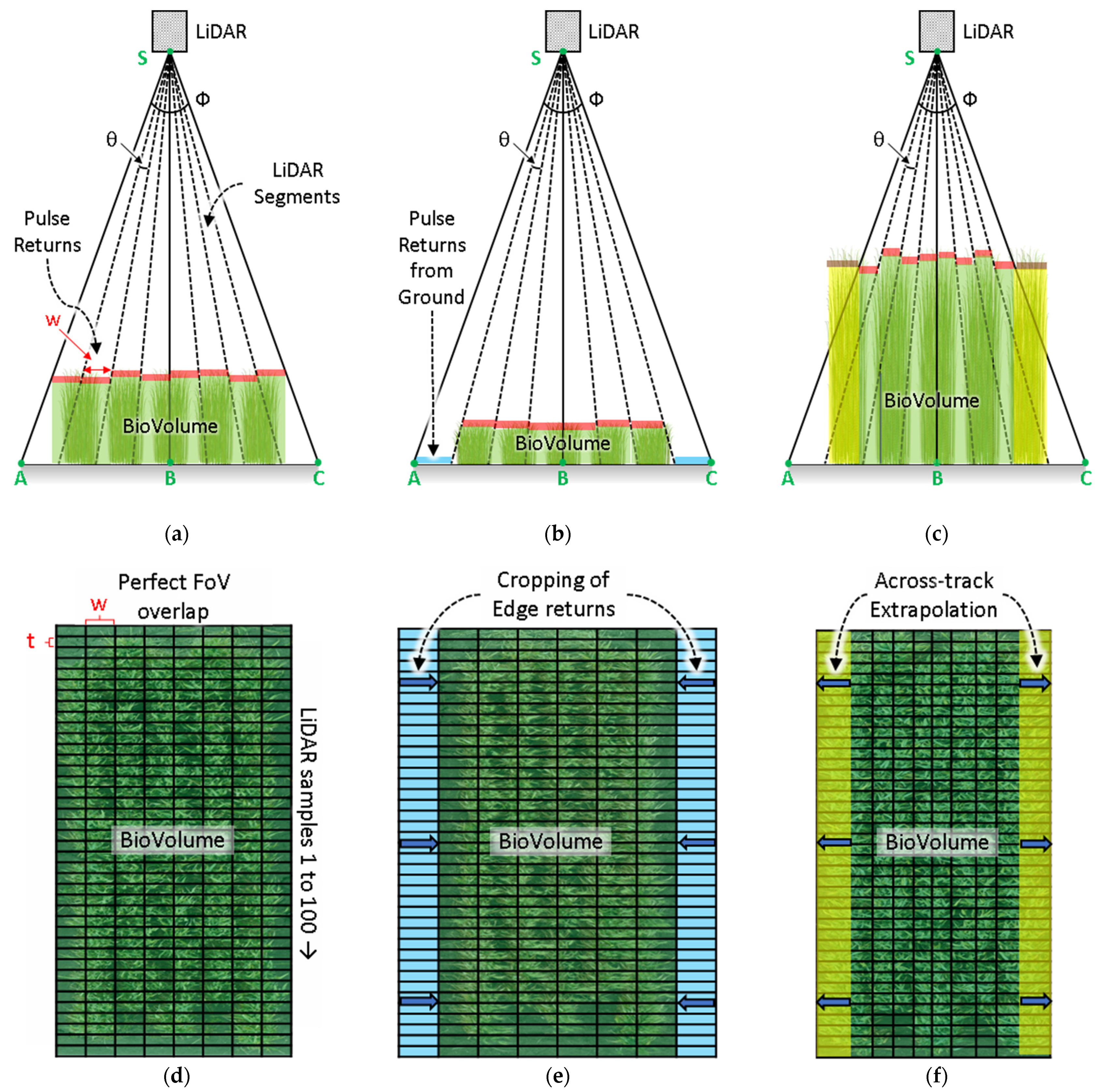
2.6. Scanning Geometry and Crop Height
2.7. Signal Processing for Plot Segemnation, Extraction of Plot Equivalent Mean Crop Height, and BioVolume
2.8. Evaluation of the CBM Sensor in Measuring Crop Height, Fresh Weight, and Dry Weight
3. Results
3.1. Laboratory Sensor Calibration Results
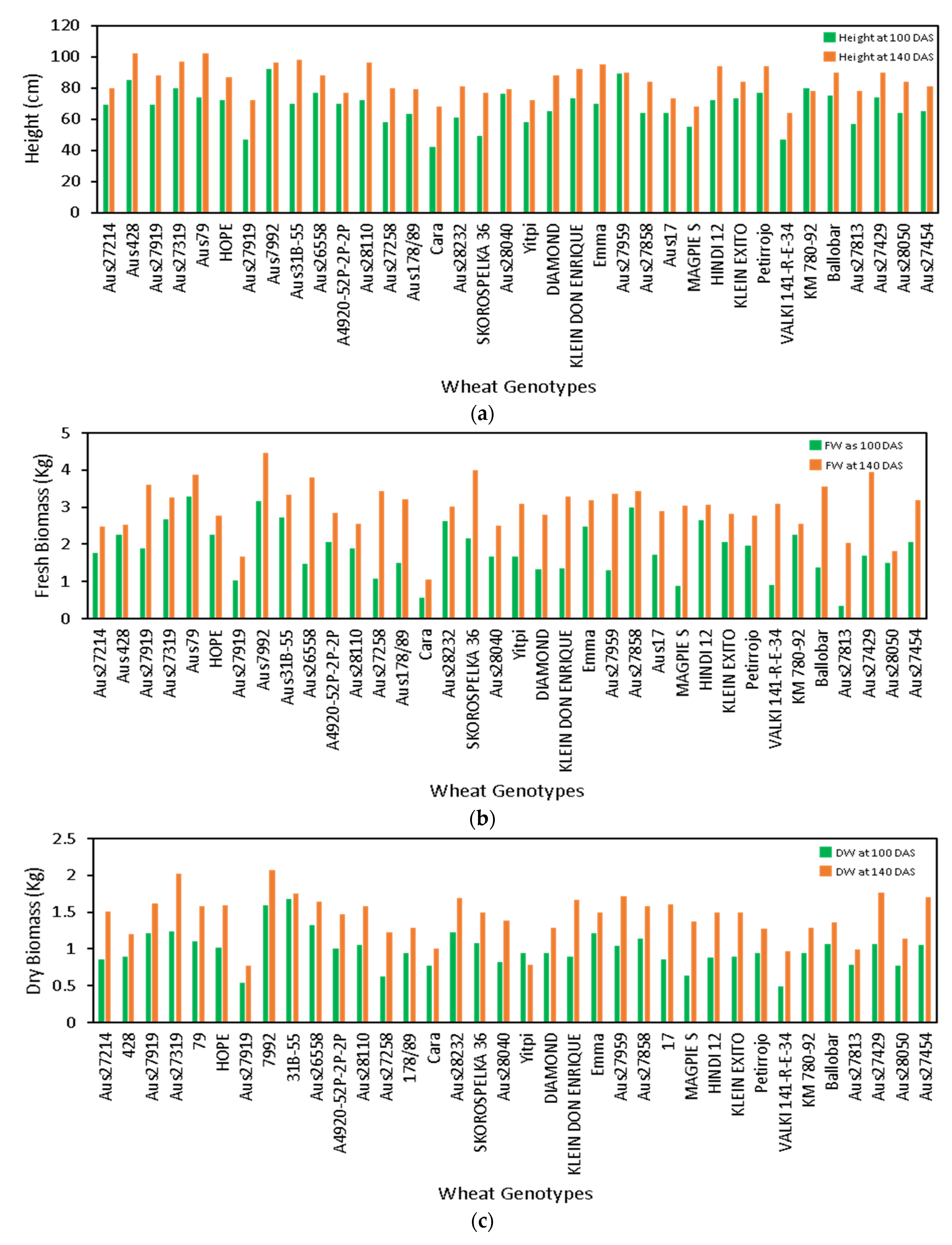
3.2. Estimation of Crop Height, FW, and DW in Field Trial
3.3. Phenotypic Screening of Wheat Genotypes
4. Discussion
4.1. Developmental Aspects of CBM Sensor
4.2. Effect of Sensing Geometry and Crop Canopy Effect
4.3. Estimation of Crop Biomass and Height
4.4. Benefits of CBM in Plant Phenomics and Precision Agriculture
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, J.; Zhao, C.; Huang, W. Fundamental and Application of Quantitative Remote Sensing in Agriculture; Science China Press: Beijing, China, 2008. [Google Scholar]
- Huang, J.X.; Sedano, F.; Huang, Y.B.; Ma, H.Y.; Li, X.L.; Liang, S.L.; Tian, L.Y.; Zhang, X.D.; Fan, J.L.; Wu, W.B. Assimilating a synthetic Kalman filter leaf area index series into the WOFOST model to improve regional winter wheat yield estimation. Agric. For. Meterol. 2016, 216, 188–202. [Google Scholar] [CrossRef]
- Tilly, N.; Aasen, H.; Bareth, G. Fusion of Plant Height and Vegetation Indices for the Estimation of Barley Biomass. Remote Sens. 2015, 7, 11449–11480. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.F.; Nicolas, T.; Wang, J.H.; Philippe, V.; Huang, W.J.; Li, B.G. New index for crop canopy fresh biomass estimation. Guang Pu Xue Yu Guang Pu Fen Xi 2010, 30, 512–517. [Google Scholar]
- Wang, X.; Singh, D.; Marla, S.; Morris, G.; Poland, J. Field-based high-throughput phenotyping of plant height in sorghum using different sensing technologies. Plant Methods 2018, 14, 53. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Harguindeguy, N.; Díaz, S.; Garnier, E.; Lavorel, S.; Poorter, H.; Jaureguiberry, P.; Bret-Harte, M.S.; Cornwell, W.K.; Craine, J.M.; Gurvich, D.E.; et al. New handbook for standardised measurement of plant functional traits worldwide. Aust. J. Bot. 2013, 61, 167–234. [Google Scholar] [CrossRef]
- Shakoor, N.; Lee, S.; Mockler, T.C. High throughput phenotyping to accelerate crop breeding and monitoring of diseases in the field. Curr. Opin. Plant Biol. 2017, 38, 184–192. [Google Scholar] [CrossRef]
- Araus, J.L.; Cairns, J.E. Field high-throughput phenotyping: The new crop breeding frontier. TrendsPlant Sci. 2014, 19, 52–61. [Google Scholar] [CrossRef]
- Virlet, N.; Sabermanesh, K.; Sadeghi-Tehran, P.; Hawkesford, M.J. Field Scanalyzer: An automated robotic field phenotyping platform for detailed crop monitoring. Funct. Plant Biol. 2016, 44, 143–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Underwood, J.; Wendel, A.; Schofield, B.; McMurray, L.; Kimber, R. Efficient in-field plant phenomics for row-crops with an autonomous ground vehicle. J. Field Robot. 2017, 34, 1061–1083. [Google Scholar] [CrossRef]
- Deery, D.; Jimenez-Berni, J.; Jones, H.; Sirault, X.; Furbank, R. Proximal Remote Sensing Buggies and Potential Applications for Field-Based Phenotyping. Agronomy 2014, 4, 349–379. [Google Scholar] [CrossRef] [Green Version]
- Jimenez-Berni, J.A.; Deery, D.M.; Rozas-Larraondo, P.; Condon, A.T.G.; Rebetzke, G.J.; James, R.A.; Bovill, W.D.; Furbank, R.T.; Sirault, X.R.R. High Throughput Determination of Plant Height, Ground Cover, and Above-Ground Biomass in Wheat with LiDAR. Front. Plant Sci. 2018, 9, 237. [Google Scholar] [CrossRef] [Green Version]
- Sytar, O.; Brestic, M.; Zivcak, M.; Olsovska, K.; Kovar, M.; Shao, H.; He, X. Applying hyperspectral imaging to explore natural plant diversity towards improving salt stress tolerance. Sci. Total Environ. 2017, 578, 90–99. [Google Scholar] [CrossRef]
- Lu, H.; Tang, L.; Whitham, S.A.; Mei, Y. A Robotic Platform for Corn Seedling Morphological Traits Characterization. Sensors 2017, 17, 2082. [Google Scholar] [CrossRef] [Green Version]
- Cai, J.; Kumar, P.; Chopin, J.; Miklavcic, S.J. Land-based crop phenotyping by image analysis: Accurate estimation of canopy height distributions using stereo images. PLoS ONE 2018, 13, e0196671. [Google Scholar] [CrossRef] [PubMed]
- van der Heijden, G.; Song, Y.; Horgan, G.; Polder, G.; Dieleman, A.; Bink, M.; Palloix, A.; van Eeuwijk, F.; Glasbey, C. SPICY: Towards automated phenotyping of large pepper plants in the greenhouse. Funct. Plant Biol. 2012, 39, 870–877. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, B.P.; Spangenberg, G.; Kant, S. Fusion of Spectral and Structural Information from Aerial Images for Improved Biomass Estimation. Remote Sens. 2020, 12, 3164. [Google Scholar] [CrossRef]
- Klose, R.; Penlington, J.; Ruckelshausen, A. Usability study of 3D time-of-flight cameras for automatic plant phenotyping. Bornimer Agrartech. Ber. 2009, 69, 12. [Google Scholar]
- Escola, A.; Planas, S.; Rosell, J.R.; Pomar, J.; Camp, F.; Solanelles, F.; Gracia, F.; Llorens, J.; Gil, E. Performance of an ultrasonic ranging sensor in apple tree canopies. Sensors 2011, 11, 2459–2477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrade-Sanchez, P.; Gore, M.A.; Heun, J.T.; Thorp, K.R.; Carmo-Silva, A.E.; French, A.N.; Salvucci, M.E.; White, J.W. Development and evaluation of a field-based high-throughput phenotyping platform. Funct. Plant Biol. 2013, 41, 68–79. [Google Scholar] [CrossRef] [Green Version]
- Yuan, W.; Li, J.; Bhatta, M.; Shi, Y.; Baenziger, P.S.; Ge, Y. Wheat Height Estimation Using LiDAR in Comparison to Ultrasonic Sensor and UAS. Sensors 2018, 18, 3731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barker, J.; Zhang, N.Q.; Sharon, J.; Steeves, R.; Wang, X.; Wei, Y.; Poland, J. Development of a field-based high-throughput mobile phenotyping platform. Comput. Electron. Agric. 2016, 122, 74–85. [Google Scholar] [CrossRef] [Green Version]
- Fricke, T.; Richter, F.; Wachendorf, M. Assessment of forage mass from grassland swards by height measurement using an ultrasonic sensor. Comput. Electron. Agric. 2011, 79, 142–152. [Google Scholar] [CrossRef]
- McCormick, R.F.; Truong, S.K.; Mullet, J.E. 3D Sorghum Reconstructions from Depth Images Identify QTL Regulating Shoot Architecture. Plant Physiol. 2016, 172, 823–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Li, C.Y.; Paterson, A.H. High throughput phenotyping of cotton plant height using depth images under field conditions. Comput. Electron. Agric. 2016, 130, 57–68. [Google Scholar] [CrossRef]
- Lin, Y. LiDAR: An important tool for next-generation phenotyping technology of high potential for plant phenomics? Comput. Electron. Agric. 2015, 119, 61–73. [Google Scholar] [CrossRef]
- French, A.; Gore, M.; Thompson, A. Cotton Phenotyping with Lidar from a Track-Mounted Platform; SPIE: Washington, DC, USA, 2016; Volume 9866. [Google Scholar]
- Singh, K.K.; Chen, G.; Vogler, J.B.; Meentemeyer, R.K. When Big Data are Too Much: Effects of LiDAR Returns and Point Density on Estimation of Forest Biomass. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 9, 3210–3218. [Google Scholar] [CrossRef]
- Pagnutti, M.; Ryan, R.E.; Cazenavette, G.; Gold, M.; Harlan, R.; Leggett, E.; Pagnutti, J. Laying the foundation to use Raspberry Pi 3 V2 camera module imagery for scientific and engineering purposes. J. Electron. Imaging 2017, 26, 013014. [Google Scholar] [CrossRef] [Green Version]
- Rehman, T.U.; Zhang, L.B.; Wang, L.J.; Ma, D.D.; Maki, H.; Sanchez-Gallego, J.A.; Mickelbart, M.V.; Jin, J. Automated leaf movement tracking in time-lapse imaging for plant phenotyping. Comput. Electron. Agric. 2020, 175, 105623. [Google Scholar] [CrossRef]
- Tausen, M.; Clausen, M.; Moeskjaer, S.; Shihavuddin, A.; Dahl, A.B.; Janss, L.; Andersen, S.U. Greenotyper: Image-Based Plant Phenotyping Using Distributed Computing and Deep Learning. Front. Plant Sci. 2020, 11, 1181. [Google Scholar] [CrossRef]
- Mutka, A.M.; Fentress, S.J.; Sher, J.W.; Berry, J.C.; Pretz, C.; Nusinow, D.A.; Bart, R. Quantitative, Image-Based Phenotyping Methods Provide Insight into Spatial and Temporal Dimensions of Plant Disease. Plant Physiol. 2016, 172, 650–660. [Google Scholar] [CrossRef] [Green Version]
- Tovar, J.C.; Hoyer, J.S.; Lin, A.; Tielking, A.; Callen, S.T.; Elizabeth Castillo, S.; Miller, M.; Tessman, M.; Fahlgren, N.; Carrington, J.C.; et al. Raspberry Pi-powered imaging for plant phenotyping. Appl. Plant Sci. 2018, 6, e1031. [Google Scholar] [CrossRef] [PubMed]
- Sangjan, W.; Carter, A.H.; Pumphrey, M.O.; Jitkov, V.; Sankaran, S. Development of a Raspberry Pi-Based Sensor System for Automated In-Field Monitoring to Support Crop Breeding Programs. Inventions 2021, 6, 42. [Google Scholar] [CrossRef]
- Valle, B.; Simonneau, T.; Boulord, R.; Sourd, F.; Frisson, T.; Ryckewaert, M.; Hamard, P.; Brichet, N.; Dauzat, M.; Christophe, A. PYM: A new, affordable, image-based method using a Raspberry Pi to phenotype plant leaf area in a wide diversity of environments. Plant Methods 2017, 13, 98. [Google Scholar] [CrossRef] [Green Version]
- Colmer, J.; O’Neill, C.M.; Wells, R.; Bostrom, A.; Reynolds, D.; Websdale, D.; Shiralagi, G.; Lu, W.; Lou, Q.; Le Cornu, T.; et al. SeedGerm: A cost-effective phenotyping platform for automated seed imaging and machine-learning based phenotypic analysis of crop seed germination. New Phytol. 2020, 228, 778–793. [Google Scholar] [CrossRef] [PubMed]
- Vasisht, D.; Kapetanovic, Z.; Won, J.; Jin, X.; Chandra, R.; Sinha, S.; Kapoor, A.; Sudarshan, M.; Stratman, S. Farmbeats: An iot platform for data-driven agriculture. In Proceedings of the 14th Symposium on Networked Systems Design and Implementation (17), Boston, MA, USA, 27–29 March 2017; pp. 515–529. [Google Scholar]
- Sharma, B.; Ritchie, G.L. High-Throughput Phenotyping of Cotton in Multiple Irrigation Environments. Crop Sci. 2015, 55, 958–969. [Google Scholar] [CrossRef] [Green Version]
- Pittman, J.J.; Arnall, D.B.; Interrante, S.M.; Moffet, C.A.; Butler, T.J. Estimation of Biomass and Canopy Height in Bermudagrass, Alfalfa, and Wheat Using Ultrasonic, Laser, and Spectral Sensors. Sensors 2015, 15, 2920–2943. [Google Scholar] [CrossRef] [Green Version]
- Farooque, A.A.; Chang, Y.K.; Zaman, Q.U.; Groulx, D.; Schumann, A.W.; Esau, T.J. Performance evaluation of multiple ground based sensors mounted on a commercial wild blueberry harvester to sense plant height, fruit yield and topographic features in real-time. Comput. Electron. Agric. 2013, 91, 135–144. [Google Scholar] [CrossRef]
- Chang, Y.K.; Zaman, Q.U.; Rehman, T.U.; Farooque, A.A.; Esau, T.; Jameel, M.W. A real-time ultrasonic system to measure wild blueberry plant height during harvesting. Biosyst. Eng. 2017, 157, 35–44. [Google Scholar] [CrossRef]
- Barmeier, G.; Mistele, B.; Schmidhalter, U. Referencing laser and ultrasonic height measurements of barleycultivars by using a herbometre as standard. Crop Pasture Sci. 2016, 67, 1215–1222. [Google Scholar] [CrossRef]
- Sun, S.P.; Li, C.Y.; Paterson, A.H. In-Field High-Throughput Phenotyping of Cotton Plant Height Using LiDAR. Remote Sens. 2017, 9, 377. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Li, C. Height estimation for blueberry bushes using LiDAR based on a field robotic platform. In Proceedings of the 2016 ASABE International Meeting, St. Joseph, MI, USA, 17 July 2016; p. 1. [Google Scholar]
- Sun, S.; Li, C.; Paterson, A.H.; Jiang, Y.; Xu, R.; Robertson, J.S.; Snider, J.L.; Chee, P.W. In-field High Throughput Phenotyping and Cotton Plant Growth Analysis Using LiDAR. Front. Plant Sci. 2018, 9, 217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greaves, H.E.; Vierling, L.A.; Eitel, J.U.H.; Boelman, N.T.; Magney, T.S.; Prager, C.M.; Griffin, K.L. Estimating aboveground biomass and leaf area of low-stature Arctic shrubs with terrestrial LiDAR. Remote Sens. Environ. 2015, 164, 26–35. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Banerjee, B.P.; Spangenberg, G.; Kant, S. CBM: An IoT Enabled LiDAR Sensor for In-Field Crop Height and Biomass Measurements. Biosensors 2022, 12, 16. https://doi.org/10.3390/bios12010016
Banerjee BP, Spangenberg G, Kant S. CBM: An IoT Enabled LiDAR Sensor for In-Field Crop Height and Biomass Measurements. Biosensors. 2022; 12(1):16. https://doi.org/10.3390/bios12010016
Chicago/Turabian StyleBanerjee, Bikram Pratap, German Spangenberg, and Surya Kant. 2022. "CBM: An IoT Enabled LiDAR Sensor for In-Field Crop Height and Biomass Measurements" Biosensors 12, no. 1: 16. https://doi.org/10.3390/bios12010016
APA StyleBanerjee, B. P., Spangenberg, G., & Kant, S. (2022). CBM: An IoT Enabled LiDAR Sensor for In-Field Crop Height and Biomass Measurements. Biosensors, 12(1), 16. https://doi.org/10.3390/bios12010016






