A Microfluidic Platform Revealing Interactions between Leukocytes and Cancer Cells on Topographic Micropatterns
Abstract
1. Introduction
2. Materials and Methods
2.1. Device Fabrication
2.2. Cell Preparation
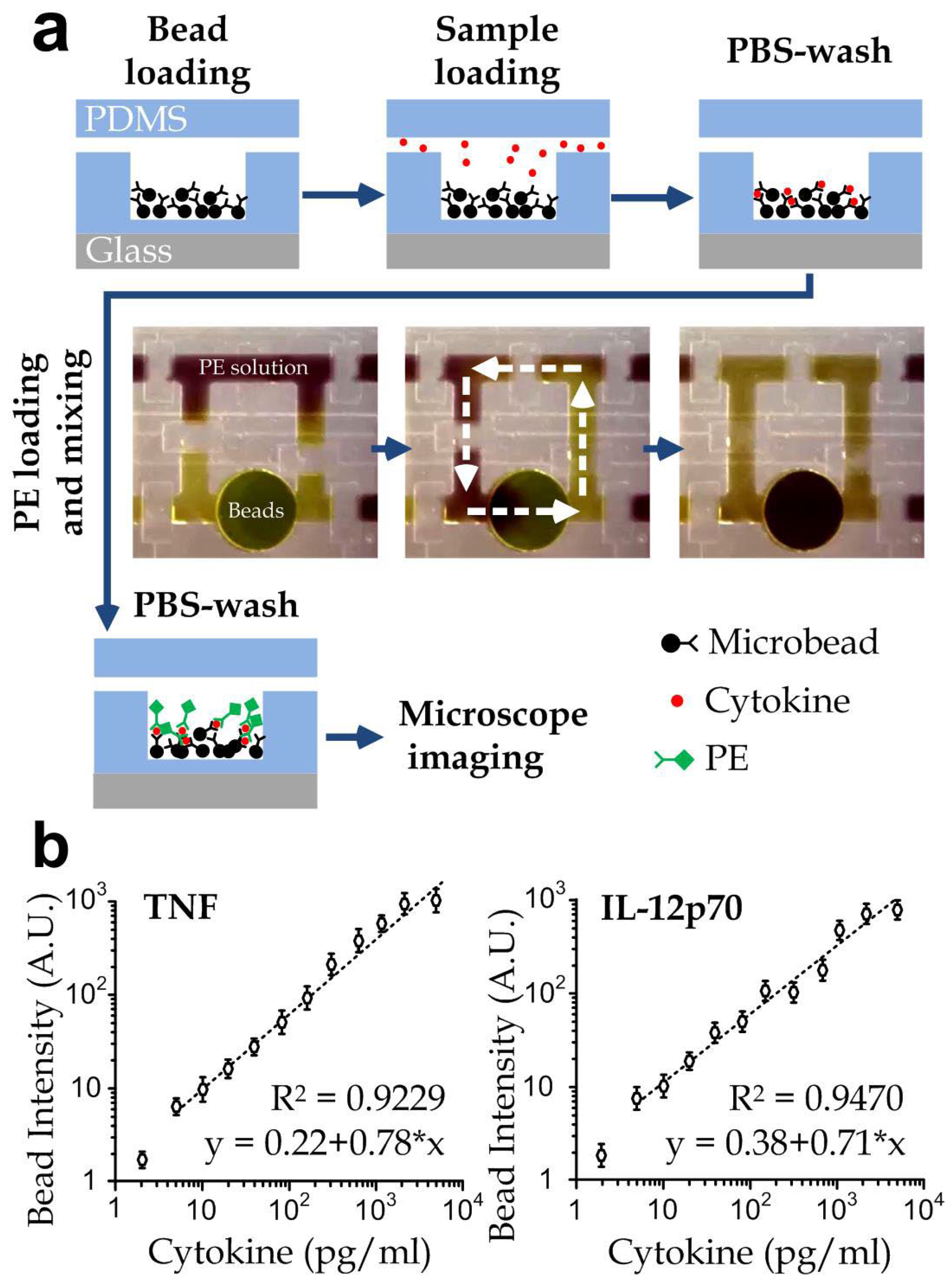
2.3. Automated Microscope and Cytokine Quantification
2.4. Statistics
3. Results and Discussion
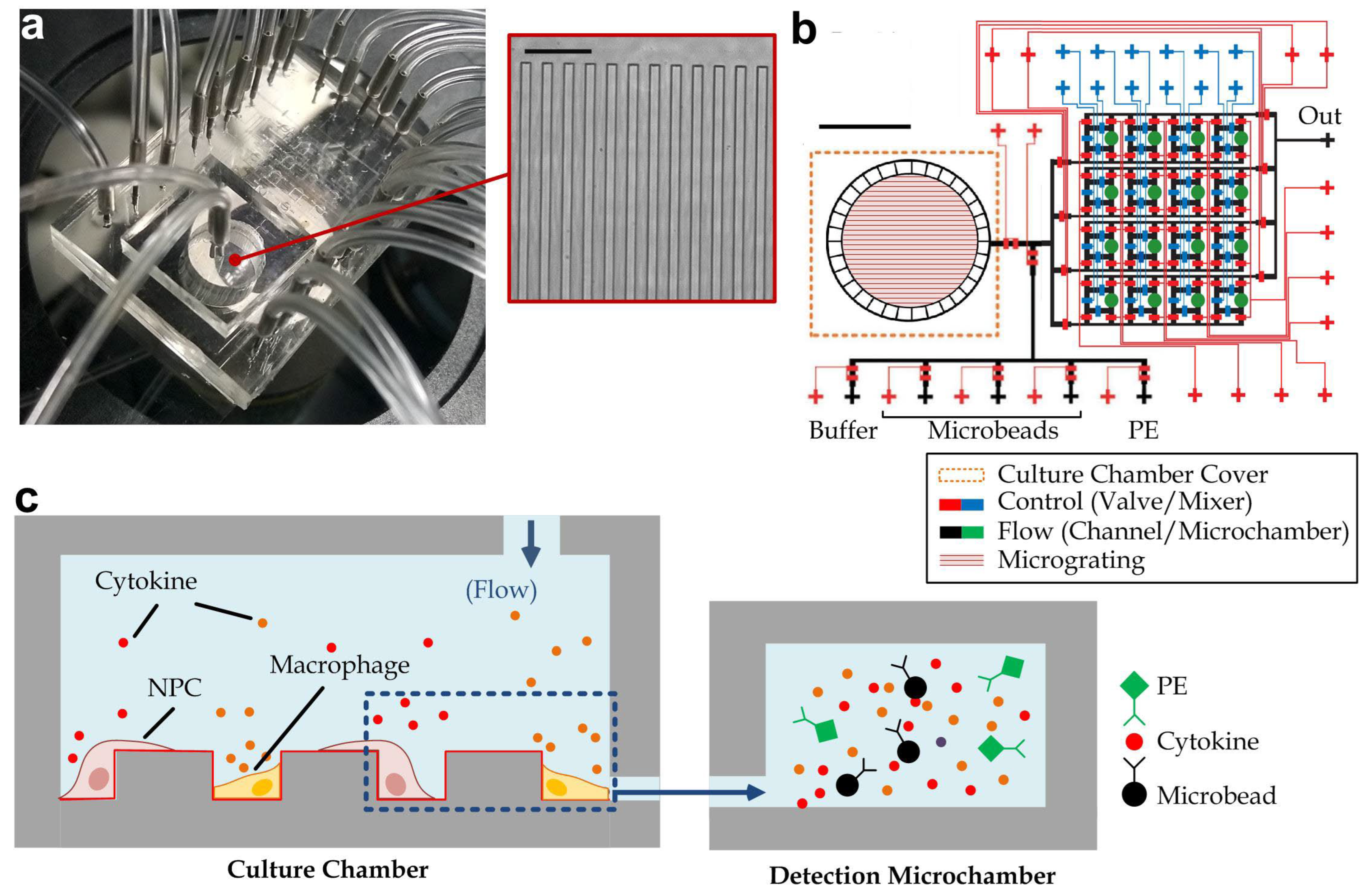
3.1. Device Design and Operation
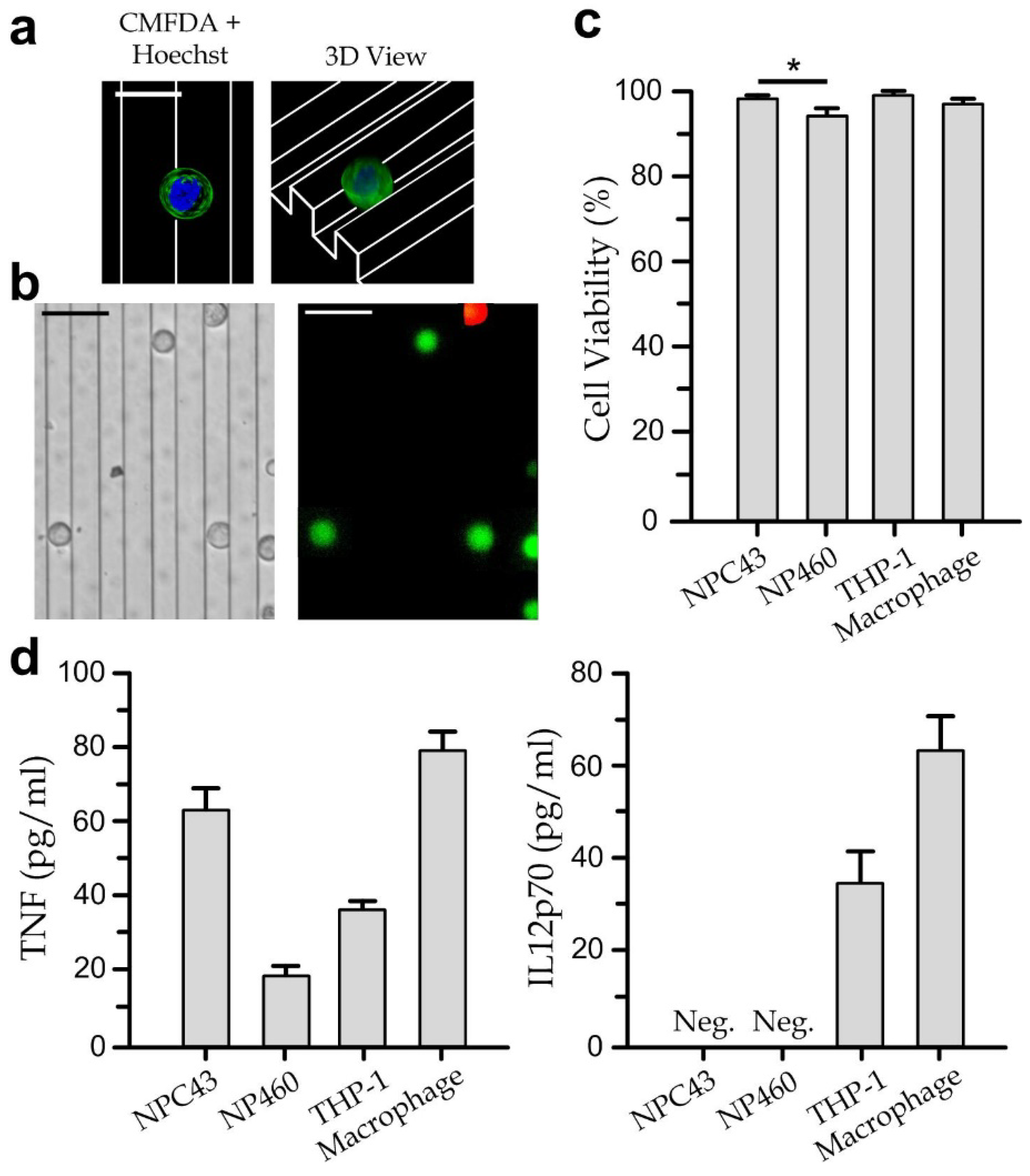
3.2. Viability and Cytokine Secretion of Single Cell-Type Cultures
3.3. Cytokine Secretion Dynamics of Immune Cells Co-Cultured with Nasopharyngeal Cells
3.4. Cell Migration
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Low, H.B.; Png, C.W.; Li, C.; Wang, D.Y.; Wong, S.B.J.; Zhang, Y. Monocyte-derived factors including PLA2G7 induced by macrophage-nasopharyngeal carcinoma cell interaction promote tumor cell invasiveness. Oncotarget 2016, 7, 55473. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.W.M.; Ng, W.T.; Chan, Y.H.; Sze, H.; Chan, C.; Lam, T.H. The battle against nasopharyngeal cancer. Radiother. Oncol. 2012, 104, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Tsang, J.; Lee, V.H.F.; Kwong, D.L.W. Novel therapy for nasopharyngeal carcinoma—Where are we. Oral Oncol. 2014, 50, 798–801. [Google Scholar] [CrossRef] [PubMed]
- Koon, H.-K.; Lo, K.-W.; Leung, K.-N.; Lung, M.L.; Chang, C.C.-K.; Wong, R.N.-S.; Leung, W.-N.; Mak, N.-K. Photodynamic therapy-mediated modulation of inflammatory cytokine production by Epstein–Barr virus-infected nasopharyngeal carcinoma cells. Cell. Mol. Immunol. 2010, 7, 323–326. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Magalhães, L.M.; Viana, A.; Chiari, E.; Galvão, L.M.; Gollob, K.J.; Dutra, W.O. Differential activation of human monocytes and lymphocytes by distinct strains of Trypanosoma cruzi. PLoS Negl. Trop. Dis. 2015, 9, e0003816. [Google Scholar] [CrossRef]
- Beutler, B.A. The role of tumor necrosis factor in health and disease. J. Rheumatol. Suppl. 1999, 57, 16–21. [Google Scholar] [PubMed]
- Gaudreault, E.; Fiola, S.; Olivier, M.; Gosselin, J. Epstein-Barr virus induces MCP-1 secretion by human monocytes via TLR2. J. Virol. 2007, 81, 8016–8024. [Google Scholar] [CrossRef]
- Chensue, S.W.; Warmington, K.S.; Ruth, J.H.; Sanghi, P.S.; Lincoln, P.; Kunkel, S.L. Role of monocyte chemoattractant protein-1 (MCP-1) in Th1 (mycobacterial) and Th2 (schistosomal) antigen-induced granuloma formation: Relationship to local inflammation, Th cell expression, and IL-12 production. J. Immunol. 1996, 157, 4602–4608. [Google Scholar]
- Zergoun, A.A.; Draleau, K.S.; Chettibi, F.; Touil-Boukoffa, C.; Djennaoui, D.; Merghoub, T.; Bourouba, M. Plasma secretome analyses identify IL-8 and nitrites as predictors of poor prognosis in nasopharyngeal carcinoma patients. Cytokine 2022, 153, 155852. [Google Scholar] [CrossRef]
- Picard, C.; Bobby Gaspar, H.; Al-Herz, W.; Bousfiha, A.; Casanova, J.L.; Chatila, T.; Crow, Y.J.; Cunningham-Rundles, C.; Etzioni, A.; Franco, J.L.; et al. International Union of Immunological Societies: 2017 Primary Immunodeficiency Diseases Committee Report on Inborn Errors of Immunity. J. Clin. Immunol. 2018, 38, 96–128. [Google Scholar] [CrossRef]
- Al-Herz, W.; Bousfiha, A.; Casanova, J.L.; Chatila, T.; Conley, M.E.; Cunningham-Rundles, C.; Etzioni, A.; Franco, J.L.; Gaspar, H.B.; Holland, S.M.; et al. Primary immunodeficiency diseases: An update on the classification from the international union of immunological societies expert committee for primary immunodeficiency. Front. Immunol. 2014, 5, 162. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, R.; Patel, V.; Chikkaveeraiah, B.V.; Munge, B.S.; Cheong, S.C.; Zain, R.B.; Abraham, M.T.; Dey, D.K.; Gutkind, J.S.; Rusling, J.F. Ultrasensitive detection of cancer biomarkers in the clinic by use of a nanostructured microfluidic array. Anal. Chem. 2012, 84, 6249–6255. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Park, J.K. Microarray-integrated optoelectrofluidic immunoassay system. Biomicrofluidics 2016, 10, 034106. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Jiang, L.; Su, X.; Qin, J.; Lin, B. Microvalves actuated sandwich immunoassay on an integrated microfluidic system. Electrophoresis 2009, 30, 2481–2487. [Google Scholar] [CrossRef] [PubMed]
- Hung, L.Y.; Chang, J.C.; Tsai, Y.C.; Huang, C.C.; Chang, C.P.; Yeh, C.S.; Lee, G.B. Magnetic nanoparticle-based immunoassay for rapid detection of influenza infections by using an integrated microfluidic system. Nanomedicine 2014, 10, 819–829. [Google Scholar] [CrossRef]
- Han, Q.; Bradshaw, E.M.; Nilsson, B.; Hafler, D.A.; Love, J.C. Multidimensional analysis of the frequencies and rates of cytokine secretion from single cells by quantitative microengraving. Lab Chip 2010, 10, 1391–1400. [Google Scholar] [CrossRef]
- Baraket, A.; Lee, M.; Zine, N.; Sigaud, M.; Bausells, J.; Errachid, A. A fully integrated electrochemical biosensor platform fabrication process for cytokines detection. Biosens. Bioelectron. 2017, 93, 170–175. [Google Scholar] [CrossRef]
- Salminen, T.; Juntunen, E.; Talha, S.M.; Pettersson, K. High-sensitivity lateral flow immunoassay with a fluorescent lanthanide nanoparticle label. J. Immunol. Methods 2019, 465, 39–44. [Google Scholar] [CrossRef]
- Soares, R.R.G.; Santos, D.R.; Pinto, I.F.; Azevedo, A.M.; Aires-Barros, M.R.; Chu, V.; Conde, J.P. Multiplexed microfluidic fluorescence immunoassay with photodiode array signal acquisition for sub-minute and point-of-need detection of mycotoxins. Lab Chip 2018, 18, 1569–1580. [Google Scholar] [CrossRef]
- Kim, J.; Staunton, J.R.; Tanner, K. Independent Control of Topography for 3D Patterning of the ECM Microenvironment. Adv. Mater. 2016, 28, 132–137. [Google Scholar] [CrossRef]
- Jeon, H.; Tsui, J.H.; Jang, S.I.; Lee, J.H.; Park, S.; Mun, K.; Boo, Y.C.; Kim, D.H. Combined effects of substrate topography and stiffness on endothelial cytokine and chemokine secretion. ACS Appl. Mater. Interfaces 2015, 7, 4525–4532. [Google Scholar] [CrossRef] [PubMed]
- Lam, B.P.; Cheung, S.K.C.; Lam, Y.W.; Pang, S.W. Microenvironmental topographic cues influence migration dynamics of nasopharyngeal carcinoma cells from tumour spheroids. RSC Adv. 2020, 10, 28975–28983. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ren, J.; Zhang, R.; Hu, S.; Pang, S.W.; Lam, R.H. Spreading and Migration of Nasopharyngeal Normal and Cancer Cells on Microgratings. ACS Appl. Bio Mater. 2021, 4, 3224–3231. [Google Scholar] [CrossRef] [PubMed]
- Andersson, A.-S.; Bäckhed, F.; von Euler, A.; Richter-Dahlfors, A.; Sutherland, D.; Kasemo, B. Nanoscale features influence epithelial cell morphology and cytokine production. Biomaterials 2003, 24, 3427–3436. [Google Scholar] [CrossRef]
- Cui, X.; Liu, Y.; Hu, D.; Qian, W.; Tin, C.; Sun, D.; Chen, W.; Lam, R.H.W. A fluorescent microbead-based microfluidic immunoassay chip for immune cell cytokine secretion quantification. Lab Chip 2018, 18, 522–531. [Google Scholar] [CrossRef]
- Liu, Y.; Li, J.; Hu, D.; Lam, J.H.M.; Sun, D.; Pang, S.W.; Lam, R.H.W. Microfluidic implementation of functional cytometric microbeads for improved multiplexed cytokine quantification. Biomicrofluidics 2018, 12, 044112. [Google Scholar] [CrossRef]
- Lin, W.; Yip, Y.L.; Jia, L.; Deng, W.; Zheng, H.; Dai, W.; Ko, J.M.Y.; Lo, K.W.; Chung, G.T.Y.; Yip, K.Y.; et al. Establishment and characterization of new tumor xenografts and cancer cell lines from EBV-positive nasopharyngeal carcinoma. Nat. Commun. 2018, 9, 4663. [Google Scholar] [CrossRef]
- Priante, G.; Bordin, L.; Musacchio, E.; Clari, G.; Baggio, B. Fatty acids and cytokine mRNA expression in human osteoblastic cells: A specific effect of arachidonic acid. Clin. Sci. 2002, 102, 403–409. [Google Scholar] [CrossRef]
- Yip, H.M.; Li, J.C.S.; Xie, K.; Cui, X.; Prasad, A.; Gao, Q.N.; Leung, C.C.; Lam, R.H.W. Automated Long-Term Monitoring of Parallel Microfluidic Operations Applying a Machine Vision-Assisted Positioning Method. Sci. World J. 2014, 2014, 608184. [Google Scholar] [CrossRef]
- Liao, Q.; Zeng, Z.; Guo, X.; Li, X.; Wei, F.; Zhang, W.; Li, X.; Chen, P.; Liang, F.; Xiang, B.; et al. LPLUNC1 suppresses IL-6-induced nasopharyngeal carcinoma cell proliferation via inhibiting the Stat3 activation. Oncogene 2014, 33, 2098–2109. [Google Scholar] [CrossRef]
- Sharma, D.; Jia, W.; Long, F.; Pati, S.; Chen, Q.; Qyang, Y.; Lee, B.; Choi, C.K.; Zhao, F. Polydopamine and collagen coated micro-grated polydimethylsiloxane for human mesenchymal stem cell culture. Bioact. Mater. 2019, 4, 142–150. [Google Scholar] [CrossRef] [PubMed]
- McKee, T.J.; Perlman, G.; Morris, M.; Komarova, S.V. Extracellular matrix composition of connective tissues: A systematic review and meta-analysis. Sci. Rep. 2019, 9, 10542. [Google Scholar] [CrossRef] [PubMed]
- Gosselin, J.; Menezes, J.; D’Addario, M.; Hiscott, J.; Flamand, L.; Lamoureux, G.; Oth, D. Inhibition of tumor necrosis factor-α transcription by Epstein-Barr virus. Eur. J. Immunol. 1991, 21, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Miao, T.; Shen, X.; Bao, L.; Zhang, C.; Yan, C.; Wei, W.; Chen, J.; Xiao, L.; Sun, C. EB virus-induced ATR activation accelerates nasopharyngeal carcinoma growth via M2-type macrophages polarization. Cell Death Dis. 2020, 11, 742. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.-F.; Hong, Y.-X.; Feng, G.-J.; Zhang, G.-F.; Rogers, H.; Lewis, M.A.; Williams, D.W.; Xia, Z.-F.; Song, B.; Wei, X.-Q. Lipopolysaccharide-induced M2 to M1 macrophage transformation for IL-12p70 production is blocked by Candida albicans mediated up-regulation of EBI3 expression. PLoS ONE 2013, 8, e63967. [Google Scholar] [CrossRef]
- Cheng, Z.; Zhu, W.; Li, H.; He, X. Macrophages promote the migration of neural stem cells into mouse spinal cord injury site. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi Chin. J. Cell. Mol. Immunol. 2016, 32, 1174–1177. [Google Scholar]
- Dubrulle, F.; Souillard, R.; Hermans, R. Extension patterns of nasopharyngeal carcinoma. Eur. Radiol. 2007, 17, 2622–2630. [Google Scholar] [CrossRef]
- Li, P.; Kaslan, M.; Lee, S.H.; Yao, J.; Gao, Z. Progress in exosome isolation techniques. Theranostics 2017, 7, 789. [Google Scholar] [CrossRef]
- Chen, W.; Huang, N.T.; Oh, B.; Lam, R.H.; Fan, R.; Cornell, T.T.; Shanley, T.P.; Kurabayashi, K.; Fu, J. Surface-Micromachined Microfiltration Membranes for Efficient Isolation and Functional Immunophenotyping of Subpopulations of Immune Cells. Adv. Healthc. Mater. 2013, 2, 965–975. [Google Scholar] [CrossRef]
- Nguyen, T.; Sarkar, T.; Tran, T.; Moinuddin, S.M.; Saha, D.; Ahsan, F. Multilayer Soft Photolithography Fabrication of Microfluidic Devices Using a Custom-Built Wafer-Scale PDMS Slab Aligner and Cost-Efficient Equipment. Micromachines 2022, 13, 1357. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, X.; Liu, L.; Li, J.; Liu, Y.; Liu, Y.; Hu, D.; Zhang, R.; Huang, S.; Jiang, Z.; Wang, Y.; et al. A Microfluidic Platform Revealing Interactions between Leukocytes and Cancer Cells on Topographic Micropatterns. Biosensors 2022, 12, 963. https://doi.org/10.3390/bios12110963
Cui X, Liu L, Li J, Liu Y, Liu Y, Hu D, Zhang R, Huang S, Jiang Z, Wang Y, et al. A Microfluidic Platform Revealing Interactions between Leukocytes and Cancer Cells on Topographic Micropatterns. Biosensors. 2022; 12(11):963. https://doi.org/10.3390/bios12110963
Chicago/Turabian StyleCui, Xin, Lelin Liu, Jiyu Li, Yi Liu, Ya Liu, Dinglong Hu, Ruolin Zhang, Siping Huang, Zhongning Jiang, Yuchao Wang, and et al. 2022. "A Microfluidic Platform Revealing Interactions between Leukocytes and Cancer Cells on Topographic Micropatterns" Biosensors 12, no. 11: 963. https://doi.org/10.3390/bios12110963
APA StyleCui, X., Liu, L., Li, J., Liu, Y., Liu, Y., Hu, D., Zhang, R., Huang, S., Jiang, Z., Wang, Y., Qu, Y., Pang, S. W., & Lam, R. H. W. (2022). A Microfluidic Platform Revealing Interactions between Leukocytes and Cancer Cells on Topographic Micropatterns. Biosensors, 12(11), 963. https://doi.org/10.3390/bios12110963








