Recent Advances in Electrochemical Biosensors for Monitoring Animal Cell Function and Viability
Abstract
1. Introduction
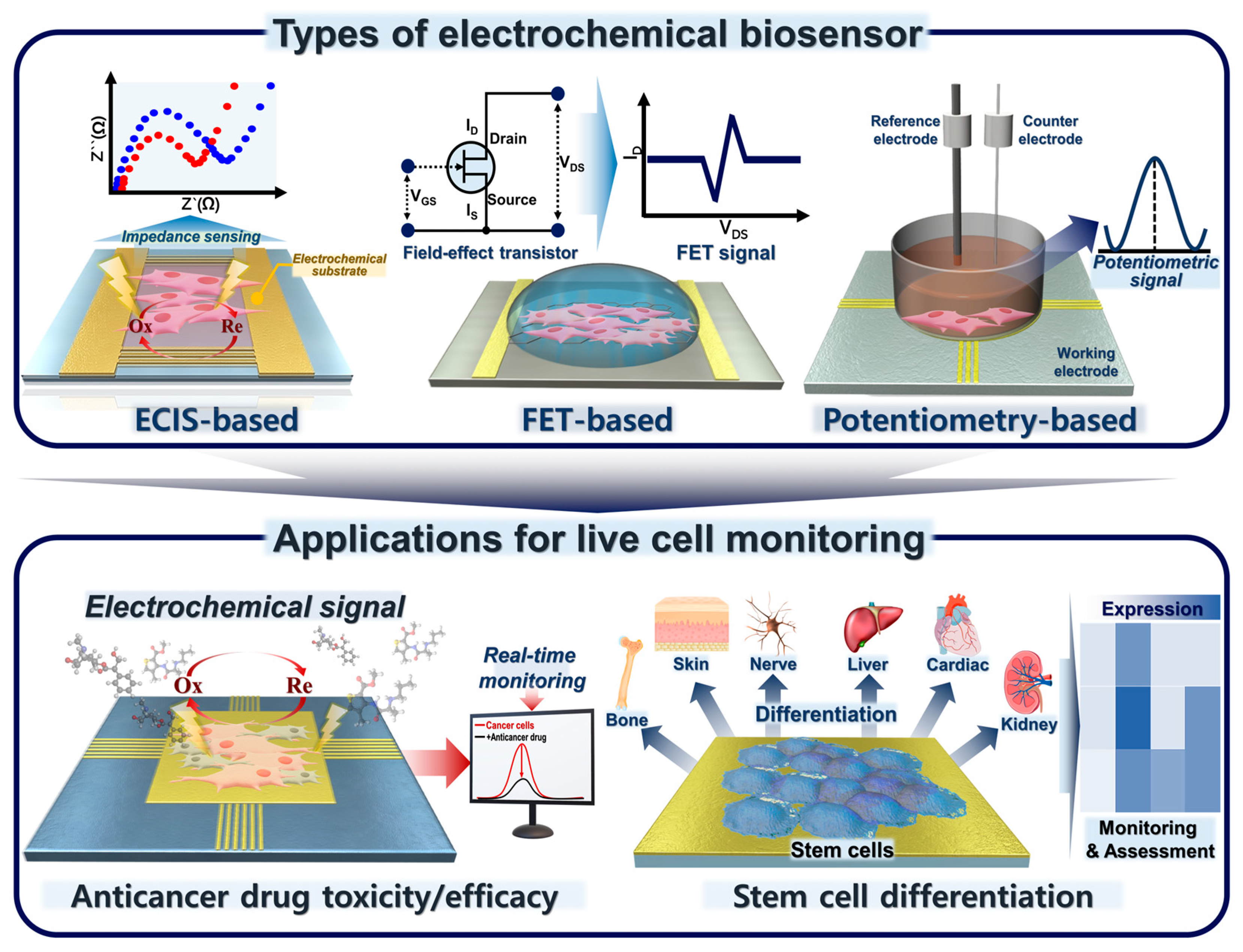
2. Various Types of Electrochemical Biosensors for Live-Cell Detection
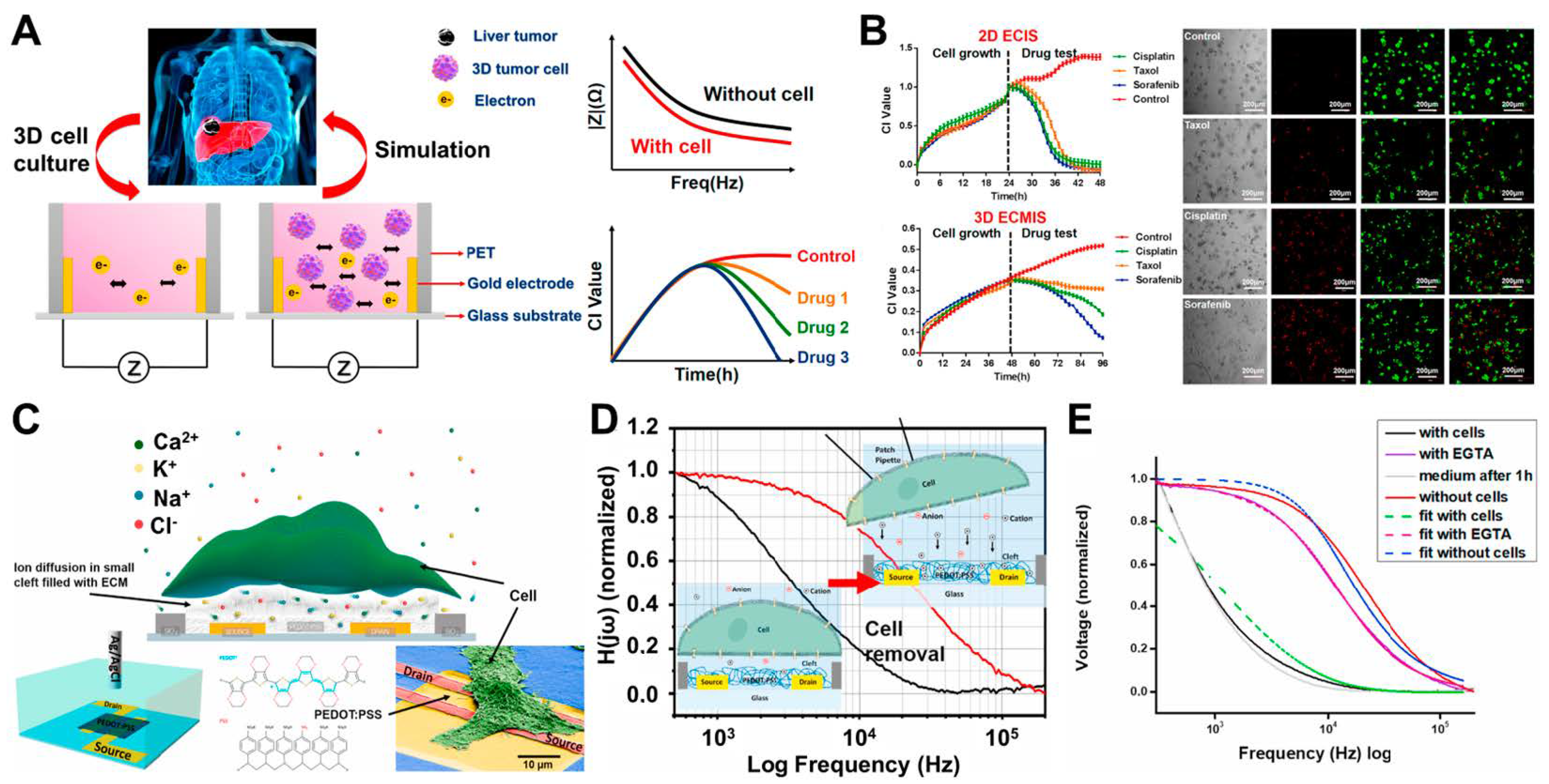
2.1. ECIS
2.2. FET-Based Electrochemical Biosensors
2.3. Potentiometric-Based Electrochemical Biosensors
3. Application for Live-Cell Monitoring Based on Electrochemical Biosensing
3.1. Cancer Cell-Based Sensing and Assessment of Anticancer Drug Candidates
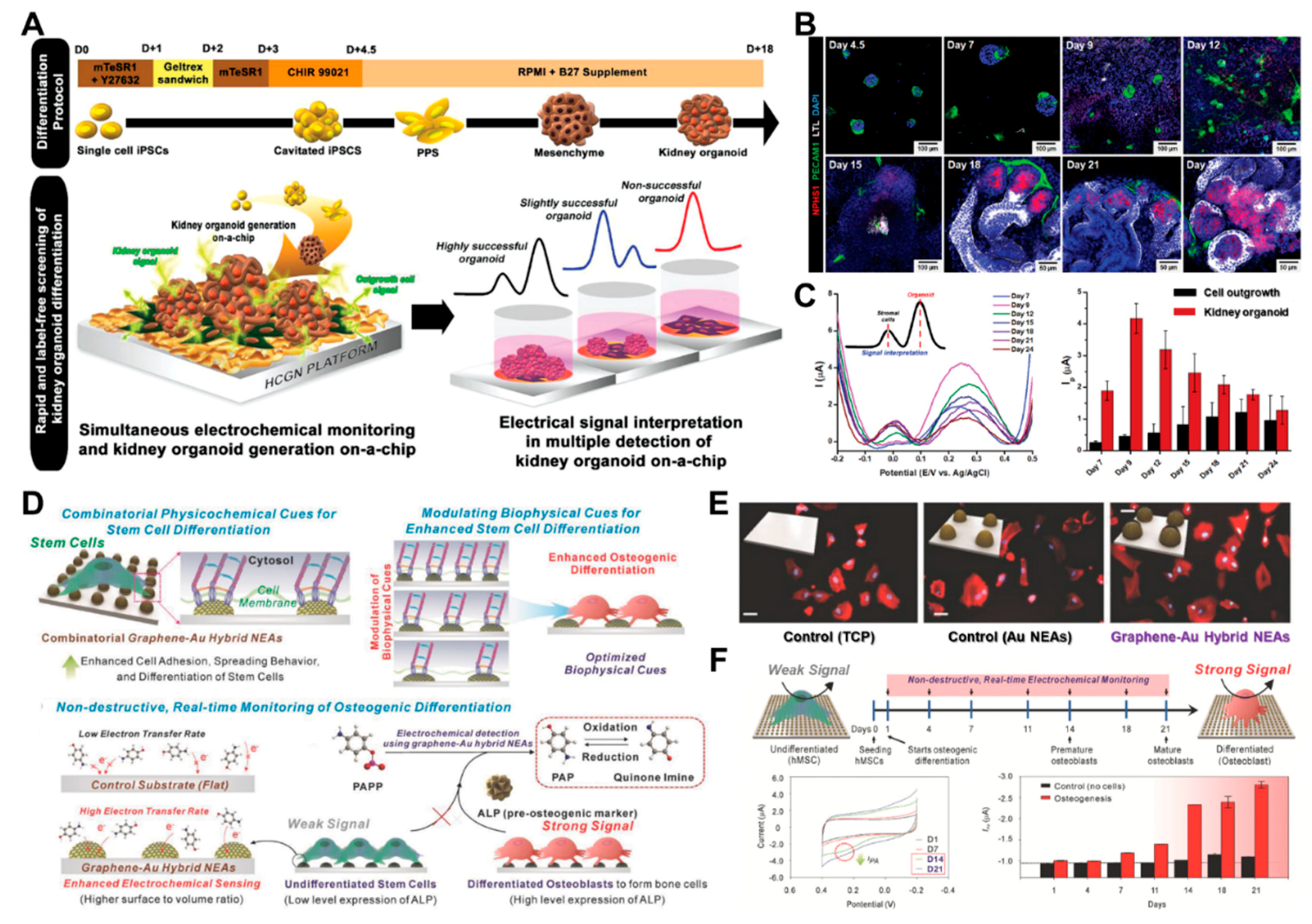
3.2. Stem Cell-Based Sensing and Monitoring for Differentiation
4. Conclusions
5. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Rubis, G.; Krishnan, S.R.; Bebawy, M. Liquid biopsies in cancer diagnosis, monitoring, and prognosis. Trends Pharmacol. Sci. 2019, 40, 172–186. [Google Scholar] [CrossRef]
- Patchsung, M.; Jantarug, K.; Pattama, A.; Aphicho, K.; Suraritdechachai, S.; Meesawat, P.; Sappakhaw, K.; Leelahakorn, N.; Ruenkam, T.; Wongsatit, T. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1140–1149. [Google Scholar] [CrossRef] [PubMed]
- Ginsburg, O.; Yip, C.H.; Brooks, A.; Cabanes, A.; Caleffi, M.; Dunstan Yataco, J.A.; Gyawali, B.; McCormack, V.; McLaughlin de Anderson, M.; Mehrotra, R. Breast cancer early detection: A phased approach to implementation. Cancer 2020, 126, 2379–2393. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H.; Seo, J.-M.; Shin, K.-J.; Yang, S.-G. Design and clinical developments of aptamer-drug conjugates for targeted cancer therapy. Biomater. Res. 2021, 25, 42. [Google Scholar] [CrossRef]
- Urbanczyk, M.; Layland, S.L.; Schenke-Layland, K. The role of extracellular matrix in biomechanics and its impact on bioengineering of cells and 3D tissues. Matrix Biol. 2020, 85–86, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Bielajew, B.J.; Hu, J.C.; Athanasiou, K.A. Collagen: Quantification, biomechanics and role of minor subtypes in cartilage. Nat. Rev. Mater. 2020, 5, 730–747. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Sharma, A.; Saji, J.; Umapathi, A.; Kumar, S.; Daima, H.K. Smart nanomaterials for cancer diagnosis and treatment. Nano Converg. 2022, 9, 21. [Google Scholar] [CrossRef]
- Ding, L.; Cao, J.; Lin, W.; Chen, H.; Xiong, X.; Ao, H.; Yu, M.; Lin, J.; Cui, Q. The roles of cyclin-dependent kinases in cell-cycle progression and therapeutic strategies in human breast cancer. Int. J. Mol. Sci. 2020, 21, 1960. [Google Scholar] [CrossRef]
- Yaku, K.; Okabe, K.; Hikosaka, K.; Nakagawa, T. NAD metabolism in cancer therapeutics. Front. Oncol. 2018, 8, 622. [Google Scholar] [CrossRef] [PubMed]
- Madl, C.M.; Heilshorn, S.C.; Blau, H.M. Bioengineering strategies to accelerate stem cell therapeutics. Nature 2018, 557, 335–342. [Google Scholar] [CrossRef]
- Harrell, C.R.; Fellabaum, C.; Jovicic, N.; Djonov, V.; Arsenijevic, N.; Volarevic, V. Molecular mechanisms responsible for therapeutic potential of mesenchymal stem cell-derived secretome. Cells 2019, 8, 467. [Google Scholar] [CrossRef]
- Du, X.; Zhang, Z.; Zheng, X.; Zhang, H.; Dong, D.; Zhang, Z.; Liu, M.; Zhou, J. An electrochemical biosensor for the detection of epithelial-mesenchymal transition. Nat. Commun. 2020, 11, 192. [Google Scholar] [CrossRef]
- Song, Z.; Ma, Y.; Chen, M.; Ambrosi, A.; Ding, C.; Luo, X. Electrochemical biosensor with enhanced antifouling capability for COVID-19 nucleic acid detection in complex biological media. Anal. Chem. 2021, 93, 5963–5971. [Google Scholar] [CrossRef] [PubMed]
- Karimi-Maleh, H.; Orooji, Y.; Karimi, F.; Alizadeh, M.; Baghayeri, M.; Rouhi, J.; Tajik, S.; Beitollahi, H.; Agarwal, S.; Gupta, V.K. A critical review on the use of potentiometric based biosensors for biomarkers detection. Biosens. Bioelectron. 2021, 184, 113252. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Ahn, J.J.; Kim, S.J.; Yu, S.Y.; Koh, E.J.; Kim, S.H.; Sung, H.S.; Huh, J.W.; Hwang, S.Y. POCT detection of 14 respiratory viruses using multiplex RT-PCR. BioChip J. 2021, 15, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Cho, H.-Y.; Shin, M.; Choi, H.K.; Lee, T.; Choi, J.-W. Flexible electrochemical biosensors for healthcare monitoring. J. Mater. Chem. B 2020, 8, 7303–7318. [Google Scholar] [CrossRef]
- Sabaté del Río, J.; Henry, O.Y.; Jolly, P.; Ingber, D.E. An antifouling coating that enables affinity-based electrochemical biosensing in complex biological fluids. Nat. Nanotechnol. 2019, 14, 1143–1149. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, A.D.; Takemura, K.; Li, T.-C.; Suzuki, T.; Park, E.Y. Electrical pulse-induced electrochemical biosensor for hepatitis E virus detection. Nat. Commun. 2019, 10, 3737. [Google Scholar] [CrossRef]
- Huang, X.; Zhu, Y.; Kianfar, E. Nano biosensors: Properties, applications and electrochemical techniques. J. Mater. Res. Technol. 2021, 12, 1649–1672. [Google Scholar] [CrossRef]
- Khatib, M.; Zohar, O.; Haick, H. Self-healing soft sensors: From material design to implementation. Adv. Mater. 2021, 33, 2004190. [Google Scholar] [CrossRef] [PubMed]
- Kadam, U.S.; Hong, J.C. Recent advances in aptameric biosensors designed to detect toxic contaminants from food, water, human fluids, and the environment. Trends Environ. Anal. Chem. 2022, 36, e00184. [Google Scholar] [CrossRef]
- Kadam, U.S.; Trinh, K.H.; Kumar, V.; Lee, K.W.; Cho, Y.; Can, M.-H.T.; Lee, H.; Kim, Y.; Kim, S.; Kang, J. Identification and structural analysis of novel malathion-specific DNA aptameric sensors designed for food testing. Biomaterials 2022, 287, 121617. [Google Scholar] [CrossRef]
- Wang, A.; Ding, Y.; Li, L.; Duan, D.; Mei, Q.; Zhuang, Q.; Cui, S.; He, X. A novel electrochemical enzyme biosensor for detection of 17β-estradiol by mediated electron-transfer system. Talanta 2019, 192, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Zheng, Y.; Zhang, P.; Zhang, H.; Xu, Y.; Zhou, J.; Zhang, H.; Karimi-Maleh, H.; Lai, G.; Zhao, S. Development of an electrochemical biosensor for phylogenetic analysis of Amaryllidaceae based on the enhanced electrochemical fingerprint recorded from plant tissue. Biosens. Bioelectron. 2020, 159, 112212. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Liu, Y.; Pi, J.; Lu, N.; Zhang, R.; Chen, W.; Zhang, Z.; Xing, D. A novel artificial peroxisome candidate based on nanozyme with excellent catalytic performance for biosensing. Biosens. Bioelectron. 2022, 196, 113686. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Han, Y.; Suhito, I.R.; Choi, Y.; Kwon, M.; Son, H.; Kim, H.-R.; Kim, T.-H. Raman spectroscopy-based 3D analysis of odontogenic differentiation of human dental pulp stem cell spheroids. Anal. Chem. 2021, 93, 9995–10004. [Google Scholar] [CrossRef] [PubMed]
- Suhito, I.R.; Han, Y.; Ryu, Y.-S.; Son, H.; Kim, T.-H. Autofluorescence-Raman Mapping Integration analysis for ultra-fast label-free monitoring of adipogenic differentiation of stem cells. Biosens. Bioelectron. 2021, 178, 113018. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.-R.; Zhu, Y.-X.; Xu, K.-F.; Pan, G.-Y.; Liu, X.; Qiao, Y.; Wu, F.-G. Efficient cell surface labelling of live zebrafish embryos: Wash-free fluorescence imaging for cellular dynamics tracking and nanotoxicity evaluation. Chem. Sci. 2019, 10, 4062–4068. [Google Scholar] [CrossRef]
- Zhang, J.-F.; Liu, B.; Hong, I.; Mo, A.; Roth, R.H.; Tenner, B.; Lin, W.; Zhang, J.Z.; Molina, R.S.; Drobizhev, M. An ultrasensitive biosensor for high-resolution kinase activity imaging in awake mice. Nat. Chem. Biol. 2021, 17, 39–46. [Google Scholar] [CrossRef]
- Ding, N.; Yuan, Z.; Zhang, X.; Chen, J.; Zhou, S.; Deng, Y. Programmable cross-ribosome-binding sites to fine-tune the dynamic range of transcription factor-based biosensor. Nucleic Acids Res. 2020, 48, 10602–10613. [Google Scholar] [CrossRef]
- Kim, K.H.; Yang, M.; Song, Y.; Kim, C.H.; Jung, Y.M.; Bae, N.-H.; Chang, S.-J.; Lee, S.J.; Kim, Y.T.; Choi, B.G. Touchable 3D hierarchically structured polyaniline nanoweb for capture and detection of pathogenic bacteria. Nano Converg. 2021, 8, 30. [Google Scholar] [CrossRef]
- Farooqi, H.M.U.; Kang, B.; Khalid, M.A.U.; Salih, A.R.C.; Hyun, K.; Park, S.H.; Huh, D.; Choi, K.H. Real-time monitoring of liver fibrosis through embedded sensors in a microphysiological system. Nano Converg. 2021, 8, 3. [Google Scholar] [CrossRef]
- Sciuto, E.L.; Petralia, S.; van der Meer, J.R.; Conoci, S. Miniaturized electrochemical biosensor based on whole-cell for heavy metal ions detection in water. Biotechnol. Bioeng. 2021, 118, 1456–1465. [Google Scholar] [CrossRef]
- Suhito, I.R.; Koo, K.-M.; Kim, T.-H. Recent advances in electrochemical sensors for the detection of biomolecules and whole cells. Biomedicines 2020, 9, 15. [Google Scholar] [CrossRef]
- Mohankumar, P.; Ajayan, J.; Mohanraj, T.; Yasodharan, R. Recent developments in biosensors for healthcare and biomedical applications: A review. Measurement 2021, 167, 108293. [Google Scholar] [CrossRef]
- Dai, Y.; Liu, C.C. Recent advances on electrochemical biosensing strategies toward universal point-of-care systems. Angew. Chem. 2019, 131, 12483–12496. [Google Scholar] [CrossRef]
- Qian, L.; Durairaj, S.; Prins, S.; Chen, A. Nanomaterial-based electrochemical sensors and biosensors for the detection of pharmaceutical compounds. Biosens. Bioelectron. 2021, 175, 112836. [Google Scholar] [CrossRef]
- Adhikari, B.-R.; Govindhan, M.; Chen, A. Carbon nanomaterials based electrochemical sensors/biosensors for the sensitive detection of pharmaceutical and biological compounds. Sensors 2015, 15, 22490–22508. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, J.G.; Rebelo, P.; Freitas, M.; Nouws, H.P.; Delerue-Matos, C. Breast cancer biomarker (HER2-ECD) detection using a molecularly imprinted electrochemical sensor. Sens. Actuators B Chem. 2018, 273, 1008–1014. [Google Scholar] [CrossRef]
- Soussi, I.; Mazouz, Z.; Collart-Dutilleul, P.Y.; Echabaane, M.; Martin, M.; Cloitre, T.; M’ghaieth, R.; Cuisinier, F.J.; Cunin, F.; Gergely, C. Electrochemical and optical investigation of dental pulp stem cell adhesion on modified porous silicon scaffolds. Colloids Surf. B Biointerfaces 2019, 181, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Dai, Y.; Chen, H.; Tang, Y.; Chen, X.; Wang, Y.; Zhao, J.; Zhu, X. Integration of fluorescence imaging and electrochemical biosensing for both qualitative location and quantitative detection of cancer cells. Biosens. Bioelectron. 2019, 130, 132–138. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, H.; Xu, G.; Guan, M.; Zhang, Q.; Wang, Z.; Dong, Z.; Chen, W.; Yang, X.; Qiao, A. A multiplexed electrochemical quantitative polymerase chain reaction platform for single-base mutation analysis. Biosens. Bioelectron. 2022, 214, 114496. [Google Scholar] [CrossRef]
- Wang, X.; Su, J.; Zeng, D.; Liu, G.; Liu, L.; Xu, Y.; Wang, C.; Liu, X.; Wang, L.; Mi, X. Gold nano-flowers (Au NFs) modified screen-printed carbon electrode electrochemical biosensor for label-free and quantitative detection of glycated hemoglobin. Talanta 2019, 201, 119–125. [Google Scholar] [CrossRef]
- Ruan, X.; Wang, Y.; Kwon, E.Y.; Wang, L.; Cheng, N.; Niu, X.; Ding, S.; Van Wie, B.J.; Lin, Y.; Du, D. Nanomaterial-enhanced 3D-printed sensor platform for simultaneous detection of atrazine and acetochlor. Biosens. Bioelectron. 2021, 184, 113238. [Google Scholar] [CrossRef]
- Ebrahimi, G.; Samadi Pakchin, P.; Shamloo, A.; Mota, A.; de la Guardia, M.; Omidian, H.; Omidi, Y. Label-free electrochemical microfluidic biosensors: Futuristic point-of-care analytical devices for monitoring diseases. Microchim. Acta 2022, 189, 252. [Google Scholar] [CrossRef]
- Campuzano, S.; Pedrero, M.; Yáñez-Sedeño, P.; Pingarrón, J.M. New challenges in point of care electrochemical detection of clinical biomarkers. Sens. Actuators B Chem. 2021, 345, 130349. [Google Scholar] [CrossRef]
- Park, C.-Y.; Min, J.-H.; Kim, Y.-S.; Song, H.-J.; Kim, J.-D. Development of Portable Electrical-cell-substrate Impedance Sensing System. Sens. Mater. 2019, 31, 253–259. [Google Scholar] [CrossRef]
- Sadighbayan, D.; Hasanzadeh, M.; Ghafar-Zadeh, E. Biosensing based on field-effect transistors (FET): Recent progress and challenges. TrAC Trends Anal. Chem. 2020, 133, 116067. [Google Scholar] [CrossRef]
- Ding, J.; Qin, W. Recent advances in potentiometric biosensors. TrAC Trends Anal. Chem. 2020, 124, 115803. [Google Scholar] [CrossRef]
- Al Mamun, M.; Wahab, Y.A.; Hossain, M.M.; Hashem, A.; Johan, M.R. Electrochemical biosensors with aptamer recognition layer for the diagnosis of pathogenic bacteria: Barriers to commercialization and remediation. TrAC Trends Anal. Chem. 2021, 145, 116458. [Google Scholar] [CrossRef]
- Rollo, S.; Rani, D.; Leturcq, R.; Olthuis, W.; Pascual García, C. High aspect ratio fin-ion sensitive field effect transistor: Compromises toward better electrochemical biosensing. Nano Lett. 2019, 19, 2879–2887. [Google Scholar] [CrossRef]
- Lei, L.; Ma, B.; Xu, C.; Liu, H. Emerging Tumor-on-Chips with Electrochemical Biosensors. TrAC Trends Anal. Chem. 2022, 153, 116640. [Google Scholar] [CrossRef]
- Shin, J.-H.; Lee, M.-J.; Choi, J.-H.; Song, J.-a.; Kim, T.-H.; Oh, B.-K. Electrochemical H2O2 biosensor based on horseradish peroxidase encapsulated protein nanoparticles with reduced graphene oxide-modified gold electrode. Nano Converg. 2020, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.; Jeong, S.; Lee, J.; Choi, H.S.; Kim, J.; Lee, H. Innovations in biomedical nanoengineering: Nanowell array biosensor. Nano Converg. 2018, 5, 9. [Google Scholar] [CrossRef] [PubMed]
- Choi, N.Y.; Lee, M.-Y.; Jeong, S. Recent Advances in 3D-Cultured Brain Tissue Models Derived from Human iPSCs. BioChip J. 2022, 16, 246–254. [Google Scholar] [CrossRef]
- Suhito, I.R.; Lee, W.; Baek, S.; Lee, D.; Min, J.; Kim, T.-H. Rapid and sensitive electrochemical detection of anticancer effects of curcumin on human glioblastoma cells. Sens. Actuators B Chem. 2019, 288, 527–534. [Google Scholar] [CrossRef]
- Suhito, I.R.; Kang, E.-S.; Kim, D.-S.; Baek, S.; Park, S.-J.; Moon, S.-H.; Luo, Z.; Lee, D.; Min, J.; Kim, T.-H. High density gold nanostructure composites for precise electrochemical detection of human embryonic stem cells in cell mixture. Colloids Surf. B Biointerfaces 2019, 180, 384–392. [Google Scholar] [CrossRef]
- Suhito, I.R.; Angeline, N.; Lee, K.H.; Kim, H.; Park, C.G.; Luo, Z.; Kim, T.H. A Spheroid-Forming Hybrid Gold Nanostructure Platform That Electrochemically Detects Anticancer Effects of Curcumin in a Multicellular Brain Cancer Model. Small 2021, 17, 2002436. [Google Scholar] [CrossRef]
- Zhou, H.; Huang, R.; Su, T.; Li, B.; Zhou, H.; Ren, J.; Li, Z. A c-MWCNTs/AuNPs-based electrochemical cytosensor to evaluate the anticancer activity of pinoresinol from Cinnamomum camphora against HeLa cells. Bioelectrochemistry 2022, 146, 108133. [Google Scholar] [CrossRef]
- Lee, S.-M.; Lee, J.-E.; Lee, Y.-K.; Yoo, D.; Seon, D.-B.; Lee, D.-W.; Kim, C.-B.; Choi, H.; Lee, K.-H. Thermal-Corrosion-Free Electrode-Integrated Cell Chip for Promotion of Electrically Stimulated Neurite Outgrowth. BioChip J. 2022, 16, 99–110. [Google Scholar] [CrossRef]
- Bagno, L.; Hatzistergos, K.E.; Balkan, W.; Hare, J.M. Mesenchymal stem cell-based therapy for cardiovascular disease: Progress and challenges. Mol. Ther. 2018, 26, 1610–1623. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, H.; Olson, E.N.; Bassel-Duby, R. Therapeutic approaches for cardiac regeneration and repair. Nat. Rev. Cardiol. 2018, 15, 585–600. [Google Scholar] [CrossRef] [PubMed]
- Suhito, I.R.; Angeline, N.; Choo, S.-S.; Woo, H.Y.; Paik, T.; Lee, T.; Kim, T.-H. Nanobiosensing platforms for real-time and non-invasive monitoring of stem cell pluripotency and differentiation. Sensors 2018, 18, 2755. [Google Scholar] [CrossRef] [PubMed]
- Birajdar, M.S.; Joo, H.; Koh, W.-G.; Park, H. Natural bio-based monomers for biomedical applications: A review. Biomater. Res. 2021, 25, 8. [Google Scholar] [CrossRef]
- Sung, Y.K.; Lee, D.R.; Chung, D.J. Advances in the development of hemostatic biomaterials for medical application. Biomater. Res. 2021, 25, 37. [Google Scholar] [CrossRef]
- Lee, H.-N.; Liang, C.; Liao, L.; Tian, W.-D. Advances in Research on Stem Cell-Based Pulp Regeneration. Tissue Eng Regen Med. 2021, 18, 931–940. [Google Scholar] [CrossRef]
- Suhito, I.R.; Kim, J.W.; Koo, K.M.; Nam, S.A.; Kim, Y.K.; Kim, T.H. In Situ Detection of Kidney Organoid Generation from Stem Cells Using a Simple Electrochemical Method. Adv. Sci. 2022, 9, 2200074. [Google Scholar] [CrossRef]
- Singh, A.; Sharma, A.; Ahmed, A.; Sundramoorthy, A.K.; Furukawa, H.; Arya, S.; Khosla, A. Recent advances in electrochemical biosensors: Applications, challenges, and future scope. Biosensors 2021, 11, 336. [Google Scholar] [CrossRef]
- Gupta, N.; Renugopalakrishnan, V.; Liepmann, D.; Paulmurugan, R.; Malhotra, B.D. Cell-based biosensors: Recent trends, challenges and future perspectives. Biosens. Bioelectron. 2019, 141, 111435. [Google Scholar] [CrossRef]
- Pan, Y.; Hu, N.; Wei, X.; Gong, L.; Zhang, B.; Wan, H.; Wang, P. 3D cell-based biosensor for cell viability and drug assessment by 3D electric cell/matrigel-substrate impedance sensing. Biosens. Bioelectron. 2019, 130, 344–351. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Hong, H.J.; Lee, H.J. Fabrication of Cell Spheroids for 3D Cell Culture and Biomedical Applications. BioChip J. 2022, 1–20. [Google Scholar] [CrossRef]
- Ling, Y.; Lyu, Q.; Zhai, Q.; Zhu, B.; Gong, S.; Zhang, T.; Dyson, J.; Cheng, W. Design of stretchable holey gold biosensing electrode for real-time cell monitoring. ACS Sens. 2020, 5, 3165–3171. [Google Scholar] [CrossRef] [PubMed]
- Sakata, T.; Sugimoto, H.; Saito, A. Live monitoring of microenvironmental pH based on extracellular acidosis around cancer cells with cell-coupled gate ion-sensitive field-effect transistor. Anal. Chem. 2018, 90, 12731–12736. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Li, K.; Liu, B.; Tu, J.; Li, T.; Li, Y.-T.; Zhang, G.-J. A pH-sensitive field-effect transistor for monitoring of cancer cell external acid environment. Talanta 2023, 252, 123764. [Google Scholar] [CrossRef] [PubMed]
- Hempel, F.; Law, J.K.Y.; Nguyen, T.C.; Lanche, R.; Susloparova, A.; Vu, X.T.; Ingebrandt, S. PEDOT: PSS organic electrochemical transistors for electrical cell-substrate impedance sensing down to single cells. Biosens. Bioelectron. 2021, 180, 113101. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Qin, Y.; Fan, W.-T.; Wu, W.-T.; Lv, S.-W.; Yan, L.-P.; Liu, Y.-L.; Huang, W.-H. Plasticizer and catalyst co-functionalized PEDOT: PSS enables stretchable electrochemical sensing of living cells. Chem. Sci. 2021, 12, 14432–14440. [Google Scholar] [CrossRef] [PubMed]
- Dang, Y.; Guan, X.; Zhou, Y.; Hao, C.; Zhang, Y.; Chen, S.; Ma, Y.; Bai, Y.; Gong, Y.; Gao, Y. Biocompatible PB/Ti3C2 hybrid nanocomposites for the non-enzymatic electrochemical detection of H2O2 released from living cells. Sens. Actuators B Chem. 2020, 319, 128259. [Google Scholar] [CrossRef]
- Yang, M.; Ren, X.; Yang, T.; Xu, C.; Ye, Y.; Sun, Z.; Kong, L.; Wang, B.; Luo, Z. Polypyrrole/sulfonated multi-walled carbon nanotubes conductive hydrogel for electrochemical sensing of living cells. Chem. Eng. J. 2021, 418, 129483. [Google Scholar] [CrossRef]
- Wang, M.; Hu, M.; Li, Z.; He, L.; Song, Y.; Jia, Q.; Zhang, Z.; Du, M. Construction of Tb-MOF-on-Fe-MOF conjugate as a novel platform for ultrasensitive detection of carbohydrate antigen 125 and living cancer cells. Biosens. Bioelectron. 2019, 142, 111536. [Google Scholar] [CrossRef]
- Du, L.; Chen, W.; Wang, J.; Cai, W.; Kong, S.; Wu, C. Folic acid-functionalized zirconium metal-organic frameworks based electrochemical impedance biosensor for the cancer cell detection. Sens. Actuators B Chem. 2019, 301, 127073. [Google Scholar] [CrossRef]
- Wang, L.; Shi, X.-H.; Zhang, Y.-F.; Liu, A.-A.; Liu, S.-L.; Wang, Z.-G.; Pang, D.-W. CdZnSeS quantum dots condensed with ordered mesoporous carbon for high-sensitive electrochemiluminescence detection of hydrogen peroxide in live cells. Electrochim. Acta 2020, 362, 137107. [Google Scholar] [CrossRef]
- Ashraf, G.; Asif, M.; Aziz, A.; Dao, A.Q.; Zhang, T.; Iftikhar, T.; Wang, Q.; Liu, H. Facet-energy inspired metal oxide extended hexapods decorated with graphene quantum dots: Sensitive detection of bisphenol A in live cells. Nanoscale 2020, 12, 9014–9023. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zheng, J.; Liang, H.; Li, C.-P. Electrochemical sensor for cancer cell detection using calix[8]arene/polydopamine/phosphorene nanocomposite based on host−guest recognition. Sens. Actuators B Chem. 2020, 317, 128193. [Google Scholar] [CrossRef]
- Cho, Y.; Pham Ba, V.A.; Jeong, J.-Y.; Choi, Y.; Hong, S. Ion-selective carbon nanotube field-effect transistors for monitoring drug effects on nicotinic acetylcholine receptor activation in live cells. Sensors 2020, 20, 3680. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Walsh, K.B.; Bayram, F.; Koley, G. Direct measurement of K+ ion efflux from neuronal cells using a graphene-based ion sensitive field effect transistor. RSC Adv. 2020, 10, 37728–37734. [Google Scholar] [CrossRef]
- Xu, S.; Zhang, C.; Jiang, S.; Hu, G.; Li, X.; Zou, Y.; Liu, H.; Li, J.; Li, Z.; Wang, X. Graphene foam field-effect transistor for ultra-sensitive label-free detection of ATP. Sens. Actuators B Chem. 2019, 284, 125–133. [Google Scholar] [CrossRef]
- Zheng, C.; Jin, X.; Li, Y.; Mei, J.; Sun, Y.; Xiao, M.; Zhang, H.; Zhang, Z.; Zhang, G.-J. Sensitive molybdenum disulfide based field effect transistor sensor for real-time monitoring of hydrogen peroxide. Sci. Rep. 2019, 9, 759. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Huan, K.; Deng, D.; Tang, L.; Wang, J.; Luo, L. Facile synthesis of ZnMn2O4@ rGO microspheres for ultrasensitive electrochemical detection of hydrogen peroxide from human breast cancer cells. ACS Appl. Mater. Interfaces 2019, 12, 3430–3437. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Rejeeth, C.; Xu, W.; Zhu, C.; Liu, X.; Wan, J.; Jiang, M.; Qian, K. Label-free electrochemical sensor for cd44 by ligand-protein interaction. Anal. Chem. 2019, 91, 7078–7085. [Google Scholar] [CrossRef] [PubMed]
- Tavakkoli, H.; Akhond, M.; Ghorbankhani, G.A.; Absalan, G. Electrochemical sensing of hydrogen peroxide using a glassy carbon electrode modified with multiwalled carbon nanotubes and zein nanoparticle composites: Application to HepG2 cancer cell detection. Microchim. Acta 2020, 187, 105. [Google Scholar] [CrossRef] [PubMed]
- Zhao, A.; She, J.; Manoj, D.; Wang, T.; Sun, Y.; Zhang, Y.; Xiao, F. Functionalized graphene fiber modified by dual nanoenzyme: Towards high-performance flexible nanohybrid microelectrode for electrochemical sensing in live cancer cells. Sens. Actuators B Chem. 2020, 310, 127861. [Google Scholar] [CrossRef]
- Jin, Z.; Wang, L.; Cao, D.; Zou, S.; Chen, C.; Kang, H.; Song, Q.; Wang, H.; Tang, Y. A new method for rapid screening of hybridoma cell clones secreting paired antibodies using sandwich cell surface fluorescence immunosorbent assay. Anal. Chim. Acta 2021, 1163, 338493. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Heo, Y.J. Biosensing technologies for chronic diseases. BioChip J. 2021, 15, 1–13. [Google Scholar] [CrossRef]
- Wrege, R.; Peter, C.; Wesling, B.N.; Rambo, C.R.; Schneider, M.C.; Galup-Montoro, C. A CMOS test chip with simple post-processing steps for dry characterization of ISFET arrays. IEEE Sens. J. 2020, 21, 4755–4763. [Google Scholar] [CrossRef]
- Thriveni, G.; Ghosh, K. Advancement and Challenges of Biosensing Using Field Effect Transistors. Biosensors 2022, 12, 647. [Google Scholar] [CrossRef]
- Navas, L.E.; Carnero, A. NAD+ metabolism, stemness, the immune response, and cancer. Signal Transduct. Target. Ther. 2021, 6, 2. [Google Scholar] [CrossRef]
- Sakata, T.; Saito, A.; Sugimoto, H. In situ measurement of autophagy under nutrient starvation based on interfacial pH sensing. Sci. Rep. 2018, 8, 8282. [Google Scholar] [CrossRef]
- Nemčeková, K.; Labuda, J. Advanced materials-integrated electrochemical sensors as promising medical diagnostics tools: A review. Mater. Sci. Eng. C 2021, 120, 111751. [Google Scholar] [CrossRef]
- Jing, L.; Xie, C.; Li, Q.; Yang, M.; Li, S.; Li, H.; Xia, F. Electrochemical biosensors for the analysis of breast cancer biomarkers: From design to application. Anal. Chem. 2021, 94, 269–296. [Google Scholar] [CrossRef]
- Daw, C.C.; Ramachandran, K.; Enslow, B.T.; Maity, S.; Bursic, B.; Novello, M.J.; Rubannelsonkumar, C.S.; Mashal, A.H.; Ravichandran, J.; Bakewell, T.M. Lactate elicits ER-mitochondrial Mg2+ dynamics to integrate cellular metabolism. Cell 2020, 183, 474–489.e17. [Google Scholar] [CrossRef] [PubMed]
- Bader, D.A.; Hartig, S.M.; Putluri, V.; Foley, C.; Hamilton, M.P.; Smith, E.A.; Saha, P.K.; Panigrahi, A.; Walker, C.; Zong, L. Mitochondrial pyruvate import is a metabolic vulnerability in androgen receptor-driven prostate cancer. Nat. Metab. 2019, 1, 70–85. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Xie, Q.; Gimple, R.C.; Zhong, Z.; Tam, T.; Tian, J.; Kidwell, R.L.; Wu, Q.; Prager, B.C.; Qiu, Z. Three-dimensional bioprinted glioblastoma microenvironments model cellular dependencies and immune interactions. Cell Res. 2020, 30, 833–853. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Su, G.-H.; Ma, D.; Xiao, Y.; Shao, Z.-M.; Jiang, Y.-Z. Technological advances in cancer immunity: From immunogenomics to single-cell analysis and artificial intelligence. Signal Transduct. Target. Ther. 2021, 6, 312. [Google Scholar] [CrossRef] [PubMed]
- Christopherson, G.T.; de Vasconcellos, J.F.; Dunn, J.C.; Griffin, D.W.; Jones, P.E.; Nesti, L.J. Three-dimensional modeling of the structural microenvironment in post-traumatic war wounds. Tissue Eng. Regen. Med. 2021, 18, 963–973. [Google Scholar] [CrossRef]
- Hur, W.; Son, S.E.; Kim, S.N.; Seong, G.H. Cell-based electrochemical cytosensor for rapid and sensitive evaluation of the anticancer effects of saponin on human malignant melanoma cells. Bioelectrochemistry 2021, 140, 107813. [Google Scholar] [CrossRef] [PubMed]
- Hideshima, S.; Hayashi, H.; Saito, S.; Tateno, H.; Momma, T.; Osaka, T. A Non-Destructive Electrical Assay of Stem Cell Differentiation Based on Semiconductor Biosensing. Anal. Sens. 2022, e202200046. [Google Scholar] [CrossRef]
- Ma, Z.; Jiang, M.; Zhu, Q.; Luo, Y.; Chen, G.; Pan, M.; Xie, T.; Huang, X.; Chen, D. A porous hollow fiber sensor for detection of cellular hydrogen peroxide release based on cell-in-lumen configuration. Sens. Actuators B Chem. 2020, 321, 128516. [Google Scholar] [CrossRef]
- Shi, X.; Xie, Y.; Chen, L.; Lu, J.; Zhang, L.; Sun, D. Combining quasi-ZIF-67 hybrid nanozyme and G-quadruplex/hemin DNAzyme for highly sensitive electrochemical sensing. Bioelectrochemistry 2023, 149, 108278. [Google Scholar] [CrossRef] [PubMed]
- Nashimoto, Y.; Mukomoto, R.; Imaizumi, T.; Terai, T.; Shishido, S.; Ino, K.; Yokokawa, R.; Miura, T.; Onuma, K.; Inoue, M. Electrochemical sensing of oxygen metabolism for a three-dimensional cultured model with biomimetic vascular flow. Biosens. Bioelectron. 2023, 219, 114808. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wang, H.; Cao, S.; Xiang, W.; Li, T.; Yang, M. Electrochemical determination of the activity and inhibition of telomerase based on the interaction of DNA with molybdate. Microchim. Acta 2019, 186, 96. [Google Scholar] [CrossRef]
- Ma, Z.; Luo, Y.; Zhu, Q.; Jiang, M.; Pan, M.; Xie, T.; Huang, X.; Chen, D. In-situ monitoring of glucose metabolism in cancer cell microenvironments based on hollow fiber structure. Biosens. Bioelectron. 2020, 162, 112261. [Google Scholar] [CrossRef]
- Lee, C.-S.; Kim, T.H. Label-free assay of protein kinase A activity and inhibition in cancer cell using electrochemically-prepared AuNP/rGO nanohybrid electrode modified with C-Kemptide. Talanta 2020, 215, 120899. [Google Scholar] [CrossRef]
- Ma, J.; Xue, L.; Zhang, M.; Li, C.; Xiang, Y.; Liu, P.; Li, G. An electrochemical sensor for Oct4 detection in human tissue based on target-induced steric hindrance effect on a tetrahedral DNA nanostructure. Biosens. Bioelectron. 2019, 127, 194–199. [Google Scholar] [CrossRef]
- Lee, J.H.; Choi, H.K.; Yang, L.; Chueng, S.T.D.; Choi, J.W.; Lee, K.B. Nondestructive real-time monitoring of enhanced stem cell differentiation using a graphene-Au hybrid nanoelectrode array. Adv. Mater. 2018, 30, 1802762. [Google Scholar] [CrossRef]
- Nasr, B.; Chatterton, R.; Yong, J.H.M.; Jamshidi, P.; D’Abaco, G.M.; Bjorksten, A.R.; Kavehei, O.; Chana, G.; Dottori, M.; Skafidas, E. Self-organized nanostructure modified microelectrode for sensitive electrochemical glutamate detection in stem cells-derived brain organoids. Biosensors 2018, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, C.; Spitz, S.; Berger, E.; Bolognin, S.; Smits, L.M.; Crepaz, P.; Rothbauer, M.; Rosser, J.M.; Marchetti-Deschmann, M.; Schwamborn, J.C. Monitoring the neurotransmitter release of human midbrain organoids using a redox cycling microsensor as a novel tool for personalized Parkinson’s disease modelling and drug screening. Analyst 2021, 146, 2358–2367. [Google Scholar] [CrossRef] [PubMed]
- Perebikovsky, A.; Hwu, A.T.; Yale, A.R.; Ghazinejad, M.; Madou, M. Nanofibrous carbon multifunctional smart scaffolds for simultaneous cell differentiation and dopamine detection. ACS Biomater. Sci. Eng. 2019, 6, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Li, G.; Tian, L.; Li, L.; Shi, Y.; Huang, T.; Li, Y.; Xu, Q. Multifunctional peptide-oligonucleotide conjugate promoted sensitive electrochemical biosensing of cardiac troponin I. Biochem. Eng. J. 2021, 174, 108104. [Google Scholar] [CrossRef]
- Hassanzadeh, P.; Atyabi, F.; Dinarvand, R. The significance of artificial intelligence in drug delivery system design. Adv. Drug Deliv. Rev. 2019, 151, 169–190. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-H.; Kim, T.-H. Recent advances in multicellular tumor spheroid generation for drug screening. Biosensors 2021, 11, 445. [Google Scholar] [CrossRef]
- Pan, Y.; Jiang, D.; Gu, C.; Qiu, Y.; Wan, H.; Wang, P. 3D microgroove electrical impedance sensing to examine 3D cell cultures for antineoplastic drug assessment. Microsyst. Nanoeng. 2020, 6, 23. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Gan, F.; Zhang, Y.; He, X.; Shen, C.; Qiu, H.; Liu, P. Selective Killing of Cancer Cells by Nonplanar Aromatic Hydrocarbon-Induced DNA Damage. Adv. Sci. 2019, 6, 1901341. [Google Scholar] [CrossRef] [PubMed]
- Post, Y.; Clevers, H. Defining adult stem cell function at its simplest: The ability to replace lost cells through mitosis. Cell Stem Cell 2019, 25, 174–183. [Google Scholar] [CrossRef]
- Zhu, X.; Zhang, B.; He, Y.; Bao, J. Liver organoids: Formation strategies and biomedical applications. Tissue Eng. Regen. Med. 2021, 18, 573–585. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; David, B.T.; Trawczynski, M.; Fessler, R.G. Advances in pluripotent stem cells: History, mechanisms, technologies, and applications. Stem Cell Rev. Rep. 2020, 16, 3–32. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Cho, A.N.; Min, S.; Kim, S.; Cho, S.W. Organoids for advanced therapeutics and disease models. Adv. Ther. 2019, 2, 1800087. [Google Scholar] [CrossRef]
- Kong, Y.; Duan, J.; Liu, F.; Han, L.; Li, G.; Sun, C.; Sang, Y.; Wang, S.; Yi, F.; Liu, H. Regulation of stem cell fate using nanostructure-mediated physical signals. Chem. Soc. Rev. 2021, 50, 12828–12872. [Google Scholar] [CrossRef] [PubMed]



| Detection Method | Cell Line | Immobilisation Strategy | LOD (Target) | Ref |
|---|---|---|---|---|
| ECIS | SH-SY5Y, ND7/23 cells | PPy/s-MWCNTs/ AuNPs hydrogel/ITO | 17 nM (Dopamine) | [78] |
| ECIS | MCF-7 | Tb-MOF-on-Fe-MOF | 58 μU/mL (CA125) | [79] |
| ECIS | HeLa | FA@UiO-66 nanocomposite/Au | 90 cells/mL | [80] |
| ECIS | HeLa, MDCK, 293T | CdZnSeS QD/OMC | 10.23 μM (H2O2) | [81] |
| ECIS | HeLa | Cu2O–CuO@GQDs | 1 nM (Bisphenol A) | [82] |
| ECIS | LNCaP | BPene@PDA−SCX8 | 36 cells/mL | [83] |
| FET | PC12 | PVC coated CNT-FET | 1 nM (Potassium ion) | [84] |
| FET | U-251 MG | Graphene-based ISFET | 1 μM (Potassium ion) | [85] |
| FET | HepG2 | graphene foam FET | 0.5 pM (ATP) | [86] |
| FET | HeLa | MoS2/RGO FET | 1 pM (H2O2) | [87] |
| Amperometric | MCF-7 | ZnMn2O4/rGO | 0.012 μM (H2O2) | [88] |
| DPV | MCF-7 | ITO/MWCNT/PDDAHA | 5.94 pg/mL (CD44) | [89] |
| LSV | HepG2 | GC/ZNBs/fMWCNT | 35 nM (H2O2) | [90] |
| Amperometric | Patient-derived cancer | MnO2-NWs@Au-NPs/GF | 1.9 μM (H2O2) | [91] |
| Detection Method | Cell Lines | Immobilisation Strategy | Anticancer Drug | Ref |
|---|---|---|---|---|
| EIS | Cervical cancer (HeLa) | c-MWCNTs/AuNPs | Pinoresinol | [59] |
| Amperometric | Lung cancer (H1299) | HRP-AuNPs-MWNT | fMLP | [107] |
| CV | Liver cancer (HepG2) | G-quadruplex/hemin/Au /QZIF-67-2/GCE | Paclitaxel | [108] |
| SECM | Patient-derived cancer organoid (colorectal) | Fibrin-collagen gel | Bortezomib | [109] |
| SWV | Cervical cancer (HeLa) | Telomerase and dNTPs | Epigallocatechin gallate (EGCG) | [110] |
| Amperometric | Lung cancer (PC9) | PHF-MWNT-PB-Gox | Osimertini | [111] |
| EIS | Cervical cancer (HeLa) | C-Kemptide-modified AuNP /rGO-GCE | H-89 | [112] |
| Detection Method | Cell Lines | Differentiated Cells | LOD (Target) | Ref |
|---|---|---|---|---|
| DPV | Human embryonic stem cell | Undifferentiated stem cell | 12,500 cells (ESC) | [57] |
| SWV | Human tissue | Cancer stem cell | 0.5–1000 ng/mL (Oct4) | [113] |
| CV | Human mesenchymal stem cell | Osteogenic differentiation | 0.03 unit/mL (ALP) | [114] |
| Amperometric | Human embryonic stem cell | Brain organoid | 0.5 mM (glutamine) | [115] |
| EIS | Human neuroepithelial stem cell | Midbrain organoid | 8 nM (epinephrine) | [116] |
| DPV | Mouse neural stem cell | Neuronal differentiation | 134 nM (dopamine) | [117] |
| DPV | Bone mesenchymal stem cell | Cardiomyogenic differentiation | 0.42 pg/mL (cTnI) | [118] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koo, K.-M.; Kim, C.-D.; Ju, F.N.; Kim, H.; Kim, C.-H.; Kim, T.-H. Recent Advances in Electrochemical Biosensors for Monitoring Animal Cell Function and Viability. Biosensors 2022, 12, 1162. https://doi.org/10.3390/bios12121162
Koo K-M, Kim C-D, Ju FN, Kim H, Kim C-H, Kim T-H. Recent Advances in Electrochemical Biosensors for Monitoring Animal Cell Function and Viability. Biosensors. 2022; 12(12):1162. https://doi.org/10.3390/bios12121162
Chicago/Turabian StyleKoo, Kyeong-Mo, Chang-Dae Kim, Fu Nan Ju, Huijung Kim, Cheol-Hwi Kim, and Tae-Hyung Kim. 2022. "Recent Advances in Electrochemical Biosensors for Monitoring Animal Cell Function and Viability" Biosensors 12, no. 12: 1162. https://doi.org/10.3390/bios12121162
APA StyleKoo, K.-M., Kim, C.-D., Ju, F. N., Kim, H., Kim, C.-H., & Kim, T.-H. (2022). Recent Advances in Electrochemical Biosensors for Monitoring Animal Cell Function and Viability. Biosensors, 12(12), 1162. https://doi.org/10.3390/bios12121162





