Detection of a Double-Stranded MGMT Gene Using Electrochemically Reduced Graphene Oxide (ErGO) Electrodes Decorated with AuNPs and Peptide Nucleic Acids (PNA)
Abstract
:1. Introduction
2. Experimental
2.1. Materials and Methods
2.2. Apparatus and Measurements
2.3. Electrode Modification
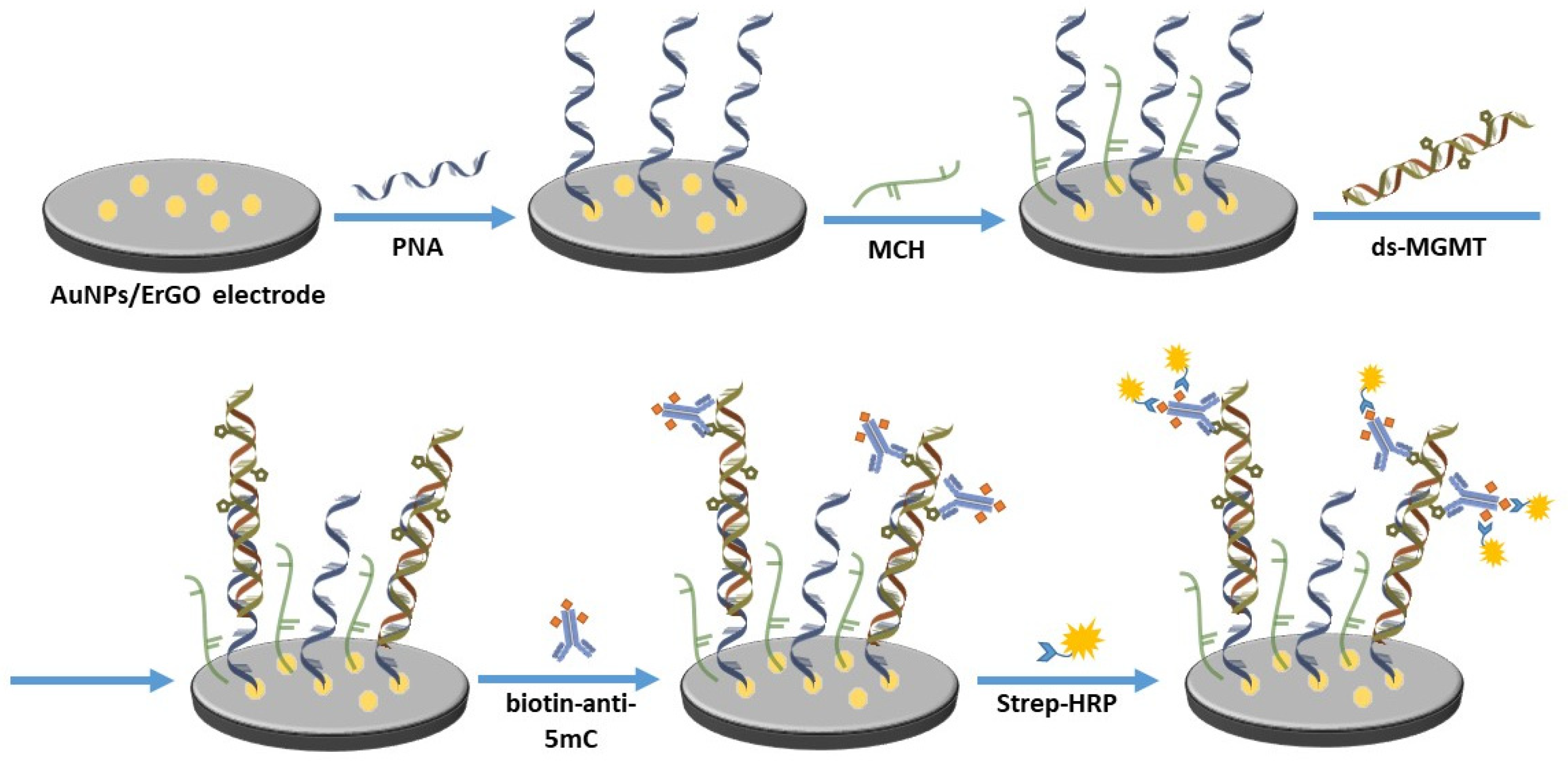
2.4. Assay Development
3. Results and Discussion
3.1. GO Reduction
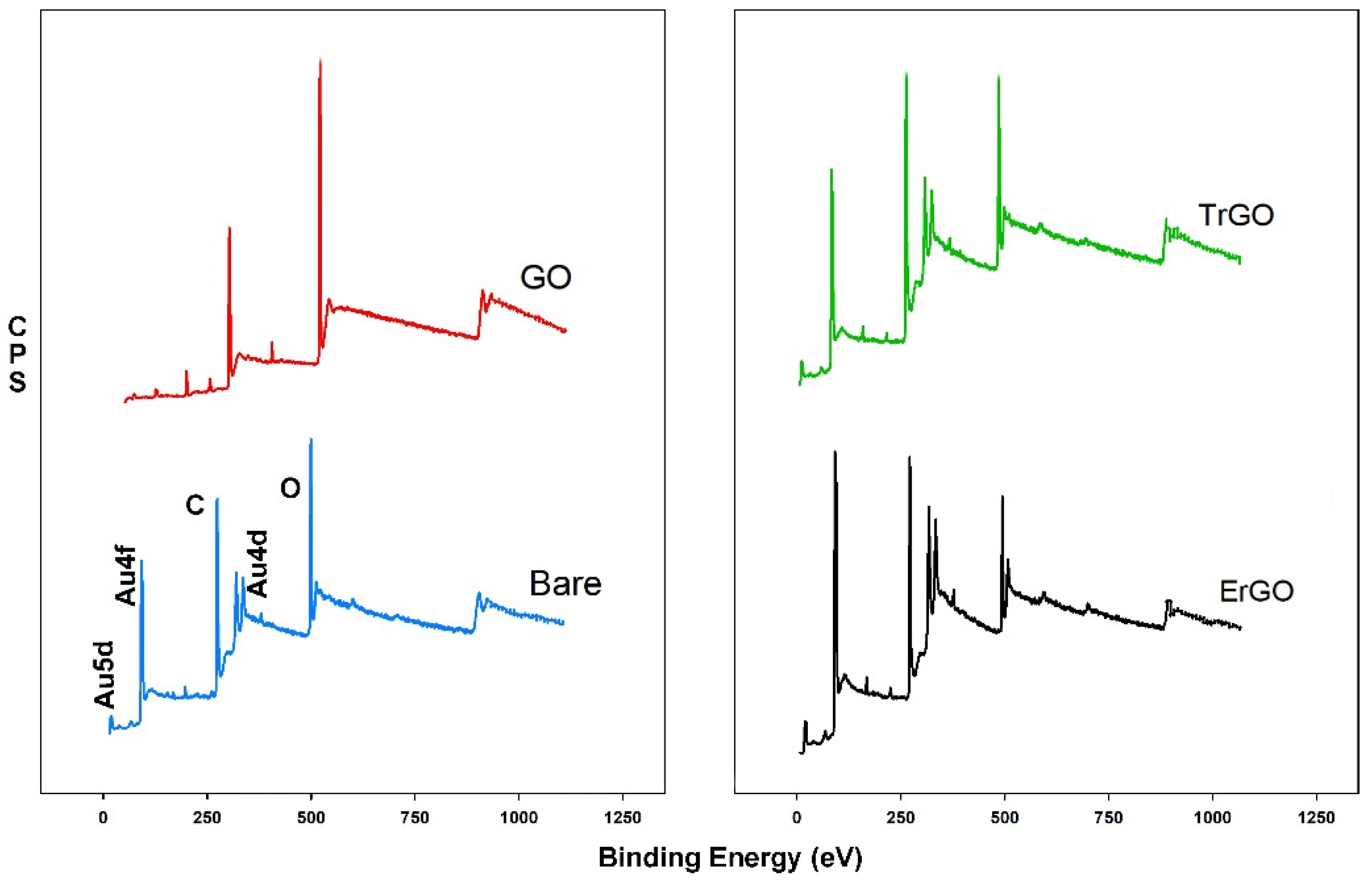
3.1.1. XPS Measurements
3.1.2. Raman Spectroscopy
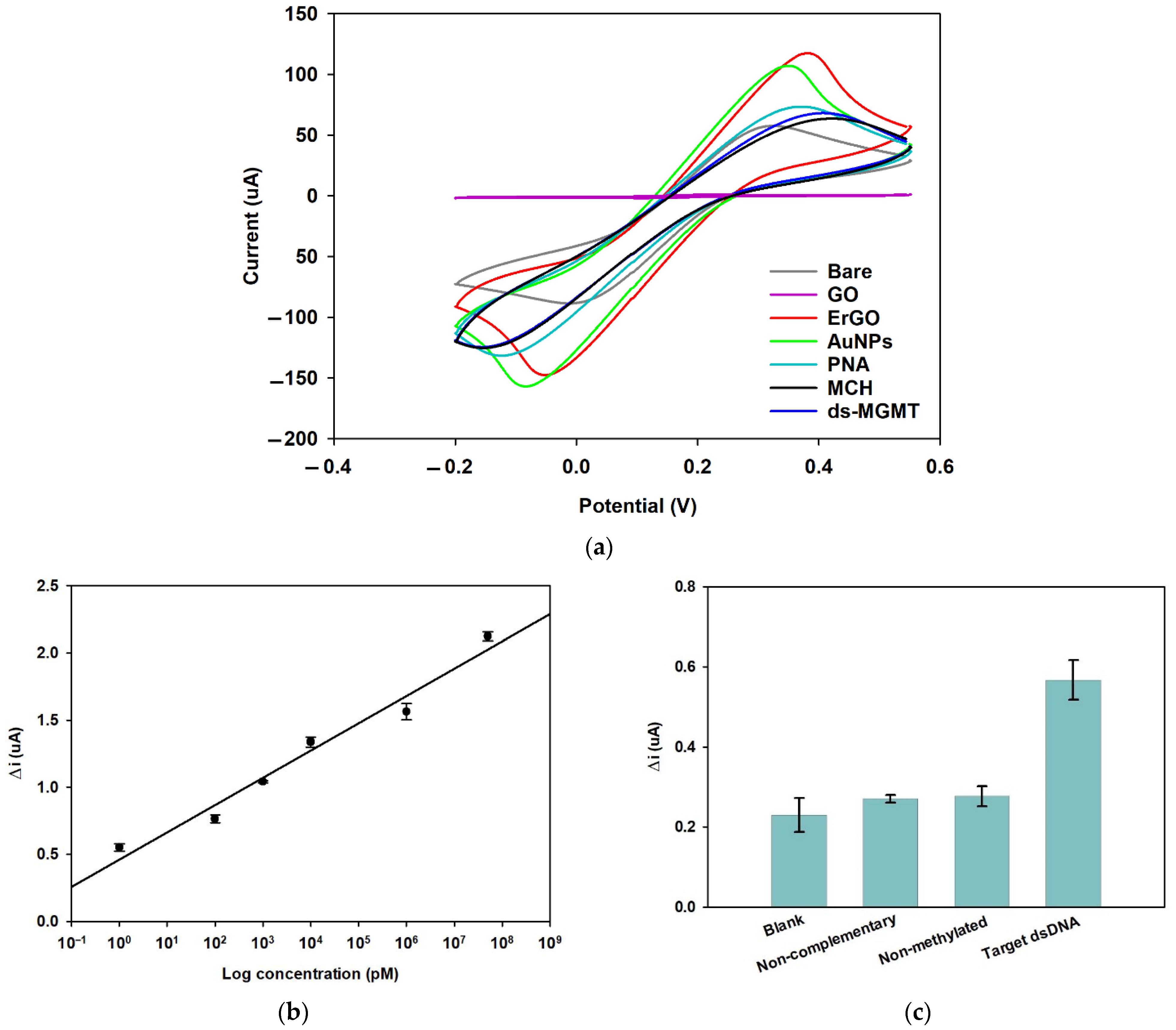
3.1.3. Cyclic Voltammetry
3.2. AuNPs
3.2.1. XPS Measurements
3.2.2. SEM and EDS
3.3. Assay Development
3.3.1. Optimization
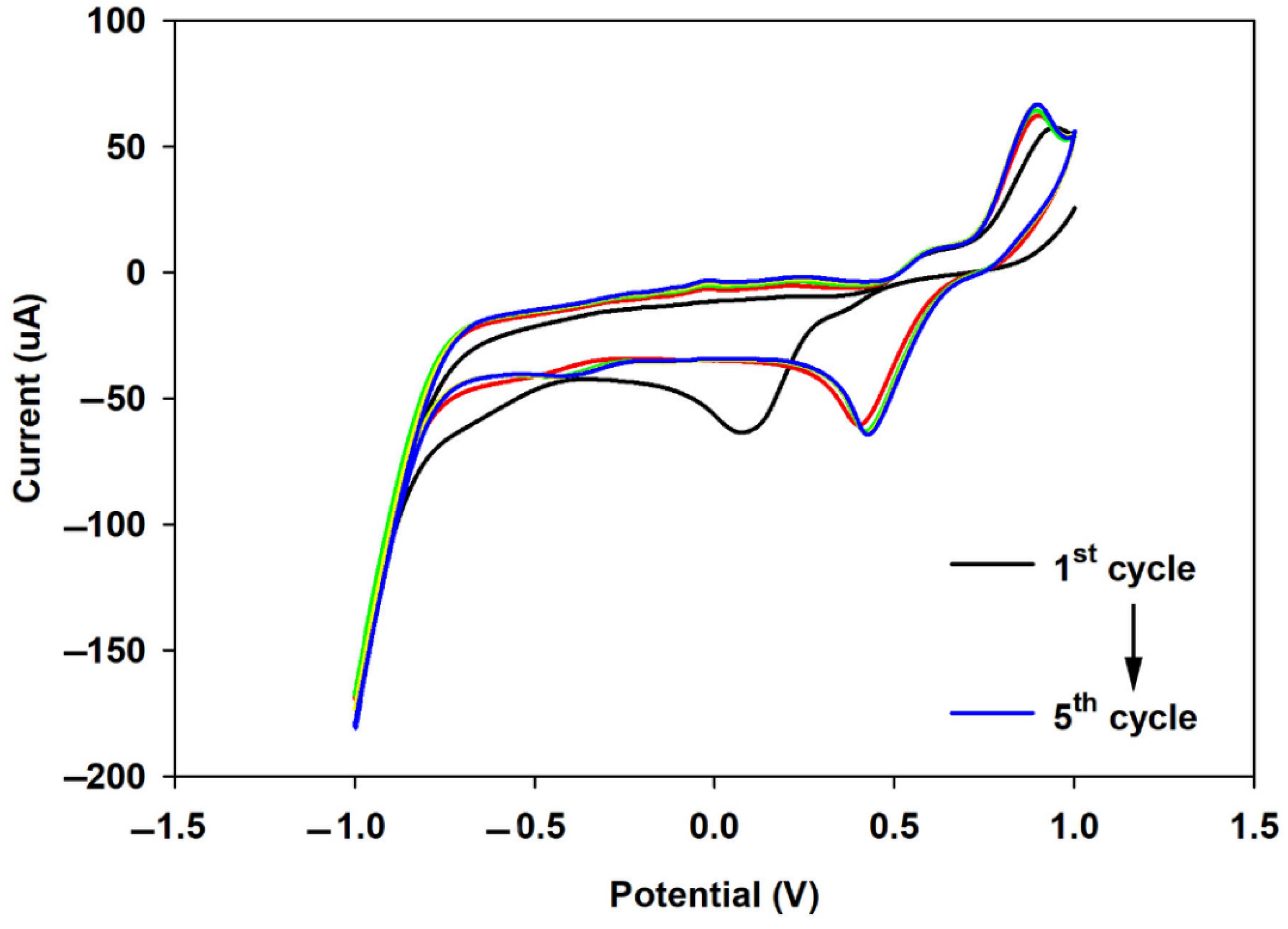
3.3.2. Cyclic Voltammetry
3.3.3. Linear Regression
3.3.4. Selectivity
3.3.5. Comparison with Other Works
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Experimental
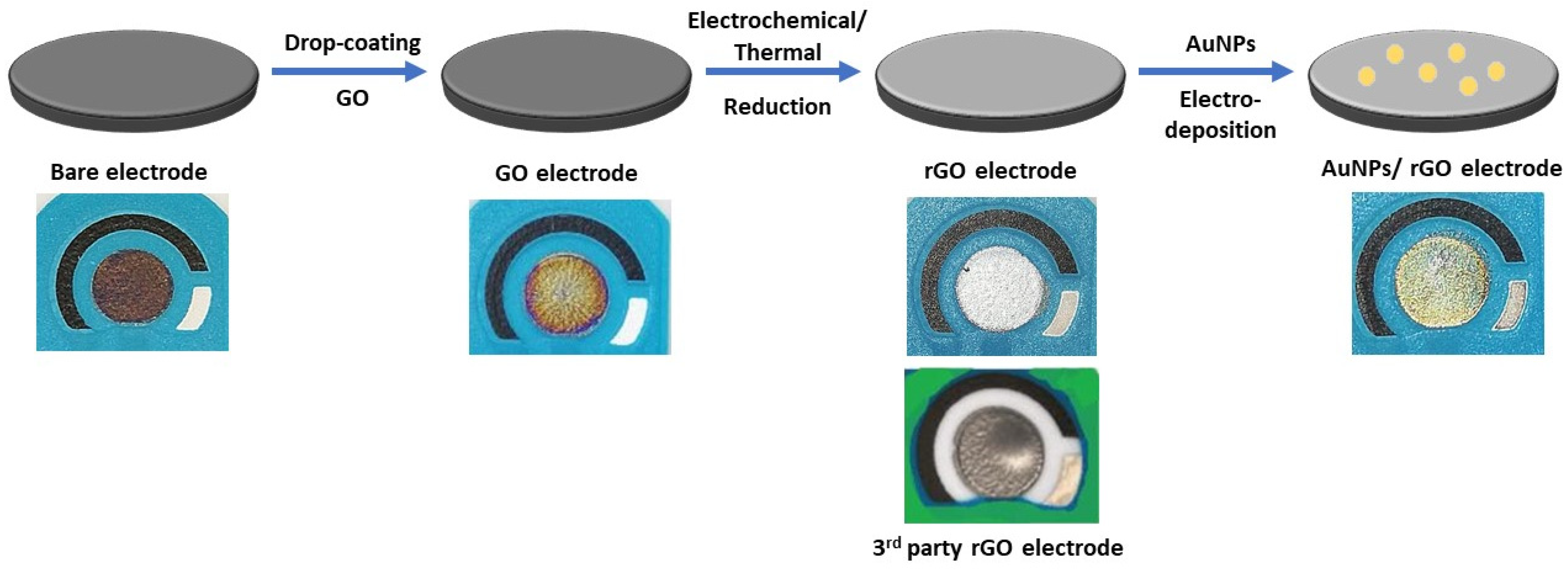
Appendix A.2. Results and Discussion
Appendix A.2.1. GO Reduction
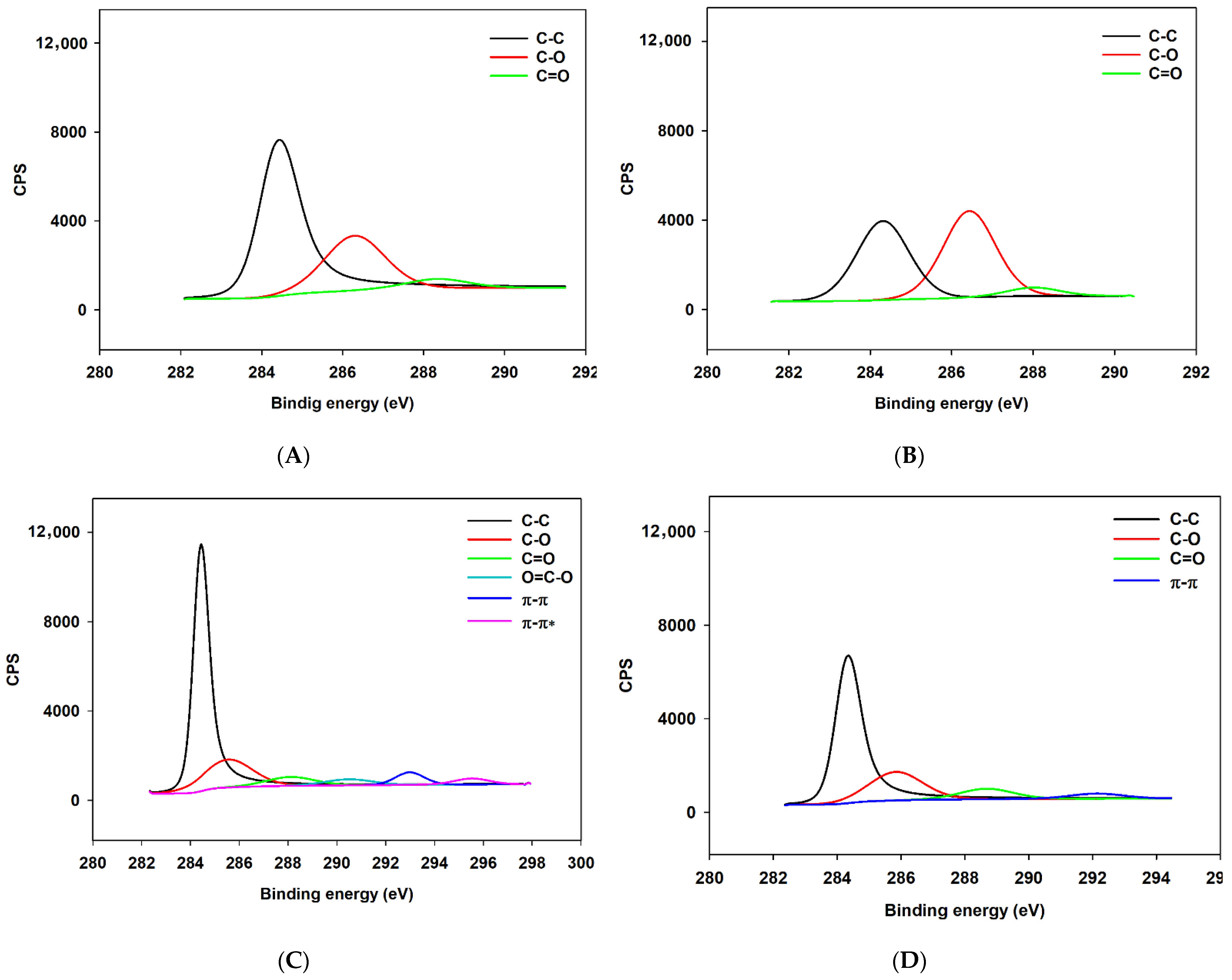
Appendix A.2.2. XPS
| Sample | Peak Name | Position | FWHM | At% |
|---|---|---|---|---|
| Bare | O1s C1s N1s Cl2p | 532.10 285.10 400.10 200.10 | 3.31 3.77 3.10 3.32 | 19.26 76.49 1.37 2.87 |
| GO | O1s C1s N1s S2p | 534.37 287.37 402.37 168.37 | 3.01 4.50 3.34 3.07 | 27.98 69.63 1.30 1.10 |
| ErGO | O1s C1s Na1s Cl2p | 533.33 285.33 1074.33 199.33 | 3.89 2.73 2.87 3.48 | 8.57 90.15 0.44 0.85 |
| TrGO | O1s C1s N1s Cl2p | 565.37 312.37 430.37 194.37 | 3.88 3.06 3.77 3.16 | 14.65 83.57 1.31 0.47 |
Appendix A.2.3. Raman
| Sample | D Peak Location (cm−1) | G Peak Location (cm−1) | ID/IG | La (nm) |
|---|---|---|---|---|
| Bare | 1340 | 1570 | 0.77 | 24.99 |
| GO | 1350 | 1570 | 0.88 | 21.84 |
| TrGO | 1340 | 1570 | 0.89 | 21.60 |
| ErGO | 1350 | 1570 | 1.15 | 16.72 |
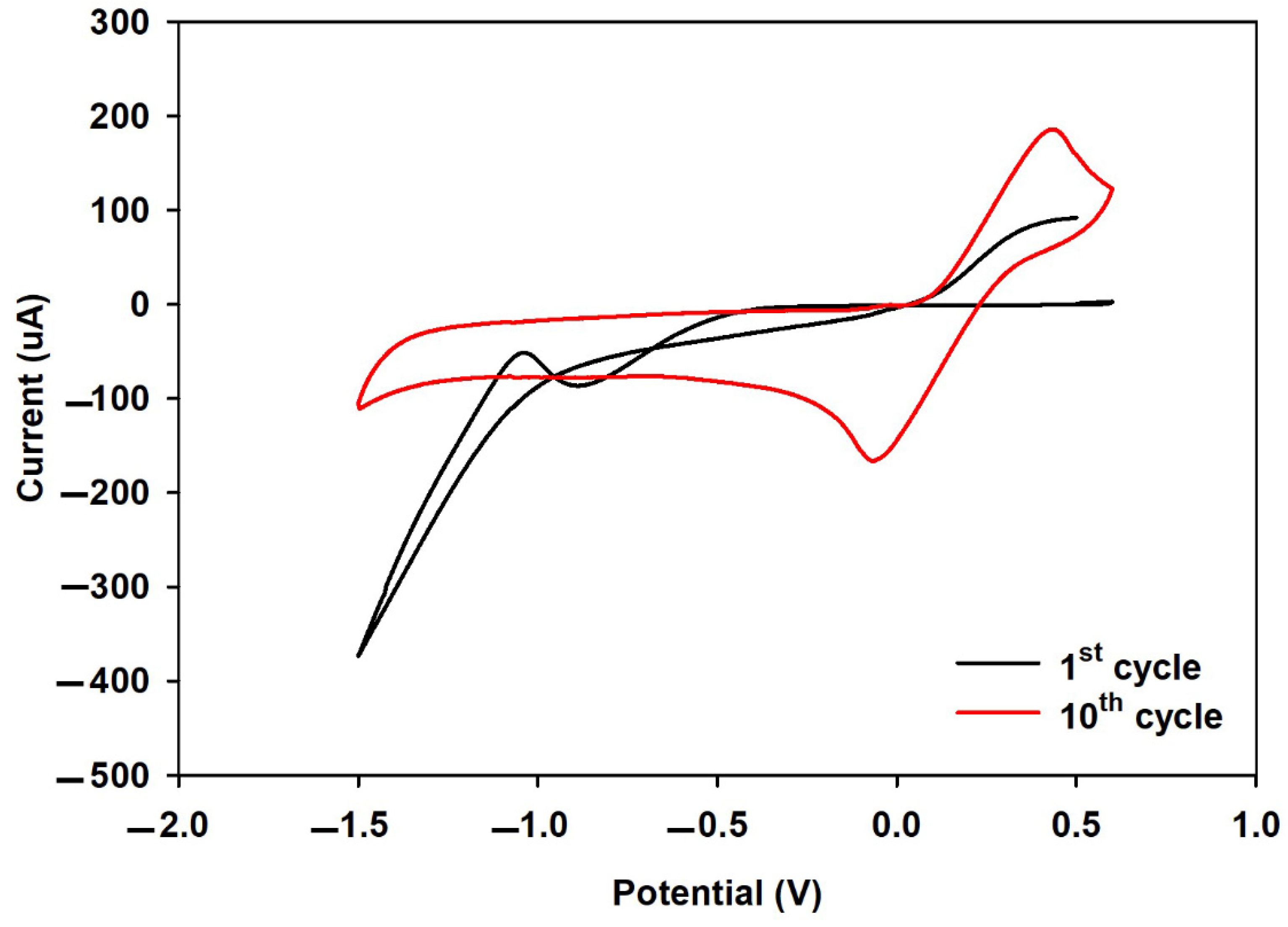
Appendix A.2.4. Cyclic Voltammetry

Appendix A.2.5. AuNPs
Appendix A.2.6. SEM and EDS
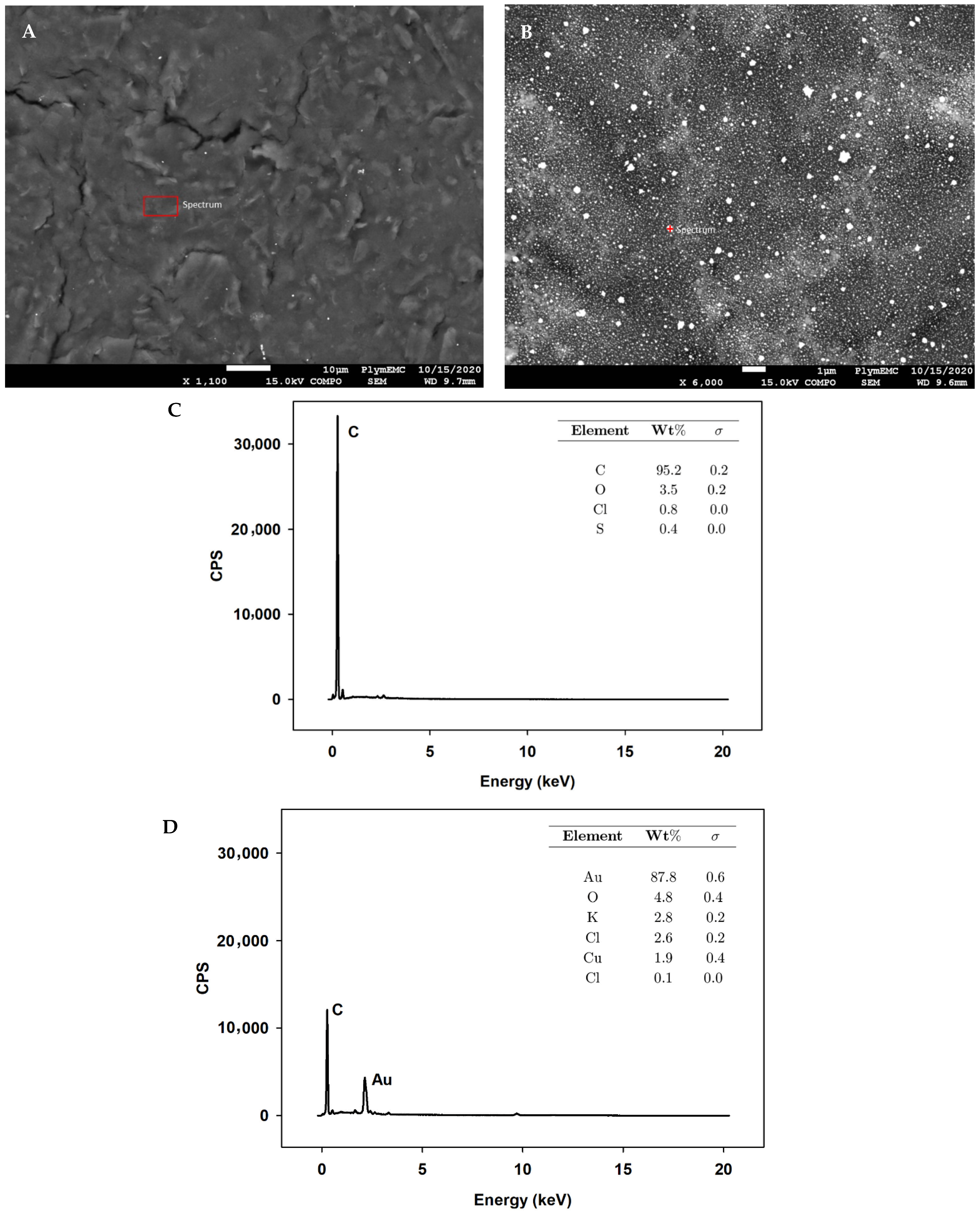
Appendix A.2.7. XPS
| Sample | Peak Name | Position | FWHM | At% |
|---|---|---|---|---|
| Bare/AuNPs | O1s C1s N1s Cl2p S2p Au4f | 532.83 284.83 401.83 199.83 168.83 83.83 | 3.20 3.61 3.27 3.37 3.25 2.50 | 19.97 72.11 1.56 1.50 0.71 4.16 |
| GO/AuNPs | O1s C1s N1s S2p | 531.89 284.89 400.89 168.89 | 3.00 4.26 2.84 3.01 | 29.40 63.83 2.76 4.00 |
| ErGO/AuNPs | O1s C1s N1s S2p Au4f | 551.00 289.99 418.99 180.99 94.99 | 3.26 2.77 3.09 2.97 2.80 | 13.25 74.10 2.89 2.38 7.38 |
| TrGO/AuNPs | O1s C1s N1s S2p Au4f | 542.79 298.79 408.79 171.79 86.79 | 3.43 2.99 3.43 2.96 2.43 | 17.65 75.06 1.75 1.46 4.08 |
Appendix A.2.8. Optimization

References
- Lam, K.; Pan, K.; Linnekamp, J.F.; Medema, J.P.; Kandimalla, R. DNA methylation-based biomarkers in colorectal cancer: A systematic review. Biochim. Biophys. Acta Rev. Cancer 2016, 1866, 106–120. [Google Scholar] [CrossRef] [PubMed]
- Mikeska, T.; Craig, J.M. DNA methylation biomarkers: Cancer and beyond. Genes 2014, 5, 821–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, P.M.; Singal, R. DNA methylation and cancer. J. Clin. Oncol. 2004, 22, 4632–4642. [Google Scholar] [CrossRef] [Green Version]
- Usadel, H.; Brabender, J.; Danenberg, K.D.; Jerónimo, C.; Harden, S.; Engles, J.; Danenberg, P.V.; Yang, S.; Sidransky, D. Quantitative adenomatous polyposis coli promoter methylation analysis in tumor tissue, serum, and plasma DNA of patients with lung cancer. Cancer Res. 2002, 62, 371–375. [Google Scholar]
- Kersting, M.; Friedl, C.; Kraus, A.; Behn, M.; Pankow, W.; Schuermann, M. Differentia frequencies of p16INK4a promoter hypermethylation, p53 mutation, and K-ras mutation in exfoliative material mark the development of lung cancer in symptomatic chronic smokers. J. Clin. Oncol. 2000, 18, 3221–3229. [Google Scholar] [CrossRef]
- Esteller, M.; Garcia-Foncillas, J.; Andion, E.; Goodman, S.N.; Hidalgo, O.F.; Vanaclocha, V.; Baylin, S.B.; Herman, J.G. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N. Engl. J. Med. 2000, 343, 1350–1354. [Google Scholar] [CrossRef]
- Silva, J.M.; Dominguez, G.; Garcia, J.M.; Gonzalez, R.; Villanueva, M.J.; Navarro, F.; Provencio, M.; San Martin, S.; Espana, P.; Bonilla, F. Presence of tumor DNA in plasma of breast cancer patients: Clinicopathological correlations. Cancer Res. 1999, 59, 3251–3256. [Google Scholar]
- Evron, E.; Dooley, W.C.; Umbricht, C.B.; Rosenthal, D.; Sacchi, N.; Gabrielson, E.; Soito, A.B.; Hung, D.T.; Ljung, B.M.; Davidson, N.E.; et al. Detection of breast cancer cells in ductal lavage fluid by methylation-specific PCR. Lancet 2001, 357, 1335–1336. [Google Scholar] [CrossRef]
- Goessl, C.; Krause, H.; Müller, M.; Heicappell, R.; Schrader, M.; Sachsinger, J.; Miller, K. Fluorescent methylation-specific polymerase chain reaction for DNA-based detection of prostate cancer in bodily fluids. Cancer Res. 2000, 60, 5941–5945. [Google Scholar]
- Lee, W.H.; Morton, R.A.; Epstein, J.I.; Brooks, J.D.; Campbell, P.A.; Bova, G.S.; Hsieh, W.S.; Isaacs, W.B.; Nelson, W.G. Cytidine methylation of regulatory sequences near the pi-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc. Natl. Acad. Sci. USA 1994, 91, 11733–11737. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharjee, R.; Moriam, S.; Umer, M.; Nguyen, N.T.; Shiddiky, M.J. DNA methylation detection: Recent developments in bisulfite free electrochemical and optical approaches. Analyst 2018, 143, 4802–4818. [Google Scholar] [CrossRef]
- Kurdyukov, S.; Bullock, M. DNA methylation analysis: Choosing the right method. Biology 2016, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Shanmuganathan, R.; Basheer, N.B.; Amirthalingam, L.; Muthukumar, H.; Kaliaperumal, R.; Shanmugam, K. Conventional and nanotechniques for DNA methylation profiling. J. Mol. Diagn. 2013, 15, 17–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Chen, F.; Zhang, D.; Zhao, Y.; Wei, J.; Wang, L.; Song, S.; Fan, C.; Zhao, Y. Single copy-sensitive electrochemical assay for circulating methylated DNA in clinical samples with ultrahigh specificity based on a sequential discrimination–amplification strategy. Chem. Sci. 2017, 8, 4764–4770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Povedano, E.; Gamella, M.; Torrente-Rodríguez, R.M.; Montiel, V.R.V.; Montero-Calle, A.; Solís-Fernández, G.; Navarro-Villoslada, F.; Pedrero, M.; Peláez-García, A.; Mendiola, M.; et al. Multiplexed magnetic beads-assisted amperometric bioplatforms for global detection of methylations in nucleic acids. Anal. Chim. Acta 2021, 1182, 338946. [Google Scholar] [CrossRef]
- Chen, X.; Huang, J.; Zhang, S.; Mo, F.; Su, S.; Li, Y.; Fang, L.; Deng, J.; Huang, H.; Luo, Z.; et al. Electrochemical biosensor for DNA methylation detection through hybridization chain-amplified reaction coupled with a tetrahedral DNA nanostructure. ACS Appl. Mater. Interfaces 2019, 11, 3745–3752. [Google Scholar] [CrossRef]
- D’Agata, R.; Giuffrida, M.C.; Spoto, G. Peptide nucleic acid-based biosensors for cancer diagnosis. Molecules 2017, 22, 1951. [Google Scholar] [CrossRef] [Green Version]
- Puiu, M.; Bala, C. Peptide-based electrochemical biosensors. In Electrochemical Biosensors; Elsevier: Amsterdam, The Netherlands, 2019; pp. 277–306. [Google Scholar]
- Karimzadeh, A.; Hasanzadeh, M.; Shadjou, N.; de la Guardia, M. Peptide based biosensors. TrAC Trends Anal. Chem. 2018, 107, 1–20. [Google Scholar] [CrossRef]
- Baker, E.S.; Hong, J.W.; Gaylord, B.S.; Bazan, G.C.; Bowers, M.T. PNA/dsDNA complexes: Site specific binding and dsDNA biosensor applications. J. Am. Chem. Soc. 2006, 128, 8484–8492. [Google Scholar] [CrossRef]
- Hamidi-Asl, E.; Raoof, J.B.; Ojani, R.; Golabi, S.M.; Hejazi, M.S. A new peptide nucleotide acid biosensor for electrochemical detection of single nucleotide polymorphism in duplex DNA via triplex structure formation. J. Iran. Chem. Soc. 2013, 10, 1075–1083. [Google Scholar] [CrossRef]
- Ahmadi, M.; Ahour, F. An electrochemical biosensor based on a graphene oxide modified pencil graphite electrode for direct detection and discrimination of double-stranded DNA sequences. Anal. Methods 2020, 12, 4541–4550. [Google Scholar] [CrossRef] [PubMed]
- Ahour, F.; Pournaghi-Azar, M.H.; Alipour, E.; Hejazi, M.S. Detection and discrimination of recombinant plasmid encoding hepatitis C virus core/E1 gene based on PNA and double-stranded DNA hybridization. Biosens. Bioelectron. 2013, 45, 287–291. [Google Scholar] [CrossRef]
- Wang, Z.; Zhou, X.; Zhang, J.; Boey, F.; Zhang, H. Direct electrochemical reduction of single layer graphene oxide and subsequent functionalization with glucose oxidase. J. Phys. Chem. C 2009, 113, 14071–14075. [Google Scholar] [CrossRef]
- Marrani, A.G.; Motta, A.; Palmieri, V.; Perini, G.; Papi, M.; Dalchiele, E.A.; Schrebler, R.; Zanoni, R. A comparative experimental and theoretical study of the mechanism of graphene oxide mild reduction by ascorbic acid and N-acetyl cysteine for biomedical applications. Mater. Adv. 2020, 1, 2745–2754. [Google Scholar] [CrossRef]
- Devadas, B.; Madhu, R.; Chen, S.M.; Yeh, H.T. Controlled electrochemical synthesis of new rare earth metal lutetium hexacyanoferrate on reduced graphene oxide and its application as a salicylic acid sensor. J. Mater. Chem. B 2014, 2, 7515–7523. [Google Scholar] [CrossRef] [PubMed]
- Devadas, B.; Rajkumar, M.; Chen, S.M.; Saraswathi, R. Electrochemically reduced graphene oxide/neodymium hexacyanoferrate modified electrodes for the electrochemical detection of paracetomol. Int. J. Electrochem. Sci. 2012, 7, 3339–3349. [Google Scholar]
- Wu, B.; Zhao, N.; Hou, S.; Zhang, C. Electrochemical synthesis of polypyrrole, reduced graphene oxide, and gold nanoparticles composite and its application to hydrogen peroxide biosensor. Nanomaterials 2016, 6, 220. [Google Scholar] [CrossRef] [Green Version]
- Schniepp, H.C.; Li, J.L.; McAllister, M.J.; Sai, H.; Herrera-Alonso, M.; Adamson, D.H.; Prud’homme, R.K.; Car, R.; Saville, D.A.; Aksay, I.A. Functionalized single graphene sheets derived from splitting graphite oxide. J. Phys. Chem. B 2006, 110, 8535–8539. [Google Scholar] [CrossRef] [Green Version]
- Ren, P.G.; Yan, D.X.; Ji, X.; Chen, T.; Li, Z.M. Temperature dependence of graphene oxide reduced by hydrazine hydrate. Nanotechnology 2010, 22, 055705. [Google Scholar] [CrossRef]
- Yang, Z.Z.; Zheng, Q.B.; Qiu, H.X.; Jing, L.; Yang, J.H. A simple method for the reduction of graphene oxide by sodium borohydride with CaCl2 as a catalyst. New Carbon Mater. 2015, 30, 41–47. [Google Scholar] [CrossRef]
- Chua, C.K.; Ambrosi, A.; Pumera, M. Graphene oxide reduction by standard industrial reducing agent: Thiourea dioxide. J. Mater. Chem. 2012, 22, 11054–11061. [Google Scholar] [CrossRef]
- Al-Gaashani, R.; Najjar, A.; Zakaria, Y.; Mansour, S.; Atieh, M. XPS and structural studies of high-quality graphene oxide and reduced graphene oxide prepared by different chemical oxidation methods. Ceram. Int. 2019, 45, 14439–14448. [Google Scholar] [CrossRef]
- Yang, D.; Velamakanni, A.; Bozoklu, G.; Park, S.; Stoller, M.; Piner, R.D.; Stankovich, S.; Jung, I.; Field, D.A.; Ventrice, C.A., Jr.; et al. Chemical analysis of graphene oxide films after heat and chemical treatments by X-ray photoelectron and Micro-Raman spectroscopy. Carbon 2009, 47, 145–152. [Google Scholar] [CrossRef]
- Krishnamoorthy, K.; Veerapandian, M.; Yun, K.; Kim, S.J. The chemical and structural analysis of graphene oxide with different degrees of oxidation. Carbon 2013, 53, 38–49. [Google Scholar] [CrossRef]
- Safarzadeh, M.; Suhail, A.; Sethi, J.; Sattar, A.; Jenkins, D.; Pan, G. A Label-Free DNA-Immunosensor Based on Aminated rGO Electrode for the Quantification of DNA Methylation. Nanomaterials 2021, 11, 985. [Google Scholar] [CrossRef]
- Lee, C.S.; Yu, S.H.; Kim, T.H. One-step electrochemical fabrication of reduced graphene oxide/gold nanoparticles nanocomposite-modified electrode for simultaneous detection of dopamine, ascorbic acid, and uric acid. Nanomaterials 2018, 8, 17. [Google Scholar] [CrossRef] [Green Version]
- Le, V.H.; Nguyen, T.H.; Nguyen, H.H.; Huynh, L.T.N.; Vo, A.L.; Nguyen, T.K.T.; Nguyen, D.T.; Lam, V.Q. Fabrication and electrochemical behavior investigation of a Pt-loaded reduced graphene oxide composite (Pt@rGO) as a High-performance cathode for dye-sensitized solar cells. Int. J. Photoenergy 2020, 2020, 8927124. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.B.; Lin, M.L.; Cong, X.; Liu, H.N.; Tan, P.H. Raman spectroscopy of graphene-based materials and its applications in related devices. Chem. Soc. Rev. 2018, 47, 1822–1873. [Google Scholar] [CrossRef] [Green Version]
- Devasenathipathy, R.; Mani, V.; Chen, S.M.; Viswanath, B.; Vasantha, V.; Govindasamy, M. Electrodeposition of gold nanoparticles on a pectin scaffold and its electrocatalytic application in the selective determination of dopamine. RSC Adv. 2014, 4, 55900–55907. [Google Scholar] [CrossRef]
- Chiang, H.C.; Wang, Y.; Zhang, Q.; Levon, K. Optimization of the electrodeposition of gold nanoparticles for the application of highly sensitive, label-free biosensor. Biosensors 2019, 9, 50. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.H.; Lin, C.W.; Liu, H.H.; Sheu, J.T.; Hung, W.H. Potential-controlled electrodeposition of gold dendrites in the presence of cysteine. Chem. Commun. 2011, 47, 2044–2046. [Google Scholar] [CrossRef]
- Marcano, D.C.; Kosynkin, D.V.; Berlin, J.M.; Sinitskii, A.; Sun, Z.; Slesarev, A.; Alemany, L.B.; Lu, W.; Tour, J.M. Improved synthesis of graphene oxide. ACS Nano 2010, 4, 4806–4814. [Google Scholar] [CrossRef] [PubMed]
- Pei, S.; Cheng, H.M. The reduction of graphene oxide. Carbon 2012, 50, 3210–3228. [Google Scholar] [CrossRef]
- Benvidi, A.; Firouzabadi, A.D.; Moshtaghiun, S.M.; Mazloum-Ardakani, M.; Tezerjani, M.D. Ultrasensitive DNA sensor based on gold nanoparticles/reduced graphene oxide/glassy carbon electrode. Anal. Biochem. 2015, 484, 24–30. [Google Scholar] [CrossRef]
- De la Llave, E.; Clarenc, R.; Schiffrin, D.J.; Williams, F.J. Organization of alkane amines on a gold surface: Structure, surface dipole, and electron transfer. J. Phys. Chem. C 2014, 118, 468–475. [Google Scholar] [CrossRef]
- Olmos-Asar, J.A.; Ludueña, M.; Mariscal, M. Monolayer protected gold nanoparticles: The effect of the headgroup—Au interaction. Phys. Chem. Chem. Phys. 2014, 16, 15979–15987. [Google Scholar] [CrossRef]
- Hu, Q.; Wang, Q.; Kong, J.; Li, L.; Zhang, X. Electrochemically mediated in situ growth of electroactive polymers for highly sensitive detection of double-stranded DNA without sequence-preference. Biosens. Bioelectron. 2018, 101, 1–6. [Google Scholar] [CrossRef]
- Saadati, A.; Hassanpour, S.; de la Guardia, M.; Mosafer, J.; Hashemzaei, M.; Mokhtarzadeh, A.; Baradaran, B. Recent advances on application of peptide nucleic acids as a bioreceptor in biosensors development. TrAC Trends Anal. Chem. 2019, 114, 56–68. [Google Scholar] [CrossRef]
- Azimzadeh, M.; Rahaie, M.; Nasirizadeh, N.; Ashtari, K.; Naderi-Manesh, H. An electrohemical nanobiosensor for plasma miRNA-155, based on graphene oxide and gold nanorod, for early detection of breast cancer. Biosens. Bioelectron. 2016, 77, 99–106. [Google Scholar] [CrossRef]
- Povedano, E.; Montiel, V.R.V.; Valverde, A.; Navarro-Villoslada, F.; Yáñez-Sedeño, P.; Pedrero, M.; Montero-Calle, A.; Barderas, R.; Peláez-García, A.; Mendiola, M.; et al. Versatile electroanalytical bioplatforms for simultaneous determination of cancer-related DNA 5-methyl-and 5-hydroxymethyl-cytosines at global and gene-specific levels in human serum and tissues. ACS Sens. 2018, 4, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Xu, L.; Gao, C.; Zhang, Y.; Yao, Q.; Zhang, G.J. A MoS2 nanosheet-based fluorescence biosensor for simple and quantitative analysis of DNA methylation. Sensors 2016, 16, 1561. [Google Scholar] [CrossRef] [PubMed]
- Khodaei, R.; Ahmady, A.; Khoshfetrat, S.M.; Kashanian, S.; Tavangar, S.M.; Omidfar, K. Voltammetric immunosensor for E-cadherin promoter DNA methylation using a Fe3O6-citric acid nanocomposite and a screen-printed carbon electrode modified with poly(vinylalcohol) and reduced graphene oxide. Microchim. Acta 2019, 186, 170. [Google Scholar] [CrossRef] [PubMed]


| Electrode | Bioreceptor | Dynamic Range/LOD | Technique | Reference |
|---|---|---|---|---|
| Gold modified with gold nanoparticles | stem-loop-tetrhedron composite DNA | 10−6–10 pM 9.326 × 10−7 pM | Chronoamperometry | [16] |
| SPCE and immuno-magnetic beads (MBs) | Anti-5mC | 4–2.5 × 102 pM 1 pM | Amperometry | [51] |
| MoS2 Nanosheets | FAM-labeled probe DNA | 100–2 × 105 pM 140 pM | Fluorescence | [52] |
| SPCE modified with rGO and polyvinyl alcohol | Anti-5mC immobilized and DNA probe conjugated with Fe3O4-citric acid nanocomposites | 7 × 10−4–140.29 pM 6.31 × 10−4 pM | DPV/EIS | [53] |
| rGO modified with ammuniom hydroxide | Anti5-mC and complementary DNA | 0.5–100 pM 0.012 pM | DPV | [36] |
| AuNPs/ErGO | PNA and anti-5mC | 1–5 × 107 pM 0.86 pM | Amperometry | This work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Safarzadeh, M.; Pan, G. Detection of a Double-Stranded MGMT Gene Using Electrochemically Reduced Graphene Oxide (ErGO) Electrodes Decorated with AuNPs and Peptide Nucleic Acids (PNA). Biosensors 2022, 12, 98. https://doi.org/10.3390/bios12020098
Safarzadeh M, Pan G. Detection of a Double-Stranded MGMT Gene Using Electrochemically Reduced Graphene Oxide (ErGO) Electrodes Decorated with AuNPs and Peptide Nucleic Acids (PNA). Biosensors. 2022; 12(2):98. https://doi.org/10.3390/bios12020098
Chicago/Turabian StyleSafarzadeh, Mina, and Genhua Pan. 2022. "Detection of a Double-Stranded MGMT Gene Using Electrochemically Reduced Graphene Oxide (ErGO) Electrodes Decorated with AuNPs and Peptide Nucleic Acids (PNA)" Biosensors 12, no. 2: 98. https://doi.org/10.3390/bios12020098
APA StyleSafarzadeh, M., & Pan, G. (2022). Detection of a Double-Stranded MGMT Gene Using Electrochemically Reduced Graphene Oxide (ErGO) Electrodes Decorated with AuNPs and Peptide Nucleic Acids (PNA). Biosensors, 12(2), 98. https://doi.org/10.3390/bios12020098





