Zip Nucleic Acid-Based Genomagnetic Assay for Electrochemical Detection of microRNA-34a
Abstract
1. Introduction
2. Materials and Methods
2.1. Instruments
2.2. Chemicals
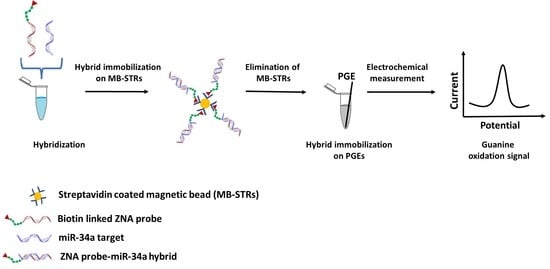
2.3. Genomagnetic Assay
2.4. Isolation of Total RNA from Cell Lysates
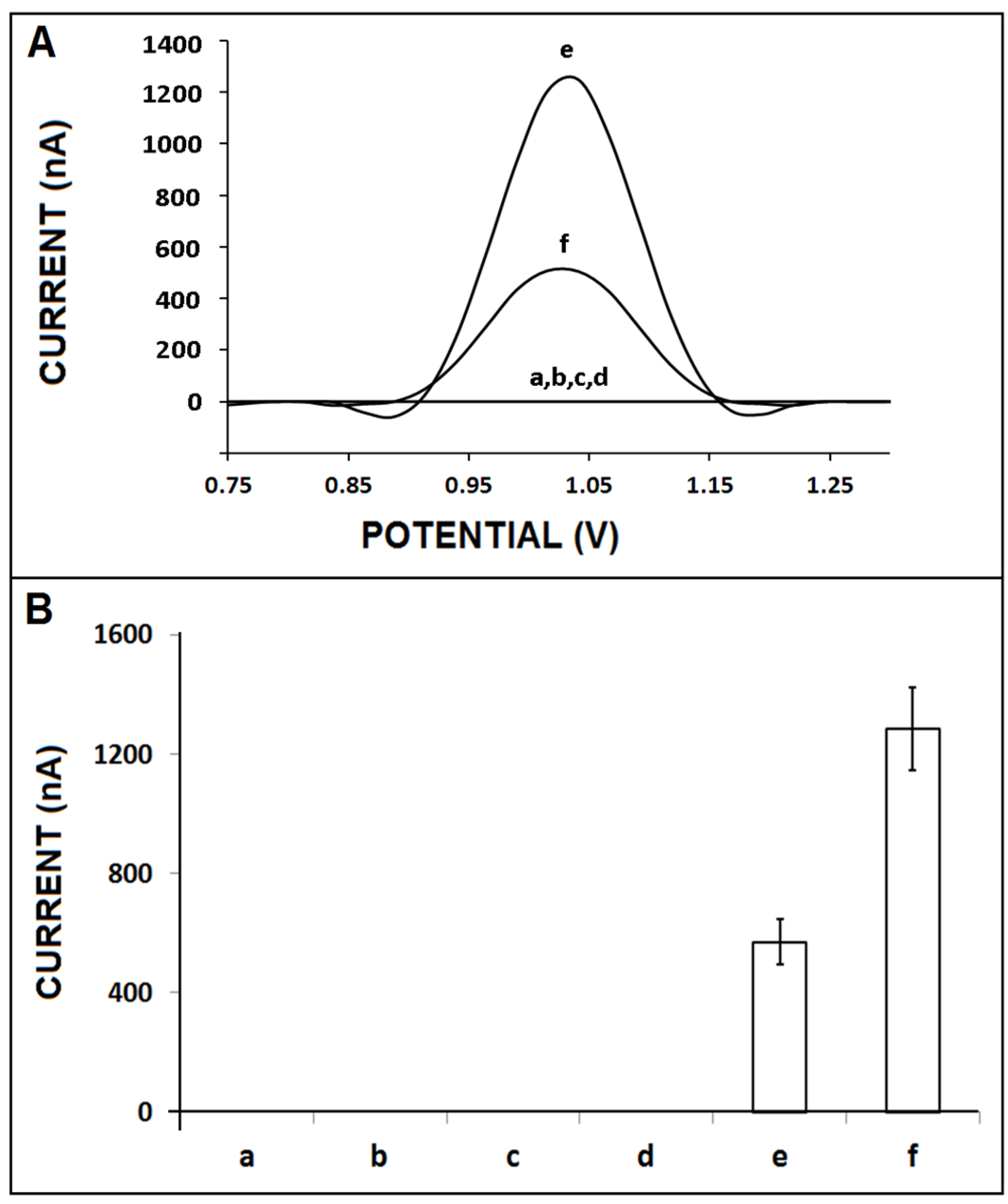
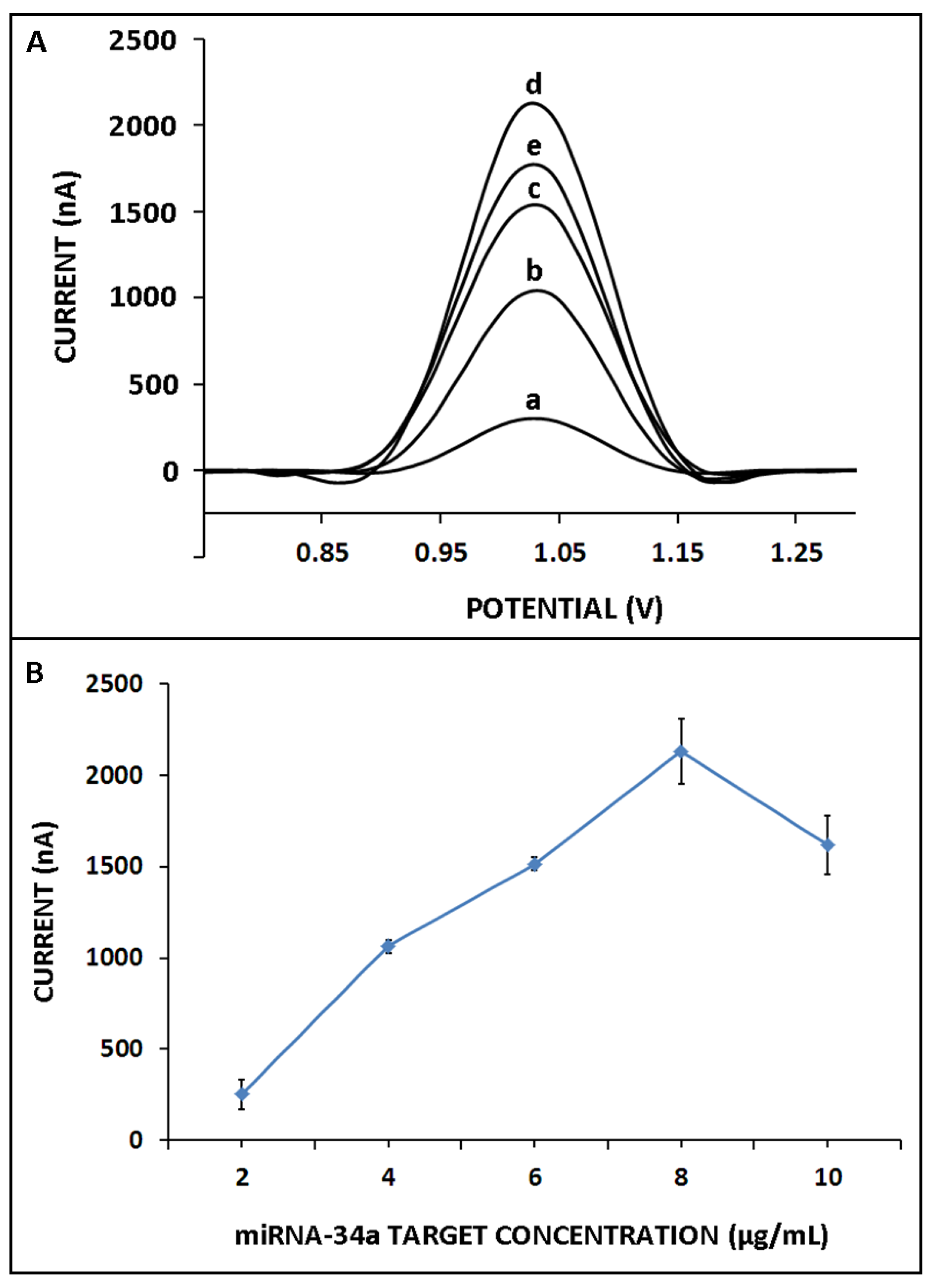
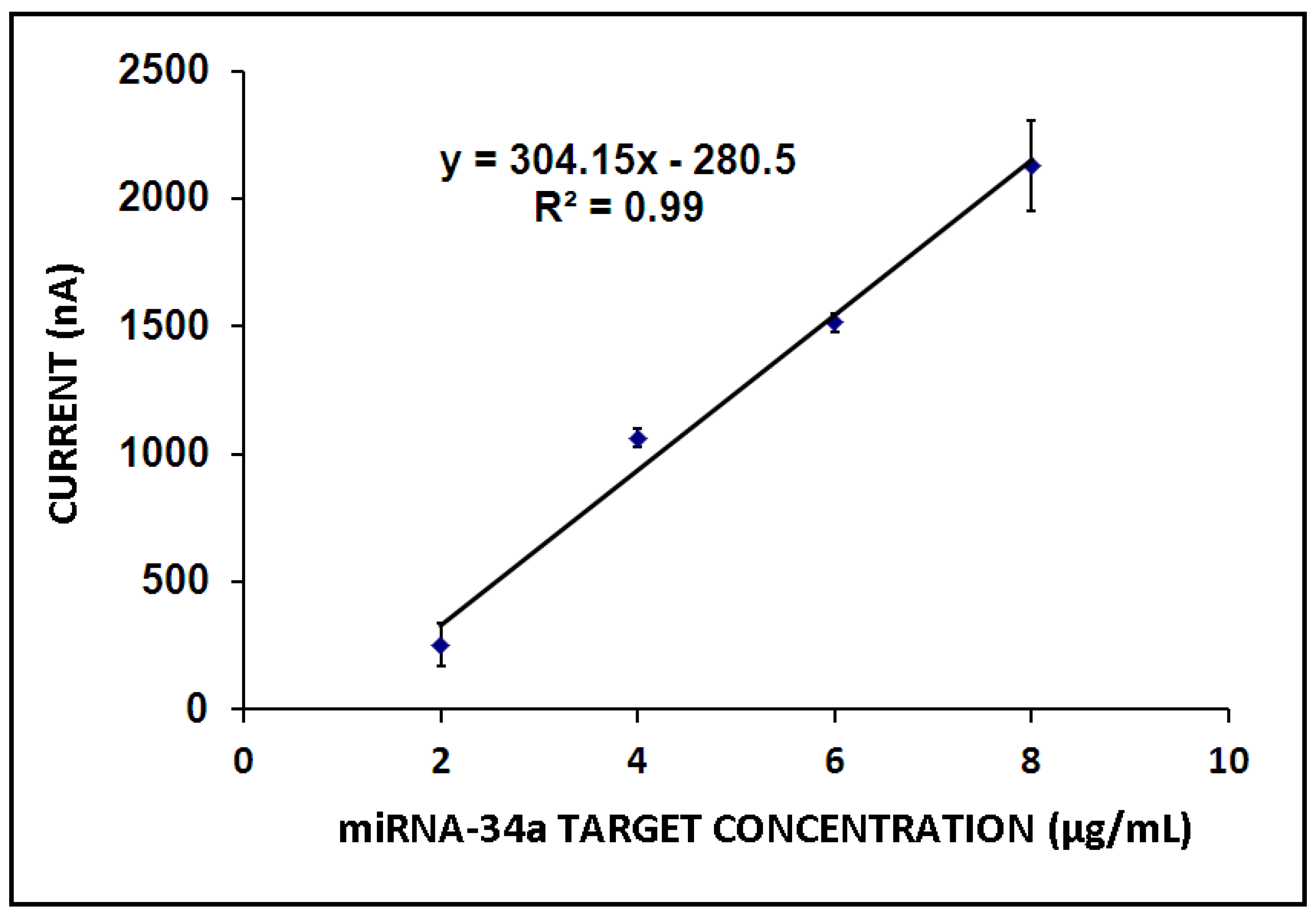
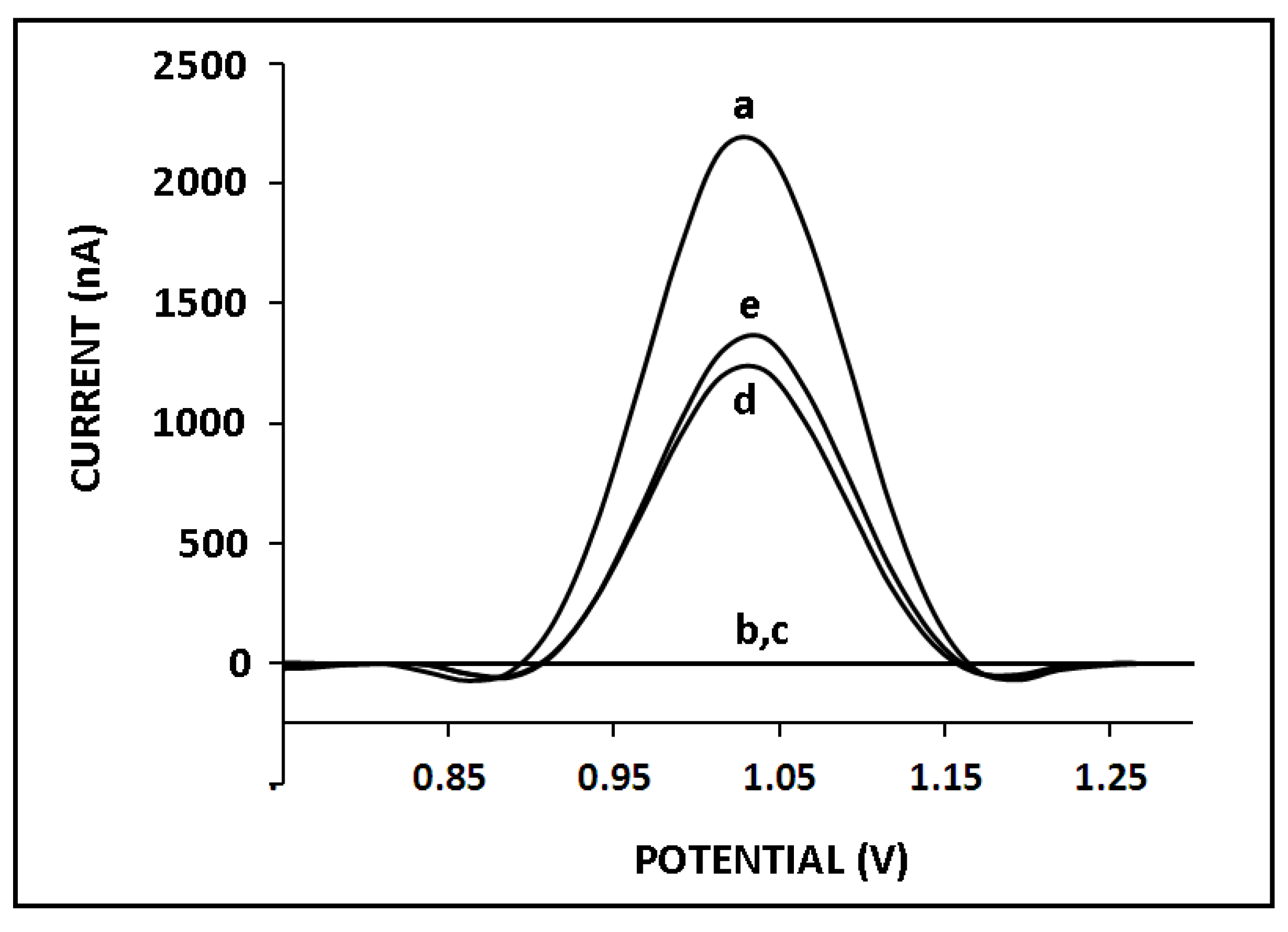
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Voirin, E.; Behr, J.P.; Kotera, M. Versatile synthesis of oligodeoxyribonucleotide—Oligospermine conjugates. Nat. Protoc. 2007, 2, 1360–1367. [Google Scholar] [CrossRef] [PubMed]
- Noir, R.; Kotera, M.; Pons, B.; Remy, J.S.; Behr, J.P. Oligonucleotide-oligospermine conjugates (zip nucleic acids): A convenient means of finely tuning hybridization temperatures. J. Am. Chem. Soc. 2008, 130, 13500–13505. [Google Scholar] [CrossRef] [PubMed]
- Pons, B.; Kotera, M.; Zuber, G.; Behr, J.P. Online synthesis of diblock cationic oligonucleotides for enhanced hybridization to their complementary sequence. ChemBioChem 2006, 7, 1173–1176. [Google Scholar] [CrossRef] [PubMed]
- Moreau, V.; Voirin, E.; Paris, C.; Kotera, M.; Nothisen, M.; Rémy, J.S.; Behr, J.P.; Erbacher, P.; Lenne-Samuel, N. Zip nucleic acids: New high affinity oligonucleotides as potent primers for PCR and reverse transcription. Nucleic Acids Res. 2009, 37, e130. [Google Scholar] [CrossRef]
- Gagnon, K.T.; Watts, J.K.; Pendergraff, H.M.; Montaillier, C.; Thai, D.; Potier, P.; Corey, D.R. Antisense and antigene inhibition of gene expression by cell-permeable oligonucleotide-oligospermine conjugates. J. Am. Chem. Soc. 2011, 133, 8404–8407. [Google Scholar] [CrossRef] [PubMed]
- Begheldo, M.; Ditengou, F.A.; Cimoli, G.; Trevisan, S.; Quaggiotti, S.; Nonis, A.; Palme, K.; Ruperti, B. Whole-mount in situ detection of microRNAs on Arabidopsis tissues using Zip Nucleic Acid probes. Anal. Biochem. 2013, 434, 60–66. [Google Scholar] [CrossRef]
- Erdem, A.; Eksin, E. Electrochemical Detection of Solution Phase Hybridization Related to Single Nucleotide Mutation by Carbon Nanofibers Enriched Electrodes. Materials 2019, 12, 3377. [Google Scholar] [CrossRef]
- Erdem, A.; Eksin, E. ZNA probe immobilized single-use electrodes for impedimetric detection of nucleic acid hybridization related to single nucleotide mutation. Anal. Chim. Acta 2019, 1071, 78–85. [Google Scholar] [CrossRef]
- Erdem, A.; Eksin, E. Zip nucleic acid based single-use biosensor for electrochemical detection of Factor V Leiden mutation. Sens. Actuators B Chem. 2019, 288, 634–640. [Google Scholar] [CrossRef]
- Erdem, A.; Eksin, E. Magnetic beads assay based on Zip nucleic acid for electrochemical detection of Factor V Leiden mutation. Int. J. Biol. Macromol. 2019, 125, 839–846. [Google Scholar] [CrossRef]
- Erdem, A.; Eksin, E. Impedimetric Sensing of Factor V Leiden Mutation by Zip Nucleic Acid Probe and Electrochemical Array. Biosensors 2020, 10, 116. [Google Scholar] [CrossRef] [PubMed]
- Xia, N.; Zhang, Y.; Wei, X.; Huang, Y.; Liu, L. An electrochemical microRNAs biosensor with the signal amplification of alkaline phosphatase and electrochemical-chemical-chemical redox cycling. Anal. Chim. Acta 2015, 878, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Jansson, M.D.; Lund, A.H. MicroRNA and cancer. Mol. Oncol. 2012, 6, 590–610. [Google Scholar] [CrossRef] [PubMed]
- Thum, T.; Gross, C.; Fiedler, J.; Fischer, T.; Kissler, S.; Bussen, M.; Galuppo, P.; Just, S.; Rottbauer, W.; Frantz, S.; et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 2008, 456, 980–984. [Google Scholar] [CrossRef] [PubMed]
- Laggerbauer, B.; Engelhardt, S. MicroRNAs as therapeutic targets in cardiovascular disease. J. Clin. Investig. 2022, 132, e159179. [Google Scholar] [CrossRef]
- Chen, J.W.; Dhahbi, J. Identification of four serum miRNAs as potential markers to screen for thirteen cancer types. PLoS ONE 2022, 17, e0269554. [Google Scholar] [CrossRef]
- Li, S.; Lei, Z.; Sun, T. The role of microRNAs in neurodegenerative diseases: A review. Cell Biol Toxicol. 2022, 20, 1–31. [Google Scholar] [CrossRef]
- El Aamri, M.; Yammouri, G.; Mohammadi, H.; Amine, A.; Korri-Youssoufi, H. Electrochemical Biosensors for Detection of MicroRNA as a Cancer Biomarker: Pros and Cons. Biosensors 2020, 10, 186. [Google Scholar] [CrossRef]
- Chen, Y.-X.; Huang, K.-J.; Niu, K.-X. Recent advances in signal amplification strategy based on oligonucleotide and nanomaterials for microRNA detection-a review. Biosens. Bioelectron. 2018, 90, 612–624. [Google Scholar] [CrossRef]
- Kim, H.Y.; Song, J.; Park, H.G.; Kang, T. Electrochemical detection of zeptomolar miRNA using an RNA-triggered Cu2+ reduction method. Sens. Actuators B 2022, 306, 131666. [Google Scholar] [CrossRef]
- Yuan, Y.-H.; Wu, Y.-D.; Chi, B.-Z.; Wen, S.-H.; Liang, R.-P.; Qiu, J.-D. Simultaneously electrochemical detection of microRNAs based on multifunctional magnetic nanoparticles probe coupling with hybridization chain reaction. Biosens. Bioelectron. 2017, 97, 325–331. [Google Scholar] [CrossRef]
- Mandli, J.; Mohammadi, H.; Amine, A. Electrochemical DNA sandwich biosensor based on enzyme amplified microRNA-21 detection and gold nanoparticles. Bioelectrochemistry 2017, 116, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Yarali, E.; Kanat, E.; Erac, Y.; Erdem, A. Ionic Liquid Modified Single-use Electrode Developed for Voltammetric Detection of miRNA-34a and its Application to Real Samples. Electroanalysis 2019, 32, 384–393. [Google Scholar] [CrossRef]
- Kalfert, D.; Ludvikova, M.; Pesta, M.; Ludvik, J.; Dostalova, L.; Kholova, I. Multifunctional Roles of miR-34a in Cancer: A Review with the Emphasis on Head and Neck Squamous Cell Carcinoma and Thyroid Cancer with Clinical Implications. Diagnostics 2020, 10, 563. [Google Scholar] [CrossRef]
- Chen, Y.-L.; Liu, X.-L.; Li, L. Prognostic value of low microRNA-34a expression in human gastrointestinal cancer: A systematic review and metaanalysis. BMC Cancer 2021, 21, 63. [Google Scholar] [CrossRef] [PubMed]
- Fawzy, M.S.; Ibrahiem, A.T.; Alsel, B.T.A.; Alghamdi, S.A.; Toraih, E.A. Analysis of microRNA-34a expression profile and rs2666433 variant in colorectal cancer: A pilot study. Sci. Rep. 2020, 10, 16940. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, Y.; Liu, R.; Kasinski, A.; Shen, H.; Slack, F.J.; Tang, D.G. MicroRNA-34a: Potent Tumor Suppressor, Cancer Stem Cell Inhibitor, and Potential Anticancer Therapeutic. Front. Cell Dev. Biol. 2021, 9, 640587. [Google Scholar] [CrossRef]
- Bazrgar, M.; Khodabakhsh, P.; Prudencio, M.; Mohagheghi, F.; Ahmadiani, A. The role of microRNA-34 family in Alzheimer’s disease: A potential molecular link between neurodegeneration and metabolic disorders. Pharmacol. Res. 2021, 172, 105805. [Google Scholar] [CrossRef]
- Zhang, H.-F.; Wang, Y.-C.; Han, Y.-D. MicroRNA-34a inhibits liver cancer cell growth by reprogramming glucose metabolism. Mol. Med. Rep. 2018, 17, 4483–4489. [Google Scholar] [CrossRef]
- Erdem, A.; Congur, G. Label-free voltammetric detection of MicroRNAs at multi-channel screen printed array of electrodes comparison to graphite sensors. Talanta 2014, 118, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Erdem, A.; Congur, G.; Eksin, E. Multi channel screen printed array of electrodes for enzyme-linked voltammetric detection of MicroRNAs. Sens. Actuators B Chem. 2013, 188, 1089–1095. [Google Scholar] [CrossRef]
- Wang, J.; Xu, D.; Erdem, A.; Polsky, R.; Salazar, M.A. Genomagnetic electrochemical assays of DNA hybridization. Talanta 2002, 56, 931–938. [Google Scholar] [CrossRef]
- Wang, J.; Kawde, A.N.; Erdem, A.; Salazar, M. Magnetic bead-based label-free electrochemical detection of DNA hybridization. Analyst 2001, 126, 2020–2024. [Google Scholar] [CrossRef]
- Erdem, A.; Ariksoysal, D.O.; Karadeniz, H.; Kara, P.; Sengonul, A.; Sayiner, A.A.; Ozsoz, M. Electrochemical genomagnetic assay for the detection of hepatitis B virus DNA in polymerase chain reaction amplicons by using disposable sensor technology. Electrochem. Commun. 2005, 7, 815–820. [Google Scholar] [CrossRef]
- Erdem, A.; Congur, G. Voltammetric aptasensor combined with magnetic beads assay developed for detection of human activated protein C. Talanta 2014, 128, 428–433. [Google Scholar] [CrossRef] [PubMed]
- Erdem, A.; Duruksu, G.; Congur, G.; Karaoz, E. Genomagnetic assay for electrochemical detection of osteogenic differentiation in mesenchymal stem cells. Analyst 2013, 138, 5424–5430. [Google Scholar] [CrossRef]
- Congur, G.; Plucnara, M.; Erdem, A.; Fojta, M. Detection of p53 Gene by Using Genomagnetic Assay Combined with Carbon Nanotube Modified Disposable Sensor Technology. Electroanalysis 2015, 27, 1579–1586. [Google Scholar] [CrossRef]
- Erdem, A.; Pividori, M.I.; Lermo, A.; Bonanni, A.; Del Valle, M.; Alegret, S. Genomagnetic assay based on label-free electrochemical detection using magneto-composite electrodes. Sens. Actuators B Chem. 2006, 114, 591–598. [Google Scholar] [CrossRef]
- Lusi, E.A.; Passamano, M.; Guarascio, P.; Scarpa, A.; Schiavo, L. Innovative electrochemical approach for an early detection of microRNAs. Anal. Chem. 2009, 81, 2819–2822. [Google Scholar] [CrossRef]
- Belluzo, M.S.; Ribone, M.É.; Lagier, C.M. Assembling amperometric biosensors for clinical diagnostics. Sensors 2008, 8, 1366. [Google Scholar] [CrossRef]
- Lucarelli, F.; Tombelli, S.; Minunni, M.; Marrazza, G.; Mascini, M. Electrochemical and piezoelectric DNA biosensors for hybridisation detection. Anal. Chim. Acta 2008, 609, 139–159. [Google Scholar] [CrossRef] [PubMed]
- Isin, D.; Eksin, E.; Erdem, A. Graphene oxide modified single-use electrodes and their application for voltammetric miRNA analysis. Mater. Sci. Eng. C 2017, 75, 1242–1249. [Google Scholar] [CrossRef] [PubMed]
- Söderlund, H. DNA hybridization: Comparison of liquid and solid phase formats. Ann. Biol. Clin. 1990, 48, 489–491. [Google Scholar]
- Liu, W.T.; Guo, H.; Wu, J.H. Effects of target length on the hybridization efficiency and specificity of rRNA-based oligonucleotide microarrays. Appl. Environ. Microbiol. 2007, 73, 73–82. [Google Scholar] [CrossRef]
- Cederquist, K.B.; Keatinh, C.D. Hybridization Efficiency of Molecular Beacons Bound to Gold Nanowires: Effect of Surface Coverage and Target Length. Langmuir 2010, 26, 18273–18280. [Google Scholar] [CrossRef]
- Miller, J.N.; Miller, J.C. Statistics and Chemometrics for Analytical Chemistry, 6th ed.; Pearson Education: Essex, UK, 2010; ISBN 978-0-273-73042-2. [Google Scholar]
- Erdem, A.; Eksin, E.; Kadikoylu, G.; Yildiz, E. Voltammetric Detection of miRNA Hybridization Based on Electroactive Indicator-Cobalt Phenanthroline. Int. J. Biol. Macromol. 2020, 158, 819–825. [Google Scholar] [CrossRef]
- Congur, G.; Erdem, A. PAMAM Dendrimer Modified Screen Printed Electrodes for Impedimetric Detection of miRNA-34a. Microchem. J. 2019, 148, 748–758. [Google Scholar] [CrossRef]
- Congur, G.; Eksin, E.; Erdem, A. Impedimetric detection of miRNA-34a using graphene oxide modifiedchemically activated graphite electrodes. Sens. Act. A 2018, 279, 493–500. [Google Scholar] [CrossRef]
- Erdem, A.; Eksin, E.; Isin, D.; Polat, D. Graphene oxide modified chemically activated graphite electrodes for detection of microRNA. Electroanalysis 2017, 2, 1350–1358. [Google Scholar] [CrossRef]
- Kesici, E.; Eksin, E.; Erdem, A. An Impedimetric biosensor based on ionic liquidmodified graphite electrodes developed for microRNA-34a detection. Sensors 2018, 18, 2868. [Google Scholar] [CrossRef]
- Mandli, J.; Amine, A. Impedimetric genosensor for miRNA-34a detection in cell lysates using polypyrrole. J. Solid. State Electrochem. 2018, 22, 1007–1014. [Google Scholar] [CrossRef]
- Bettazzi, F.; Hamid-Asl, E.; Esposito, C.L.; Quintavalle, C.; Formisano, N.; Laschi, S.; Catuogno, S.; Iaboni, M.; Marrazza, G.; Mascini, M.; et al. Electrochemical detection of miRNA-222 by use of a magnetic bead-based bioassay. Anal. Bioanal. Chem. 2013, 405, 1025–1034. [Google Scholar] [CrossRef] [PubMed]
- Kilic, T.; Topkaya, S.N.; Ozkan Ariksoysal, D.; Ozsoz, M.; Ballar, P.; Erac, Y.; Gozen, O. Electrochemical based detection of microRNA, mir21 in breast cancer cells. Biosens. Bioelectron. 2012, 38, 195–201. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Erdem, A.; Eksin, E. Zip Nucleic Acid-Based Genomagnetic Assay for Electrochemical Detection of microRNA-34a. Biosensors 2023, 13, 144. https://doi.org/10.3390/bios13010144
Erdem A, Eksin E. Zip Nucleic Acid-Based Genomagnetic Assay for Electrochemical Detection of microRNA-34a. Biosensors. 2023; 13(1):144. https://doi.org/10.3390/bios13010144
Chicago/Turabian StyleErdem, Arzum, and Ece Eksin. 2023. "Zip Nucleic Acid-Based Genomagnetic Assay for Electrochemical Detection of microRNA-34a" Biosensors 13, no. 1: 144. https://doi.org/10.3390/bios13010144
APA StyleErdem, A., & Eksin, E. (2023). Zip Nucleic Acid-Based Genomagnetic Assay for Electrochemical Detection of microRNA-34a. Biosensors, 13(1), 144. https://doi.org/10.3390/bios13010144