A Lab-Made E-Nose-MOS Device for Assessing the Bacterial Growth in a Solid Culture Medium
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Inoculum Preparation
2.2. Growth Conditions
2.3. HS-SPME-GC-MS Evaluation of VOCs Emitted by Each Bacteria
2.4. E-Nose Setup
2.4.1. Apparatus
2.4.2. Sampling and Analysis
2.4.3. Data Acquisition, Feature Extraction and Signal Treatment
2.5. Statistical Analysis
3. Results and Discussion
3.1. Evaluation of VOCs Emitted during the Bacterial Growth by HS-SPME-GC-MS
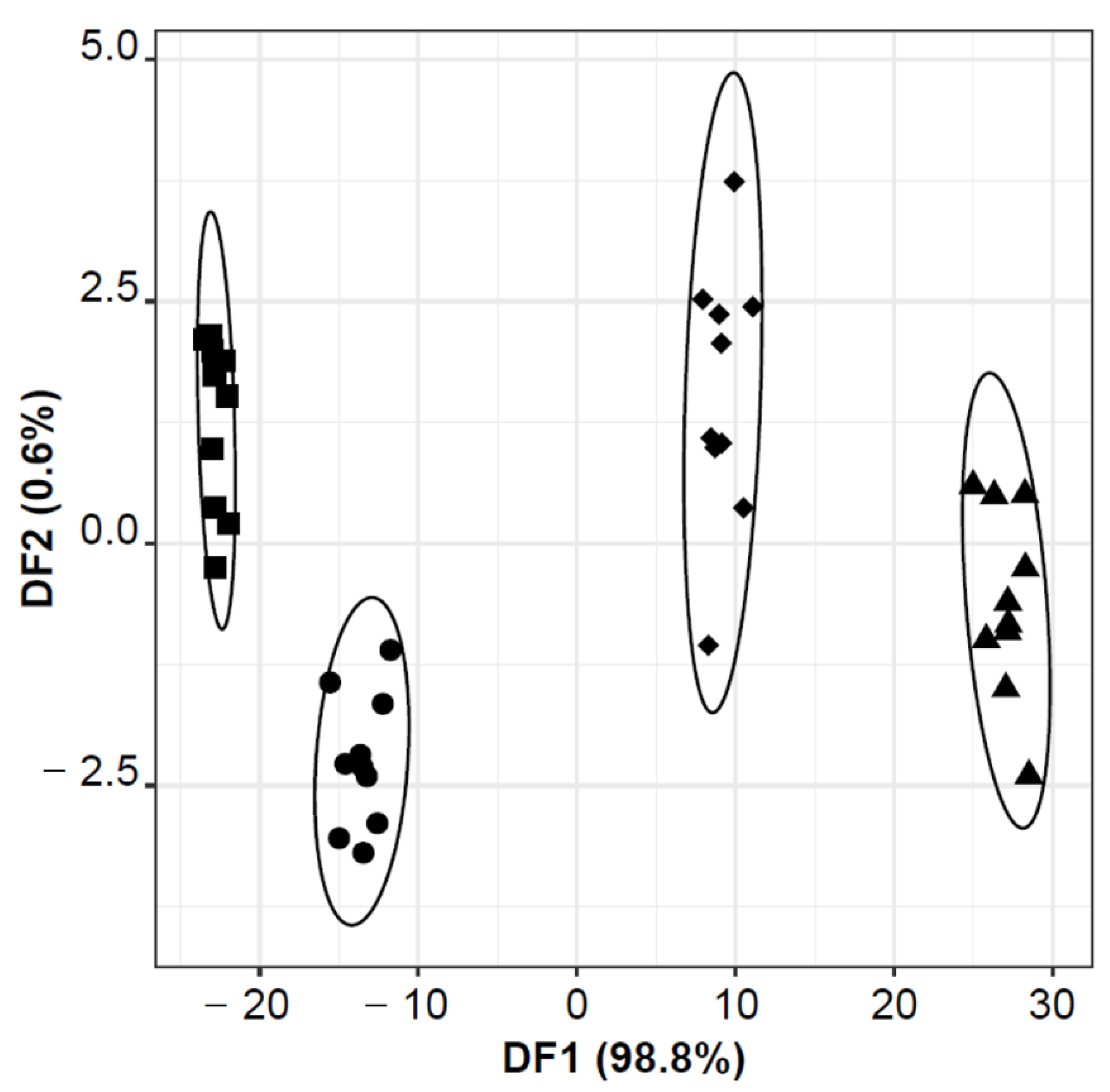
3.2. Bacterial Species Discrimination Using the E-Nose-MOS Lab-Made Device
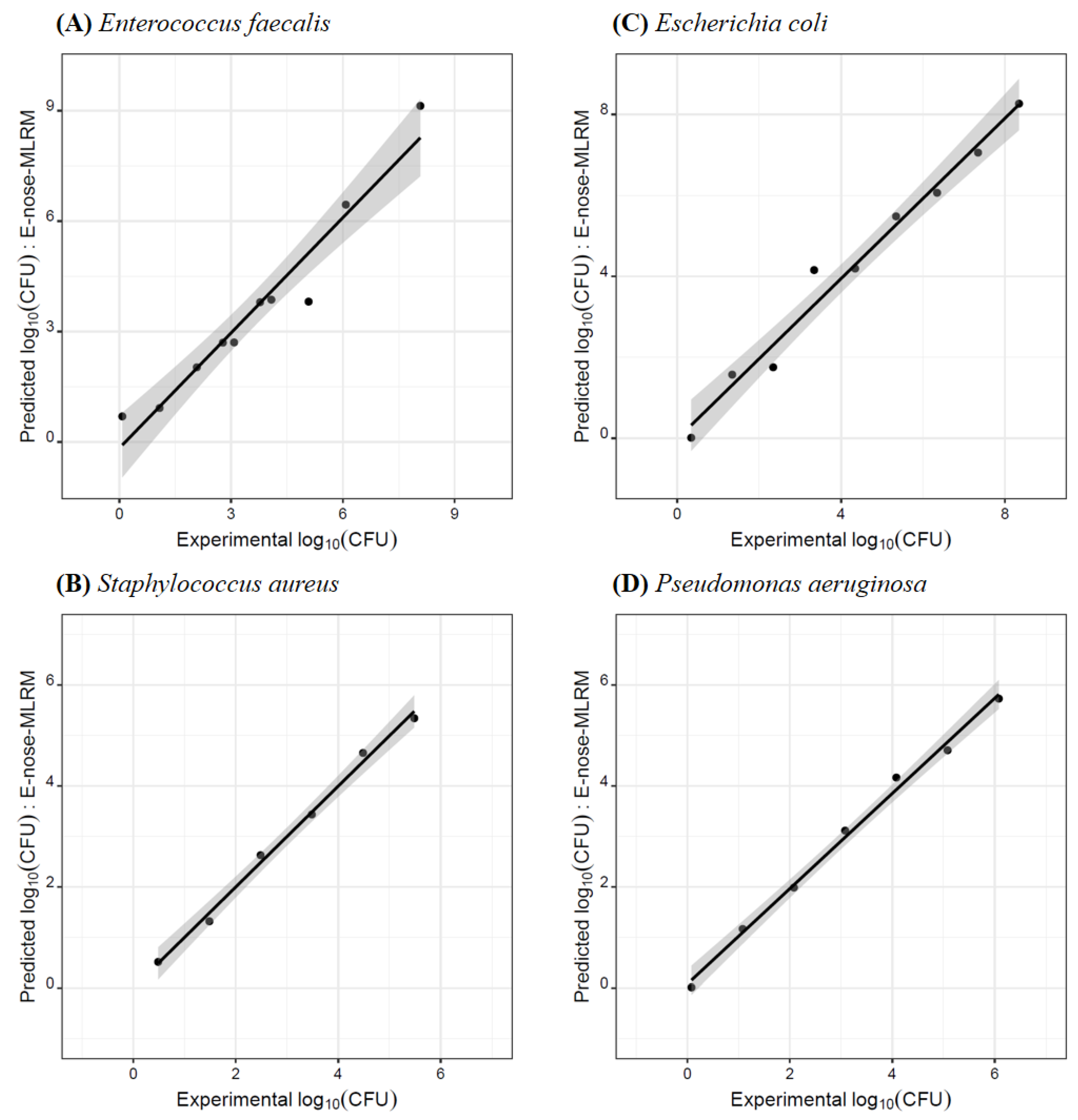
3.3. Quantification of Bacteria CFUs Using the E-Nose-MOS Lab-Made Device
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Green, G.C.; Chan, A.D.C.; Lin, M. Robust identification of bacteria based on repeated odor measurements from individual bacteria colonies. Sens. Actuators B Chem. 2014, 190, 16–24. [Google Scholar] [CrossRef]
- Bonah, E.; Huang, X.; Yi, R.; Aheto, J.H.; Osae, R.; Golly, M. Electronic nose classification and differentiation of bacterial foodborne pathogens based on support vector machine optimized with particle swarm optimization algorithm. J. Food Process Eng. 2019, 42, e13236. [Google Scholar] [CrossRef]
- Capuano, R.; Paba, E.; Mansi, A.; Marcelloni, A.M.; Chiominto, A.; Proietto, A.R.; Zampetti, E.; Macagnano, A.; Lvova, L.; Catini, A.; et al. Aspergillus species discrimination using a gas sensor array. Sensors 2020, 20, 4004. [Google Scholar] [CrossRef]
- Nieto-Arribas, P.; Seseña, S.; Poveda, J.M.; Chicón, R.; Cabezas, L.; Palop, L. Enterococcus populations in artisanal Manchego cheese: Biodiversity, technological and safety aspects. Food Microbiol. 2011, 28, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Thorn, R.M.S.; Reynolds, D.M.; Greenman, J. Multivariate analysis of bacterial volatile compound profiles for discrimination between selected species and strains in vitro. J. Microbiol. Meth. 2011, 84, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Bos, L.D.J.; Sterk, P.J.; Schultz, M.J. Volatile metabolites of pathogens: A systematic review. PLoS Pathog. 2013, 9, e1003311. [Google Scholar] [CrossRef] [Green Version]
- Boots, A.W.; Smolinska, A.; Van Berkel, J.J.B.N.; Fijten, R.R.R.; Stobberingh, E.E.; Boumans, M.L.L.; Moonen, E.J.; Wouters, E.F.M.; Dallinga, J.W.; Van Schooten, F.J. Identification of microorganisms based on headspace analysis of volatile organic compounds by gas chromatography-mass spectrometry. J. Breath Res. 2014, 8, 027106. [Google Scholar] [CrossRef]
- Zscheppank, C.; Wiegand, H.L.; Lenzen, C.; Wingender, J.; Telgheder, U. Investigation of volatile metabolites during growth of Escherichia coli and Pseudomonas aeruginosa by needle trap-GC-MS. Anal. Bioanal. Chem. 2014, 406, 6617–6628. [Google Scholar] [CrossRef]
- Tait, E.; Perry, J.D.; Stanforth, S.P.; Dean, J.R. Identification of volatile organic compounds produced by bacteria using HS-SPME-GC-MS. J. Chromatogr. Sci. 2014, 52, 363–373. [Google Scholar] [CrossRef] [Green Version]
- Bonah, E.; Huang, X.; Aheto, J.H.; Osae, R. Application of electronic nose as a non-invasive technique for odor fingerprinting and detection of bacterial foodborne pathogens: A review. J. Food Sci. Technol. 2020, 57, 1977–1990. [Google Scholar] [CrossRef]
- Kladsomboon, S.; Thippakorn, C.; Seesaard, T. Development of organic-inorganic hybrid optical gas sensors for the non-invasive monitoring of pathogenic bacteria. Sensors 2018, 18, 3189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, C.; Lu, A. A deep learning method combined with electronic nose to identify the rice origin. J. Instrum. 2022, 17, P08016. [Google Scholar] [CrossRef]
- Lin, H.; Chen, H.; Yin, C.; Zhang, Q.; Li, Z.; Shi, Y.; Men, H. Lightweight Residual Convolutional Neural Network for Soybean Classification Combined with Electronic Nose. IEEE Sens. J. 2022, 22, 11463–11473. [Google Scholar] [CrossRef]
- Lelono, D.; Triyana, K.; Hartati, S.; Istiyanto, J.E. Classification of Indonesia black teas based on quality by using electronic nose and principal component analysis. AIP Conf. Proc. 2016, 1755, 020003. [Google Scholar]
- Astantri, P.F.; Prakoso, W.S.A.; Triyana, K.; Untari, T.; Airin, C.M.; Astudi, P. Lab-Made Electronic Nose for Fast Detection of Listeria monocytogenes and Bacillus cereus. Vet. Sci. 2020, 7, 20. [Google Scholar] [CrossRef] [Green Version]
- Prakoso, W.S.A.; Astantri, P.F.; Triyana, K.; Untari, T.; Airin, C.M.; Astudi, P. Rapid Detection of Escherichia coli and Salmonella Typhimurium Using Lab-Made Electronic Nose Coupled with Chemometric Tools. Int. J. Adv. Sci. Eng. Inf. Technol. 2021, 11, 1494–1500. [Google Scholar] [CrossRef]
- Roda, B.; Mirasoli, M.; Zattoni, A.; Casale, M.; Oliveri, P.; Bigi, A.; Reschiglian, P.; Simoni, P.; Roda, A. A new analytical platform based on field-flow fractionation and olfactory sensor to improve the detection of viable and non-viable bacteria in food. Anal. Bioanal. Chem. 2016, 408, 7367–7377. [Google Scholar] [CrossRef]
- Carrillo, J.; Durán, C. Fast identification of Bacteria for Quality Control of Drinking Water through A Static Headspace Sampler Coupled to a Sensory Perception System. Biosensors 2019, 9, 23. [Google Scholar] [CrossRef] [Green Version]
- Dutta, R.; Das, A.; Stocks, N.G.; Morgan, D. Stochastic resonance-based electronic nose: A novel way to classify bacteria. Sens. Actuators B Chem. 2006, 115, 17–27. [Google Scholar] [CrossRef]
- Canhoto, O.; Magan, N. Electronic nose technology for the detection of microbial and chemical contamination of potable water. Sens. Actuators B Chem. 2005, 106, 3–6. [Google Scholar] [CrossRef]
- Seesaard, T.; Thippakorn, C.; Kerdcharoenc, T.; Kladsomboon, S. A hybrid electronic nose system for discrimination of pathogenic bacterial volatile compounds. Anal. Methods 2020, 12, 5671–5683. [Google Scholar] [CrossRef] [PubMed]
- Shauloff, N.; Morag, A.; Yaniv, K.; Singh, S.; Malishev, R.; Paz-Tal, O.; Rokach, L.; Jelinek, R. Sniffing Bacteria with a Carbon-Dot Artificial Nose. Nano-Micro Lett. 2021, 13, 112. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Gómez, J.K.; Acevedo, C.M.D.; García-Rico, R.O. Detection of the bacteria concentration level in pasteurized milk by using two different artificial multisensory methods. Sens. Bio-Sens. Res. 2021, 33, 100428. [Google Scholar] [CrossRef]
- Bordbar, M.M.; Tashkhourian, J.; Tavassoli, A.; Bahramali, E.; Hemmateenej, B. Ultrafast detection of infectious bacteria using optoelectronic nose based on metallic nanoparticles. Sens. Actuators B Chem. 2020, 319, 128262. [Google Scholar] [CrossRef]
- Shumeiko, V.; Zaken, Y.; Hidas, G.; Paltiel, Y.; Bisker, G.; Shoseyov, O. Peptide-Encapsulated Single-Wall Carbon Nanotube-Based Near-Infrared Optical Nose for Bacteria Detection and Classification. IEEE Sens. J. 2022, 22, 6277–6287. [Google Scholar] [CrossRef]
- Carrillo-Gómez, J.; Durán-Acevedo, C.; García-Rico, R. Concentration detection of the E. coli bacteria in drinking water treatment plants through an E-nose and a volatiles extraction system (VES). Water 2019, 11, 774. [Google Scholar] [CrossRef] [Green Version]
- Tonezzer, M.; Thai, N.X.; Gasperi, F.; Duy, N.V.; Biasioli, F. Quantitative Assessment of Trout Fish Spoilage with a Single Nanowire Gas Sensor in a Thermal Gradient. Nanomaterials 2021, 11, 1604. [Google Scholar] [CrossRef]
- Barbri, N.E.; Mirhisse, J.; Ionescu, R.; Bari, N.E.; Correig, X.; Bouchikhi, B.; Llobet, E. An electronic nose system based on a micro-machined gas sensor array to assess the freshness of sardines. Sens. Actuators B Chem. 2009, 141, 538–543. [Google Scholar] [CrossRef]
- Ghrissi, H.; Veloso, A.C.A.; Marx, Í.M.G.; Dias, T.; Peres, A.M. A potentiometric electronic tongue as a discrimination tool of water-food indicator/contamination bacteria. Chemosensors 2021, 9, 143. [Google Scholar] [CrossRef]
- Smith, A.C.; Hussey, M.A. Gram Stain Protocols. Am. Soc. Microbiol. 2016, 9, 14. [Google Scholar]
- Marx, Í.M.G.; Casal, S.; Rodrigues, N.; Cruz, R.; Peres, F.; Veloso, A.C.A.; Pereira, J.A.; Peres, A.M. Impact of fresh olive leaves addition during the extraction of Arbequina virgin olive oils on the phenolic and volatile profiles. Food Chem. 2022, 393, 133327. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, G.G.; Dias, L.G.; Rodrigues, N.; Marx, Í.M.G.; Veloso, A.C.A.; Pereira, J.A.; Peres, A.M. Application of a lab-made electronic nose for extra virgin olive oils commercial classification according to the perceived fruitiness intensity. Talanta 2021, 226, 122122. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, G.G.; Peres, A.M.; Estevinho, L.; Geraldes, P.; Garcia-Cabezon, C.; Martin-Pedrosa, F.; Rodriguez-Mencez, M.L.; Dias, L.G. Enose lab made with vacuum sampling: Quantitative applications. Chemosensors 2022, 10, 261. [Google Scholar] [CrossRef]
- Gila, D.M.M.; García, J.G.; Bellincontro, A.; Mencarelli, F.; Ortega, J.G. Fast tool based on electronic nose to predict olive fruit quality after harvest. Postharvest Biol. Technol. 2020, 160, 111058. [Google Scholar] [CrossRef]
- Beleites, C.; Salzer, R. Assessing and improving the stability of chemometric models in small sample size situations. Anal. Bioanal. Chem. 2008, 390, 1261–1271. [Google Scholar] [CrossRef] [PubMed]
- Poveda, J. Beneficial effects of microbial volatile organic compounds (MVOCs) in plants. Appl. Soil Ecol. 2021, 168, 104118. [Google Scholar] [CrossRef]
| Volatile Compounds | Gram-Negative Bacteria | Gram-Positive Bacteria | ||
|---|---|---|---|---|
| E. coli | P. aeruginosa | E. faecalis | S. aureus | |
| Alcohols | ||||
| 1-Butanol | 33.82 | nd | 51.68 | nd |
| 1-Nonanol | 5.42 | nd | nd | nd |
| 1-Pentanol | 53.96 | 95.53 | 869.56 | 51.09 |
| 3-Chloro-2-methyl-2-pentanol | nd | nd | nd | 11.11 |
| Ethanol | nd | nd | 695.93 | nd |
| Phenylethyl alcohol | 15.46 | nd | 23.30 | nd |
| Aldehydes | ||||
| Phenol | 6.19 | nd | 6.87 | nd |
| Alkanes | ||||
| Isocetane | 5.11 | 5.21 | 5.55 | 5.41 |
| Alkenes | ||||
| (E)-1,4-Undecadiene | nd | 70.09 | nd | nd |
| 1-Undecene | nd | 32.00 | 23.71 | 43.71 |
| Carboxylic acids | ||||
| 2-Methylbutanoic acid | nd | nd | 13.63 | nd |
| Acetic acid | nd | nd | 29.40 | nd |
| Isovaleric acid | nd | 43.40 | 37.17 | nd |
| Undecane | 9.89 | 11.45 | 16.63 | 7.22 |
| Esters | ||||
| Methyl valerate | nd | 70.17 | nd | 114.72 |
| Ketones | ||||
| 2-Tridecanone | 7.92 | nd | nd | nd |
| Phellandrenes | ||||
| α-Phellandrene | nd | nd | nd | 6.81 |
| Pyrazines | ||||
| 2,5-Dimethylpyrazine | 38.62 | 46.01 | 48.88 | 46.87 |
| 2-Ethyl-6-methyl-pyrazine | 0.52 | nd | 0.79 | nd |
| 3-Ethyl-2,5-dimethylpyrazine | 1.11 | nd | nd | nd |
| Trimethylpyrazine | nd | nd | nd | 0.59 |
| Terpenes | ||||
| Camphene | nd | nd | nd | 1.50 |
| D-Limonene | 0.74 | nd | nd | nd |
| β-Pinene | 2.56 | 4.78 | 4.71 | 3.08 |
| Others | ||||
| 2,4-Thujadiene | 0.52 | nd | 1.47 | 0.59 |
| E-7-Dodecen-1-ol acetate | nd | nd | 11.67 | nd |
| Indole | nd | 2.97 | nd | 32.75 |
| Methyl undecyl ether | nd | nd | 19.82 | nd |
| Actual Bacterium | Predicted Bacterium | Total | Sensitivity | ||||
|---|---|---|---|---|---|---|---|
| Gram-Negative | Gram-Positive | ||||||
| E. coli | P. aeruginosa | E. faecalis | S. aureus | ||||
| Gram-negative | E. coli | 9 | 0 | 0 | 1 | 10 | 90% |
| P. aeruginosa | 1 | 8 | 0 | 1 | 10 | 80% | |
| Gram-positive | E. faecalis | 0 | 1 | 9 | 0 | 10 | 90% |
| S. aureus | 0 | 0 | 0 | 10 | 10 | 100% | |
| Total | 10 | 9 | 9 | 12 | 40 | 90% | |
| Specificity | 90% | 89% | 100% | 83% | 91% | --- | |
| Microorganism | Concentration Range (log10(CFU)) a | Selected Extracted Feature Parameters b | Goodness of Fitting Parameters c | ||
|---|---|---|---|---|---|
| R2 | RMSE (log10(CFU)) | ||||
| Gram-negative | E. coli | [0.342, 8.342] | S4_LP; S6_SUM; S9_MEAN | 0.978 | 0.436 |
| P. aeruginosa | [0.079, 6.079] | S8_MAX; S9_MAX; S9_SD | 0.995 | 0.174 | |
| Gram-positive | E. faecalis | [0.079, 8.079] | S9_INT; S9_MIN; S8_MEAN; S5_SD | 0.943 | 0.602 |
| S. aureus | [0.484, 5.484] | S5_INT; S4_SUM; S4_MEAN | 0.994 | 0.158 | |
| Microorganism | R2 | Slope | Slope CI | Intercept (log10(CFU)) | Intercept CI (log10(CFU)) | |
|---|---|---|---|---|---|---|
| Gram-negative | E. coli | 0.978 | 0.990 | [0.857, 1.124] | −0.016 | [−0.690, 0.658] |
| P. aeruginosa | 0.995 | 0.943 | [0.863, 1.022] | 0.082 | [−0.211, 0.375] | |
| Gram-positive | E. faecalis | 0.943 | 1.044 | [0.834, 1.254] | 0.167 | [−1.060, 0.726] |
| S. aureus | 0.994 | 0.998 | [0.893, 1.103] | 0.004 | [−0.358, 0.366] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dias, T.; Santos, V.S.; Zorgani, T.; Ferreiro, N.; Rodrigues, A.I.; Zaghdoudi, K.; Veloso, A.C.A.; Peres, A.M. A Lab-Made E-Nose-MOS Device for Assessing the Bacterial Growth in a Solid Culture Medium. Biosensors 2023, 13, 19. https://doi.org/10.3390/bios13010019
Dias T, Santos VS, Zorgani T, Ferreiro N, Rodrigues AI, Zaghdoudi K, Veloso ACA, Peres AM. A Lab-Made E-Nose-MOS Device for Assessing the Bacterial Growth in a Solid Culture Medium. Biosensors. 2023; 13(1):19. https://doi.org/10.3390/bios13010019
Chicago/Turabian StyleDias, Teresa, Vítor S. Santos, Tarek Zorgani, Nuno Ferreiro, Ana I. Rodrigues, Khalil Zaghdoudi, Ana C. A. Veloso, and António M. Peres. 2023. "A Lab-Made E-Nose-MOS Device for Assessing the Bacterial Growth in a Solid Culture Medium" Biosensors 13, no. 1: 19. https://doi.org/10.3390/bios13010019
APA StyleDias, T., Santos, V. S., Zorgani, T., Ferreiro, N., Rodrigues, A. I., Zaghdoudi, K., Veloso, A. C. A., & Peres, A. M. (2023). A Lab-Made E-Nose-MOS Device for Assessing the Bacterial Growth in a Solid Culture Medium. Biosensors, 13(1), 19. https://doi.org/10.3390/bios13010019






