Surface-Enhanced Raman Scattering Active Core-Shell Ag NPs@Carbon Dots with Enzyme-Mimicking Activities for Label-Free Measurement Cholesterol
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemical Reagents
2.2. Instruments
2.3. Preparation of N, S-Doped Carbon Dot (DCD)
2.4. Details the Preparation of Ag NPs and Ag NPs@DCD
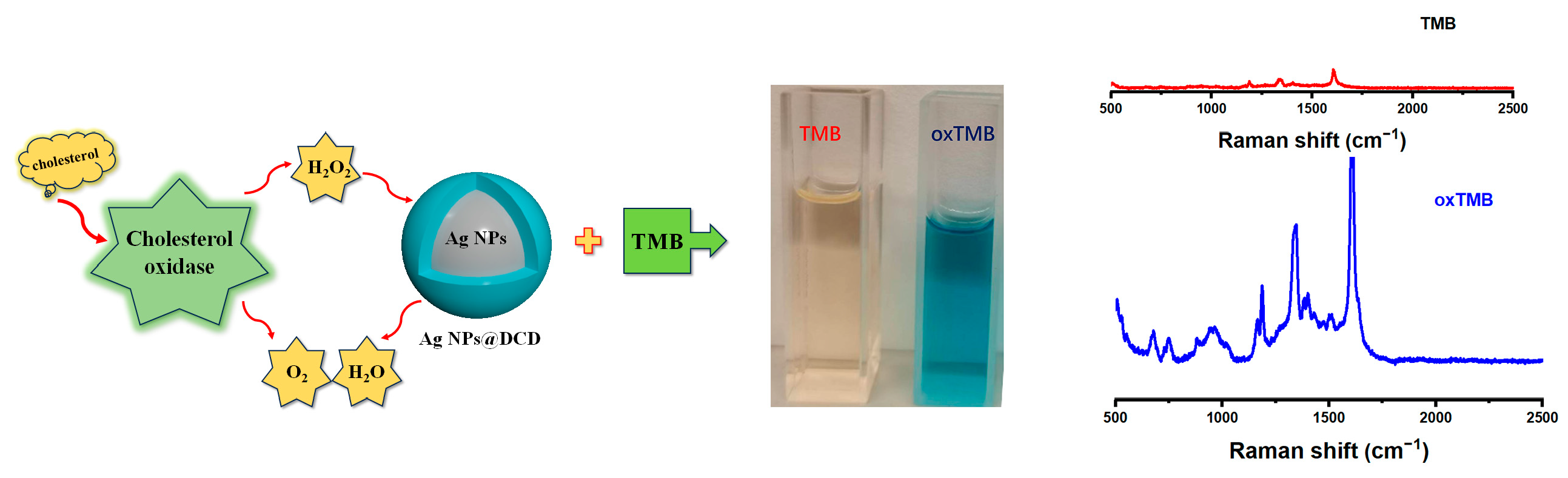
2.5. Evaluation of Enzyme-Activity of AgNPs@DCD Nanocomposites
2.6. Examination of SERS Activity of Prepared Ag NPs@DCD as SERS Substrate
2.7. SERS Detection of Cholesterol Utilizing TMB-Ag NPs@DCD System
2.8. Testing of Serum Samples
3. Result and Discussion
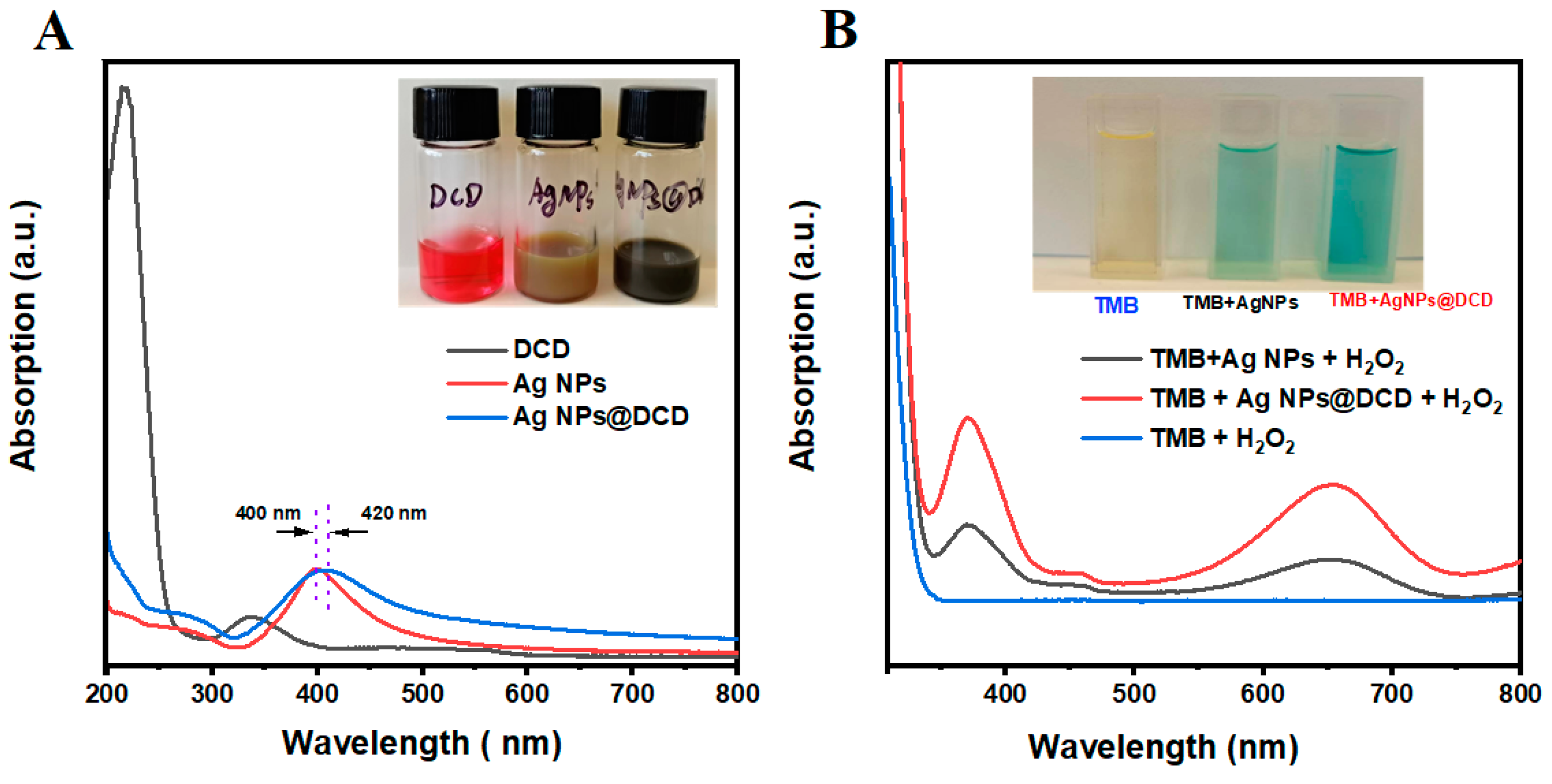
3.1. Characterization of DCD, Ag NPs and Ag NPs@DCD
3.2. Examination of Raman Performance of Ag NPs and Ag NPs@DCD
3.3. Enzyme-Mimicking Activities of Ag NPs@DCD
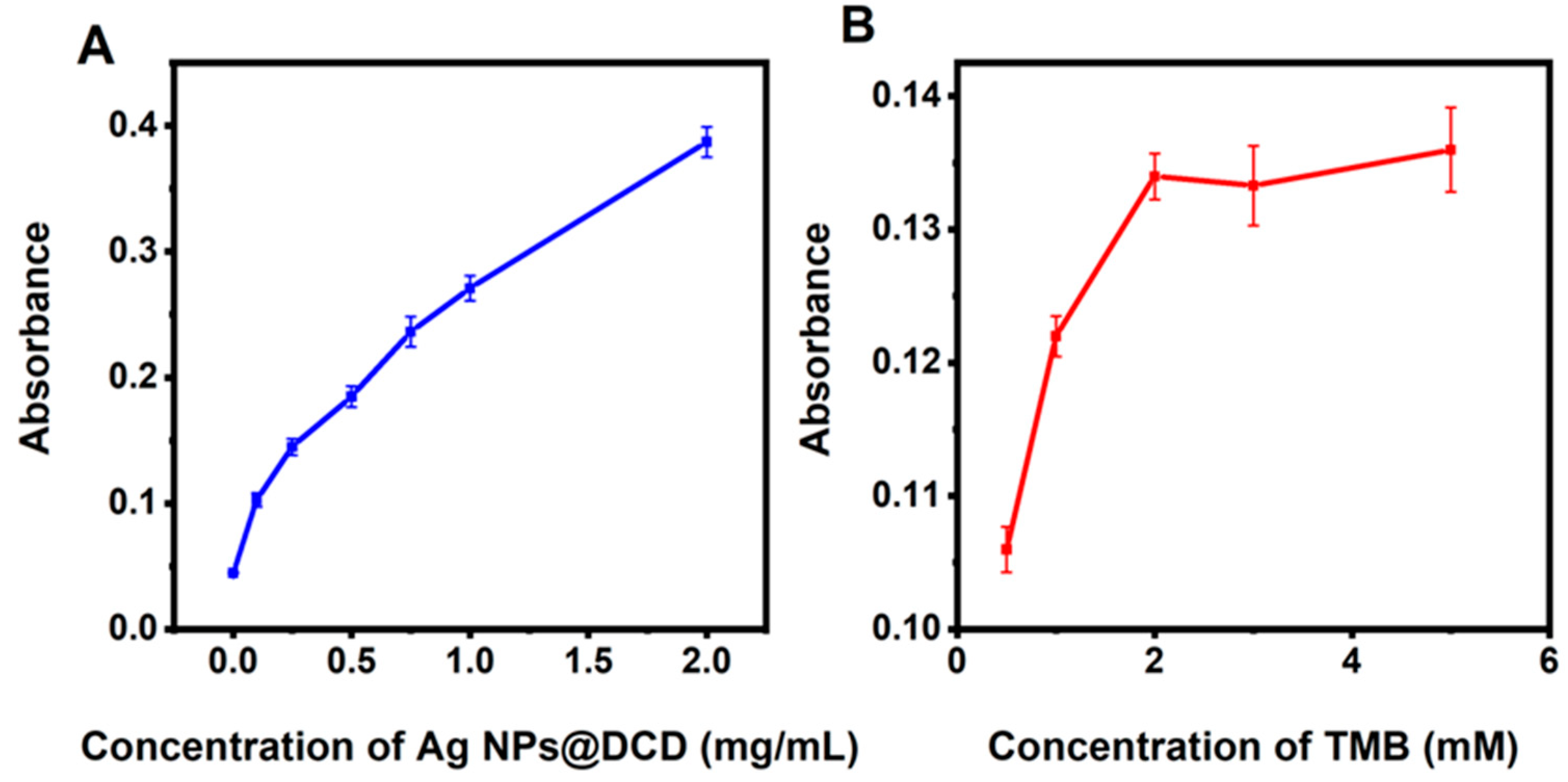
3.4. Conditions for the Experiments
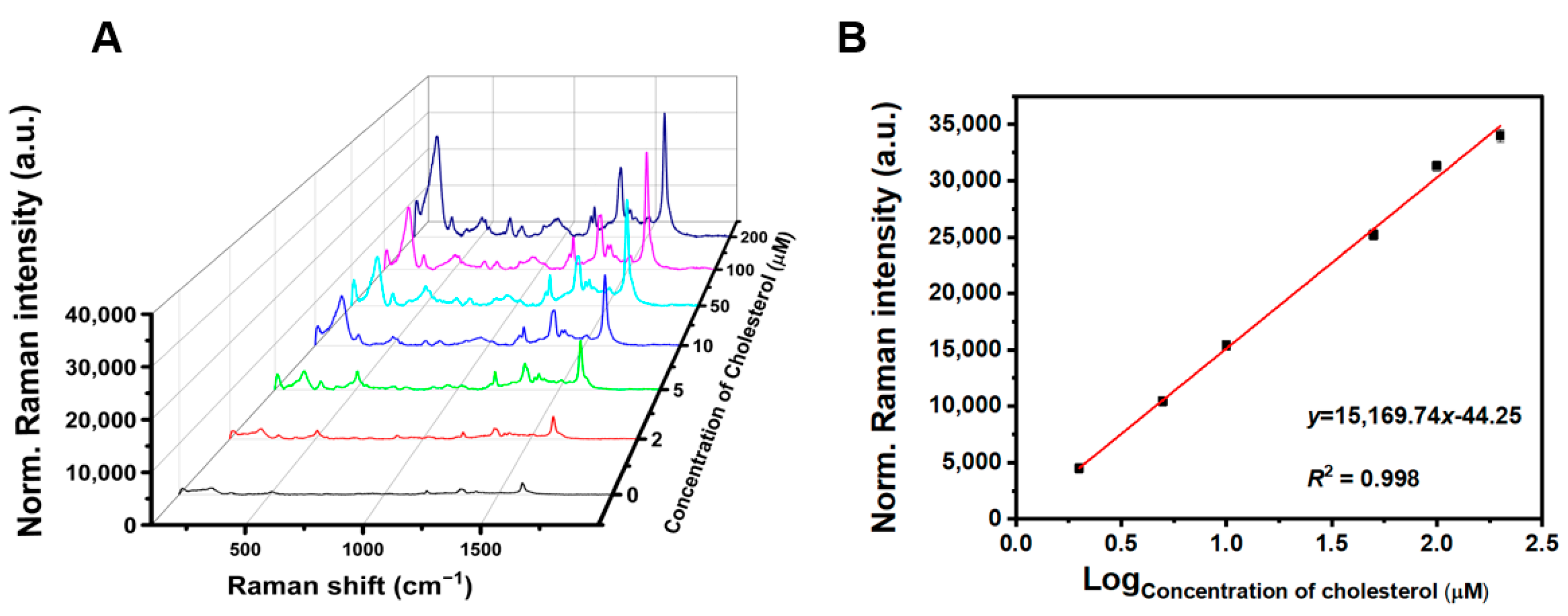
3.5. Analytical Performance of TMB-Ag NPs@DCD SERS Sensing for Cholesterol Detection
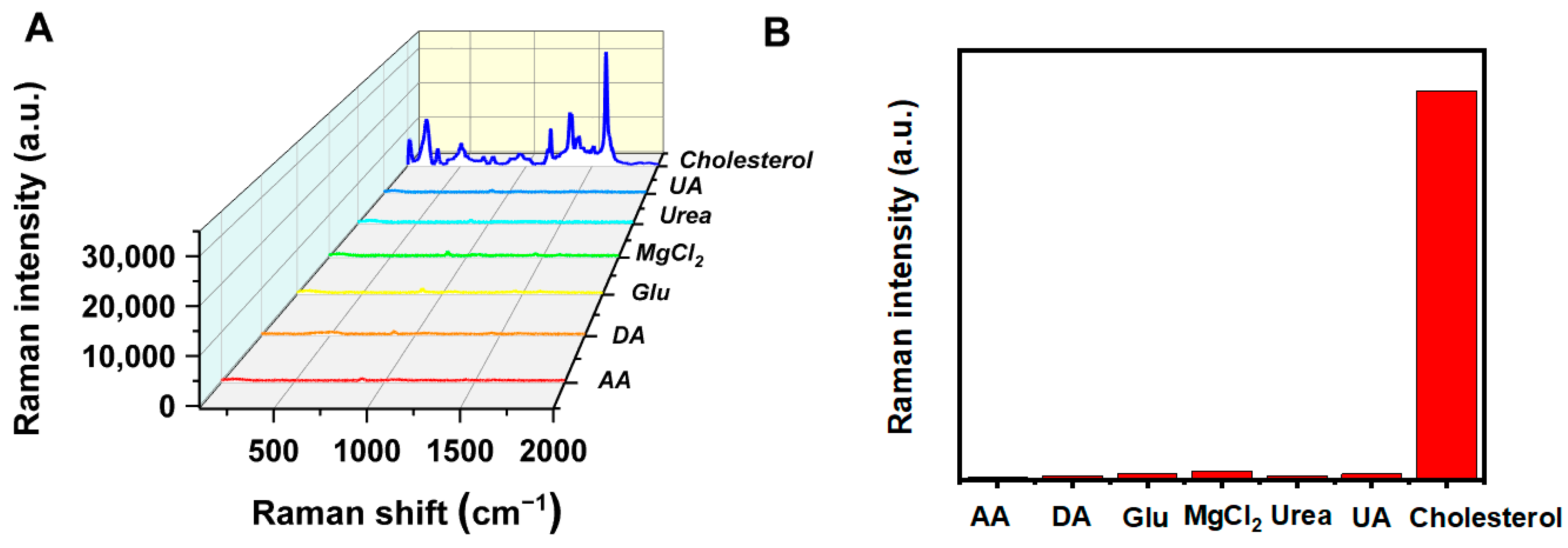
3.6. Selectivity of the TMB-Ag NPs@DCD SERS Sensing System for Cholesterol Detection
3.7. Measurement of Cholesterol in Serum Sample
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aspelund, T.; Gudnason, V. Repositioning of the global epicentre of non-optimal cholesterol. Nature 2020, 582, 73–77. [Google Scholar]
- Lim, G.B. Inclusion of remnant cholesterol improves risk prediction for ischaemic heart disease. Nat. Rev. Cardiol. 2022, 19, 504. [Google Scholar] [CrossRef]
- Allott, E.H.; Howard, L.E.; Cooperberg, M.R.; Kane, C.J.; Aronson, W.J.; Terris, M.K.; Amling, C.L.; Freedland, S.J. Serum lipid profile and risk of prostate cancer recurrence: Results from the SEARCH database. Cancer Epidem. Biomar. 2014, 23, 2349–2356. [Google Scholar] [CrossRef] [PubMed]
- Silvente-Poirot, S.; Poirot, M. Cholesterol and cancer, in the balance. Science 2014, 343, 1445–1446. [Google Scholar] [CrossRef] [PubMed]
- Brahim, S.; Narinesingh, D.; Guiseppi-Elie, A. Amperometric determination of cholesterol in serum using a biosensor of cholesterol oxidase contained within a polypyrrole–hydrogel membrane. Anal. Chim. Acta 2001, 448, 27–36. [Google Scholar] [CrossRef]
- Sigal, G.A.; Tavoni, T.M.; Silva, B.M.; Khalil-Filho, R.; Brandão, L.G.; Baracat, E.C.; Maranhão, R.C. Subclinical hyperthyroidism: Status of the cholesterol transfers to HDL and other parameters related to lipoprotein metabolism in patients submitted to thyroidectomy for thyroid cancer. Front. Endocrinol. 2020, 11, 176. [Google Scholar] [CrossRef]
- Haba, T.; Sakai, Y.; Koizumi, J.; Miyamoto, S.; Mabuchi, H.; Takeda, R. A case of heterozygous familial hypercholesterolemia associated with hyperthyroidism: Effects of triiodothyronine on low-density lipoprotein receptor and cholesterol synthesis. Metabolism 1983, 32, 1129–1132. [Google Scholar] [CrossRef]
- Hofmaenner, D.A.; Kleyman, A.; Press, A.; Bauer, M.; Singer, M. The many roles of cholesterol in sepsis: A review. Am. J. Respir. Crit. Care Med. 2022, 205, 388–396. [Google Scholar] [CrossRef]
- Zhu, J.; Ye, Z.; Fan, X.; Wang, H.; Wang, Z.; Chen, B. A highly sensitive biosensor based on Au NPs/rGO-PAMAM-Fc nanomaterials for detection of cholesterol. Int. J. Nanomed. 2019, 14, 835. [Google Scholar] [CrossRef]
- Xia, T.; Liu, G.; Wang, J.; Hou, S.; Hou, S. MXene-based enzymatic sensor for highly sensitive and selective detection of cholesterol. Biosens. Bioelectron. 2021, 183, 113243. [Google Scholar] [CrossRef]
- Kaur, G.; Tomar, M.; Gupta, V. Development of a microfluidic electrochemical biosensor: Prospect for point-of-care cholesterol monitoring. Sens. Actuators B Chem. 2018, 261, 460–466. [Google Scholar] [CrossRef]
- Srisawasdi, P.; Jearanaikoon, P.; Wetprasit, N.; Sriwanthana, B.; Kroll, M.H.; Lolekha, P.H. Application of Streptomyces and Brevibacterium cholesterol oxidase for total serum cholesterol assay by the enzymatic kinetic method. Clin. Chim. Acta. 2006, 372, 103–111. [Google Scholar] [CrossRef]
- Zhu, D.D.; Duong, P.K.; Cheah, R.H.; Liu, X.Y.; Wong, J.R.; Wang, W.J.; Guan, S.T.T.; Zheng, X.T.; Chen, P. Colorimetric microneedle patches for multiplexed transdermal detection of metabolites. Biosens. Bioelectron. 2022, 212, 114412. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; He, Y.; Xu, L.; Wei, Y. Fabrication of polydopamine/hemin-cyclodextrin supramolecular assemblies for mimicking natural peroxidases and their sensitive detection of cholesterol. J. Mol. Liq. 2021, 328, 115490. [Google Scholar] [CrossRef]
- Zheng, W.-L.; Zhang, Y.-N.; Li, L.-K.; Li, X.-G.; Zhao, Y. A plug-and-play optical fiber SPR sensor for simultaneous measurement of glucose and cholesterol concentrations. Biosens. Bioelectron. 2022, 198, 113798. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Li, N.; Li, W.; Zhang, X.; Zhang, X.; Liu, Z.; Liu, Q. 5, 10, 15, 20-tetrakis (4-carboxylphenyl) porphyrin functionalized NiCo2S4 yolk-shell nanospheres: Excellent peroxidase-like activity, catalytic mechanism and fast cascade colorimetric biosensor for cholesterol. Sens. Actuators B Chem. 2021, 326, 128850. [Google Scholar] [CrossRef]
- Vrielink, A. Cholesterol oxidase: Structure and function. In Cholesterol Binding and Cholesterol Transport Proteins: Structure and Function in Health and Disease; Springer: Berlin/Heidelberg, Germany, 2010; Volume 51, pp. 137–158. [Google Scholar]
- Song, K.; Yang, R.; Chen, X.; Wang, X.; Chen, G.; Zhao, N. The structures of CoFe2O4/PEDOT electrodes effect on the stability and specific capacity for electrochemical energy storage. Appl. Surf. Sci. 2021, 542, 148670. [Google Scholar] [CrossRef]
- Liu, D.-M.; Xu, B.; Dong, C. Recent advances in colorimetric strategies for acetylcholinesterase assay and their applications. Trends Anal. Chem. 2021, 142, 116320. [Google Scholar] [CrossRef]
- Zhu, M.; Yang, C. Blue fluorescent emitters: Design tactics and applications in organic light-emitting diodes. Chem. Soc. Rev. 2013, 42, 4963–4976. [Google Scholar] [CrossRef]
- Jiang, X.; Tan, Z.; Lin, L.; He, J.; He, C.; Thackray, B.D.; Zhang, Y.; Ye, J. Surface-Enhanced Raman Nanoprobes with Embedded Standards for Quantitative Cholesterol Detection. Small Methods 2018, 2, 1800182. [Google Scholar] [CrossRef]
- Wu, Y.; Chen, J.-Y.; He, W.-M. Surface-enhanced Raman spectroscopy biosensor based on silver nanoparticles@ metal-organic frameworks with peroxidase-mimicking activities for ultrasensitive monitoring of blood cholesterol. Sens. Actuators B Chem. 2022, 365, 131939. [Google Scholar] [CrossRef]
- Yang, S.; Cai, W.; Kong, L.; Lei, Y. Surface nanometer-scale patterning in realizing large-scale ordered arrays of metallic nanoshells with well-defined structures and controllable properties. Adv. Funct. Mater. 2010, 20, 2527–2533. [Google Scholar] [CrossRef]
- Xu, H.; Zhu, J.; Cheng, Y.; Cai, D. Functionalized UIO-66@ Ag nanoparticles substrate for rapid and ultrasensitive SERS detection of di-(2-ethylhexyl) phthalate in plastics. Sens. Actuators B Chem. 2021, 349, 130793. [Google Scholar] [CrossRef]
- Yao, J.; Quan, Y.; Gao, M.; Gao, R.; Chen, L.; Liu, Y.; Lang, J.; Shen, H.; Zhang, Y.; Yang, L. AgNPs decorated Mg-doped ZnO heterostructure with dramatic SERS activity for trace detection of food contaminants. J. Mater. Chem. C 2019, 7, 8199–8208. [Google Scholar] [CrossRef]
- Liu, C.; Wang, L.; Guo, Y.; Gao, X.; Xu, Y.; Wei, Q.; Man, B.; Yang, C. Suspended 3D AgNPs/CNT nanohybrids for the SERS application. Appl. Surf. Sci. 2019, 487, 1077–1083. [Google Scholar] [CrossRef]
- Wang, L.; Li, C.; Luo, Y.; Jiang, Z. Silver nanosol SERS quantitative analysis of ultratrace biotin coupled N-doped carbon dots catalytic amplification with affinity reaction. Food Chem. 2020, 317, 126433. [Google Scholar] [CrossRef]
- Hu, T.; Wang, H.; Zhang, L.; Tao, S. Preparation of hollow silver-polymer microspheres with a hierarchical structure for SERS. Appl. Surf. Sci. 2019, 490, 293–301. [Google Scholar] [CrossRef]
- Ren, X.; Cheshari, E.C.; Qi, J.; Li, X. Silver microspheres coated with a molecularly imprinted polymer as a SERS substrate for sensitive detection of bisphenol A. Microchim. Acta 2018, 185, 1–8. [Google Scholar] [CrossRef]
- Li, J.; Li, W.; Rao, Y.; Shi, F.; Yu, S.; Yang, H.; Min, L.; Yang, Z. Synthesis of highly ordered AgNPs-coated silica photonic crystal beads for sensitive and reproducible 3D SERS substrates. Chin. Chem. Lett. 2021, 32, 150–153. [Google Scholar] [CrossRef]
- Wang, L.; Huang, J.; Su, M.-J.; Wu, J.-D.; Liu, W. AgNPs decorated 3D bionic silicon nanograss arrays pattern with high-density hot-spots for SERS sensing via green galvanic displacement without additives. RSC Adv. 2021, 11, 27152–27159. [Google Scholar] [CrossRef]
- Su, K.; Zhang, Y.; Chen, S.; Zuo, S.; Ha, Y.; Dan, J.; Chen, W.; Sun, C.; Dai, Z.; Shi, X. Selectively encapsulating Ag nanoparticles on the surface of two-dimensional graphene for surface-enhanced Raman scattering. Appl. Surf. Sci. 2019, 492, 108–115. [Google Scholar] [CrossRef]
- Zheng, H.; Ni, D.; Yu, Z.; Liang, P. Preparation of SERS-active substrates based on graphene oxide/silver nanocomposites for rapid zdetection of l-Theanine. Food Chem. 2017, 217, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Xia, C.; Liu, B.; Feng, R.; Wang, M.; Zhu, H.; Niu, X. In situ controllable growth of Ag particles on paper for smartphone optical sensing of Hg2+ based on nanozyme activity stimulation. Talanta 2023, 253, 124055. [Google Scholar] [CrossRef]
- Ju, J.; Zhang, R.; Chen, W. Photochemical deposition of surface-clean silver nanoparticles on nitrogen-doped graphene quantum dots for sensitive colorimetric detection of glutathione. Sens. Actuators B Chem. 2016, 228, 66–73. [Google Scholar] [CrossRef]
- Mou, X.; Wu, Q.; Zhang, Z.; Liu, Y.; Zhang, J.; Zhang, C.; Chen, X.; Fan, K.; Liu, H. Nanozymes for regenerative medicine. Small Methods 2022, 6, 2200997. [Google Scholar] [CrossRef] [PubMed]
- Li, J.F.; Huang, Y.F.; Ding, Y.; Yang, Z.L.; Li, S.B.; Zhou, X.S.; Fan, F.R.; Zhang, W.; Zhou, Z.Y.; Wu, D.Y. Shell-isolated nanoparticle-enhanced Raman spectroscopy. Nature 2010, 464, 392–395. [Google Scholar] [CrossRef]
- Zhang, Y.-J.; Ze, H.; Fang, P.-P.; Huang, Y.-F.; Kudelski, A.; Fernández-Vidal, J.; Hardwick, L.J.; Lipkowski, J.; Tian, Z.-Q.; Li, J.-F. Shell-isolated nanoparticle-enhanced Raman spectroscopy. Nat. Rev. Met. Primers 2023, 3, 36. [Google Scholar] [CrossRef]
- Ju, J.; Zhang, X.; Li, L.; Regmi, S.; Yang, G.; Tang, S. Development of fluorescent lateral flow immunoassay for SARS-CoV-2-specific IgM and IgG based on aggregation-induced emission carbon dots. Front. Bioeng. Biotech. 2022, 10, 1042926. [Google Scholar] [CrossRef]
- Yuen, C.; Liu, Q. Optimization of Fe3O4@ Ag nanoshells in magnetic field-enriched surface-enhanced resonance Raman scattering for malaria diagnosis. Analyst 2013, 138, 6494–6500. [Google Scholar] [CrossRef]
- Ju, J.; Liu, W.; Perlaki, C.M.; Chen, K.; Feng, C.; Liu, Q. Sustained and cost effective silver substrate for surface enhanced Raman spectroscopy based biosensing. Sci. Rep. 2017, 7, 6917. [Google Scholar] [CrossRef]
- Sun, X.-T.; Zhang, Y.; Zheng, D.-H.; Yue, S.; Yang, C.-G.; Xu, Z.-R. Multitarget sensing of glucose and cholesterol based on Janus hydrogel microparticles. Biosens. Bioelectron. 2017, 92, 81–86. [Google Scholar] [CrossRef]
- Jin, J.; Song, W.; Wang, J.; Li, L.; Tian, Y.; Zhu, S.; Zhang, Y.; Xu, S.; Yang, B.; Zhao, B. A highly sensitive SERS platform based on small-sized Ag/GQDs nanozyme for intracellular analysis. Chem. Eng. J. 2022, 430, 132687. [Google Scholar] [CrossRef]
- Gao, L.; Zhuang, J.; Nie, L.; Zhang, J.; Zhang, Y.; Gu, N.; Wang, T.; Feng, J.; Yang, D.; Perrett, S. Intrinsic peroxidase-like activity of ferromagnetic nanoparticles. Nat. Nanotechnol. 2007, 2, 577–583. [Google Scholar] [CrossRef]
- Sun, L.; Li, S.; Ding, W.; Yao, Y.; Yang, X.; Yao, C. Fluorescence detection of cholesterol using a nitrogen-doped graphene quantum dot/chromium picolinate complex-based sensor. J. Mater. Chem. B 2017, 5, 9006–9014. [Google Scholar] [CrossRef]
- Mondal, A.; Jana, N.R. Fluorescent detection of cholesterol using β-cyclodextrin functionalized graphene. Chem. Comm. 2012, 48, 7316–7318. [Google Scholar] [CrossRef]
- Xiao, W.; Yang, Z.; Liu, J.; Chen, Z.; Li, H. Sensitive cholesterol determination by β-cyclodextrin recognition based on fluorescence enhancement of gold nanoclusters. Microchem. J. 2022, 175, 107125. [Google Scholar] [CrossRef]
- Nirala, N.R.; Abraham, S.; Kumar, V.; Bansal, A.; Srivastava, A.; Saxena, P.S. Colorimetric detection of cholesterol based on highly efficient peroxidase mimetic activity of graphene quantum dots. Sens. Actuators B Chem. 2015, 218, 42–50. [Google Scholar] [CrossRef]
- He, Y.; Niu, X.; Shi, L.; Zhao, H.; Li, X.; Zhang, W.; Pan, J.; Zhang, X.; Yan, Y.; Lan, M. Photometric determination of free cholesterol via cholesterol oxidase and carbon nanotube supported Prussian blue as a peroxidase mimic. Microchim. Acta 2017, 184, 2181–2189. [Google Scholar] [CrossRef]
- Hong, C.; Zhang, X.; Wu, C.; Chen, Q.; Yang, H.; Yang, D.; Huang, Z.; Cai, R.; Tan, W. On-site colorimetric detection of cholesterol based on polypyrrole nanoparticles. ACS App. Mater. Interfaces 2020, 12, 54426–54432. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.-N.; Sun, X.-T.; Chen, L.; Xu, Z.-R. Boron nitride nanosheet/CuS nanocomposites as mimetic peroxidase for sensitive colorimetric detection of cholesterol. Sens. Actuators B Chem. 2017, 246, 118–126. [Google Scholar] [CrossRef]
- Chen, H.-Y.; Xin, P.-L.; Xu, H.-B.; Lv, J.; Qian, R.-C.; Li, D.-W. Self-Assembled Plasmonic Nanojunctions Mediated by Host–Guest Interaction for Ultrasensitive Dual-Mode Detection of Cholesterol. ACS Sens. 2023, 8, 388–396. [Google Scholar] [CrossRef]
- Wang, S.; Chen, S.; Shang, K.; Gao, X.; Wang, X. Sensitive electrochemical detection of cholesterol using a portable paper sensor based on the synergistic effect of cholesterol oxidase and nanoporous gold. Int. J. Biol. Macromol. 2021, 189, 356–362. [Google Scholar] [CrossRef]
- Manjunatha, R.; Suresh, G.S.; Melo, J.S.; D’Souza, S.F.; Venkatesha, T.V. An amperometric bienzymatic cholesterol biosensor based on functionalized graphene modified electrode and its electrocatalytic activity towards total cholesterol determination. Talanta 2012, 99, 302–309. [Google Scholar] [CrossRef]




| Methods | Sensing System | Linear Range (μM) | LOD (μM) | Refs. |
|---|---|---|---|---|
| Fluorometry | N-GQDs/CrPic | 0–520 | 0.4 | [45] |
| b-CD-G-R6G | 5–30 | 5 | [46] | |
| CD-AuNCs | 10–100 | 5.77 | [47] | |
| Colorimetry | ChOx/GQDs-TMB-H2O2 | 20–600 | 6 | [48] |
| ChOx-PB/MWCNT | 4–100 | 3.01 | [49] | |
| PPy NPs | 10–100 | 3.5 | [50] | |
| BNNS@CuS | 10–100 | 2.9 | [51] | |
| SERS | AgNPs@MIL-101(Fe) | 1–100 | 0.36 | [21] |
| HS-β-CD@Au | 0.45–22 | 0.74 | [52] | |
| Electrochemial | NPG/SPE | 50–6000 | 8.36 | [53] |
| ChOx-FG/Gr | 50–350 | 5 | [54] | |
| SERS | TMB-Ag NPs@DCD | 2–200 | 0.8 | This work |
| Sample | Added (µM) | Found (µM) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| Serum 1 | 12.87 | 1.50 | ||
| 1 | 13.96 | 99.80 | 3.20 | |
| 10 | 21.47 | 101.90 | 4.51 | |
| 25 | 39.46 | 102.80 | 2.17 | |
| Serum 2 | 10.98 | 0.98 | ||
| 1 | 12.05 | 99.98 | 1.46 | |
| 10 | 20.65 | 102.90 | 1.58 | |
| 25 | 36.76 | 101.90 | 2.68 | |
| Serum 3 | 11.36 | 1.42 | ||
| 1 | 12.50 | 99.00 | 1.51 | |
| 10 | 21.98 | 100.80 | 2.30 | |
| 25 | 36.07 | 101.76 | 1.98 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ju, J.; Li, L.; Li, B.; Regmi, S.; Wang, T.; Xu, J.; Li, C.; Tang, S. Surface-Enhanced Raman Scattering Active Core-Shell Ag NPs@Carbon Dots with Enzyme-Mimicking Activities for Label-Free Measurement Cholesterol. Biosensors 2023, 13, 927. https://doi.org/10.3390/bios13100927
Ju J, Li L, Li B, Regmi S, Wang T, Xu J, Li C, Tang S. Surface-Enhanced Raman Scattering Active Core-Shell Ag NPs@Carbon Dots with Enzyme-Mimicking Activities for Label-Free Measurement Cholesterol. Biosensors. 2023; 13(10):927. https://doi.org/10.3390/bios13100927
Chicago/Turabian StyleJu, Jian, Lin Li, Bei Li, Sagar Regmi, Tingting Wang, Jiao Xu, Chaojie Li, and Shixing Tang. 2023. "Surface-Enhanced Raman Scattering Active Core-Shell Ag NPs@Carbon Dots with Enzyme-Mimicking Activities for Label-Free Measurement Cholesterol" Biosensors 13, no. 10: 927. https://doi.org/10.3390/bios13100927
APA StyleJu, J., Li, L., Li, B., Regmi, S., Wang, T., Xu, J., Li, C., & Tang, S. (2023). Surface-Enhanced Raman Scattering Active Core-Shell Ag NPs@Carbon Dots with Enzyme-Mimicking Activities for Label-Free Measurement Cholesterol. Biosensors, 13(10), 927. https://doi.org/10.3390/bios13100927







