CRISPR/Cas12a-Based Detection Platform for Early and Rapid Diagnosis of Scrub Typhus
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Bacterial Strains
2.3. Clinical Sample Collection and Processing
2.4. Positive Control Preparation
2.5. Isothermal Amplification with RPA
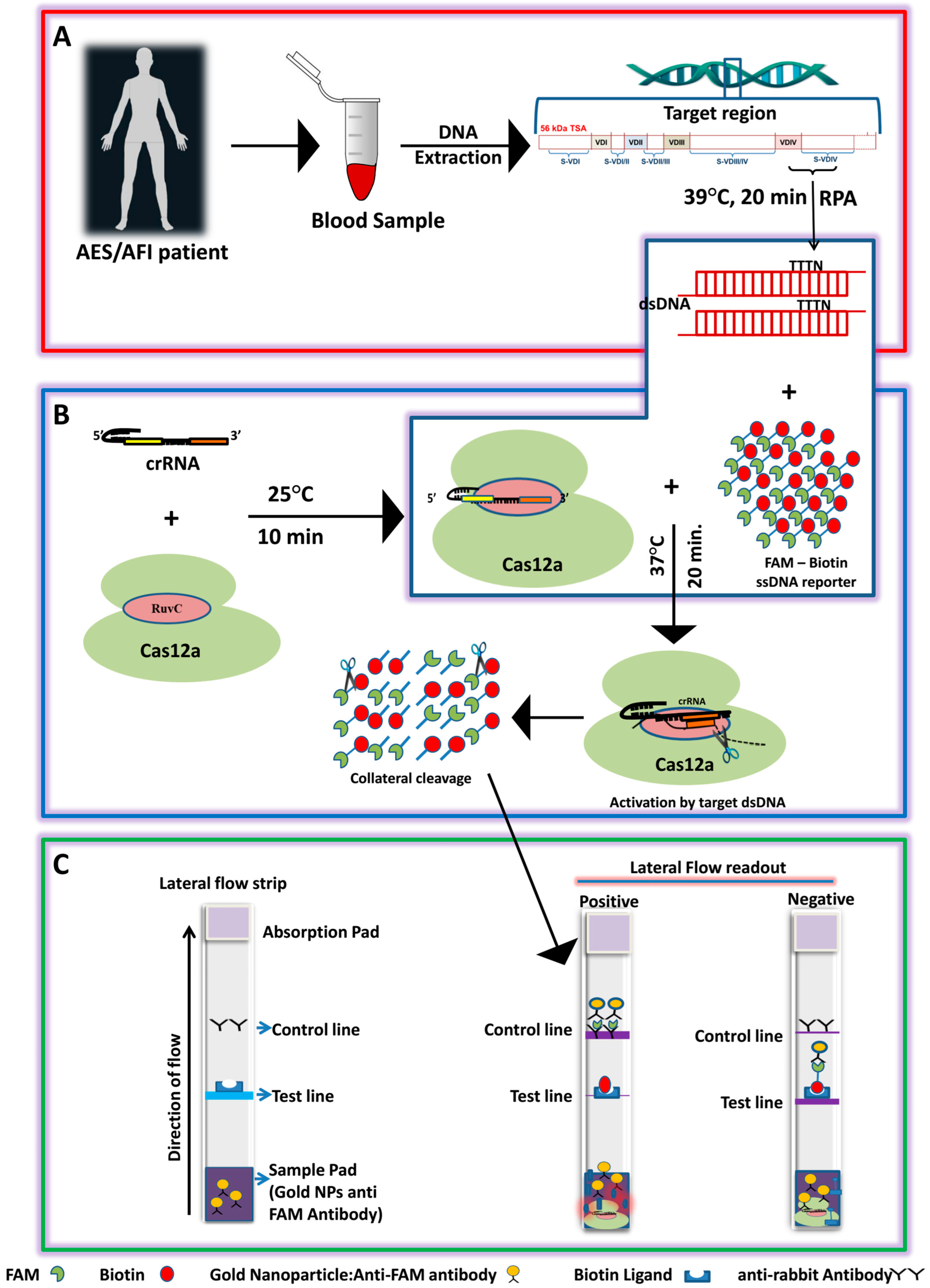
2.6. CRISPR RNA (crRNA) Synthesis and CRISPR/Cas12a-Based Detection of ST
2.7. RPA-CRISPR/Cas12a Coupled with a Lateral Flow Detection
2.8. Establishment of Sensitivity and Specificity of LoCIST
2.9. Real-Time PCR
2.10. Bioinformatical Tools
3. Results
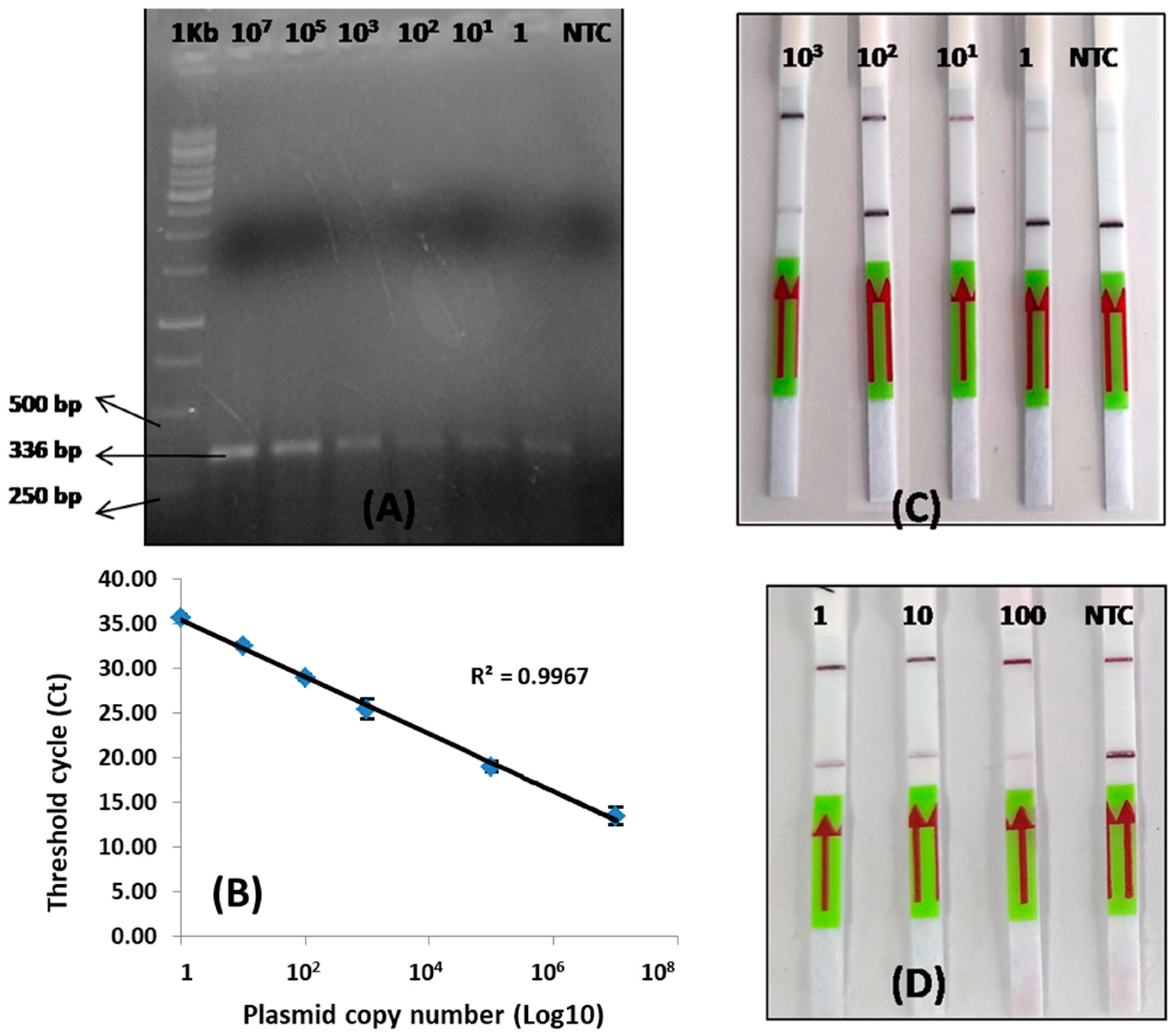
3.1. Construction of the Recombinant Plasmid
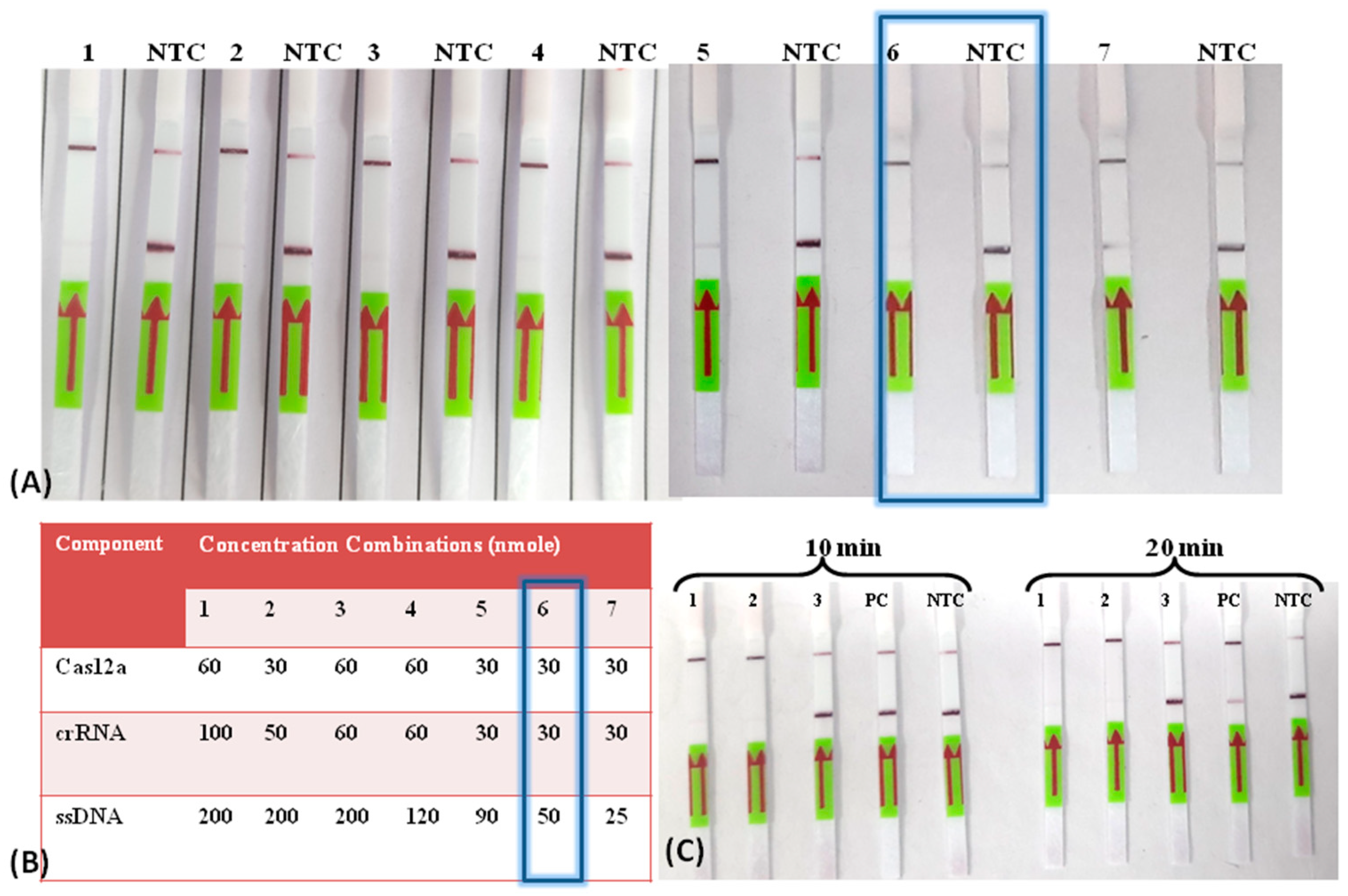
3.2. Establishment of the LoCIST Platform
3.3. Detection Limit of LoCIST
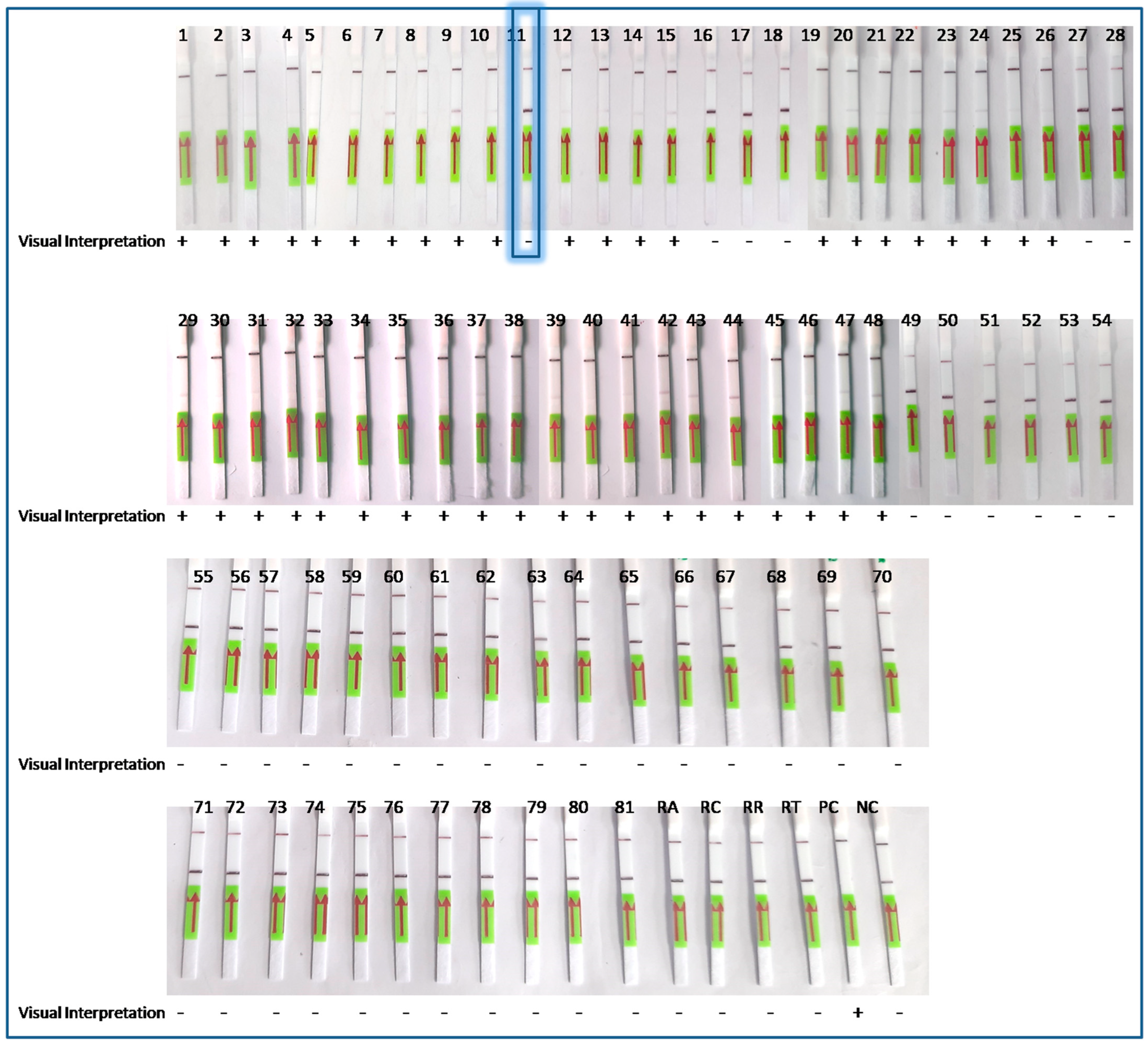
3.4. Clinical Specimens
3.5. Performance of the LoCIST with Clinical Samples
3.6. Unambiguous Detection of OT by cr56kDa2
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, G.; Walker, D.H.; Jupiter, D.; Melby, P.C.; Arcari, C.M. A review of the global epidemiology of scrub typhus. PLoS Negl. Trop. Dis. 2017, 11, e0006062. [Google Scholar] [CrossRef] [PubMed]
- Luce-Fedrow, A.; Lehman, M.L.; Kelly, D.J.; Mullins, K.; Maina, A.N.; Stewart, R.L.; Ge, H.; John, H.S.; Jiang, J.; Richards, A.L. A Review of Scrub Typhus (Orientia tsutsugamushi and Related Organisms): Then, Now, and Tomorrow. Trop. Med. Infect. Dis. 2018, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Richards, A.L. Scrub Typhus: No Longer Restricted to the Tsutsugamushi Triangle. Trop. Med. Infect. Dis. 2018, 3, 11. [Google Scholar] [CrossRef] [PubMed]
- Richards, A.L.; Jiang, J. Scrub Typhus: Historic Perspective and Current Status of the Worldwide Presence of Orientia Species. Trop. Med. Infect. Dis. 2020, 5, 49. [Google Scholar] [CrossRef] [PubMed]
- Mittal, M.; Thangaraj, J.W.V.; Rose, W.; Verghese, V.P.; Kumar, C.G.; Mittal, M.; Sabarinathan, R.; Bondre, V.; Gupta, N.; Murhekar, M.V. Scrub Typhus as a Cause of Acute Encephalitis Syndrome, Gorakhpur, Uttar Pradesh, India. Emerg. Infect. Dis. 2017, 23, 1414–1416. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.; Prakash, S.; Tripathi, P.K.; Chauhan, A.; Gupta, S.; Sharma, U.; Jaiswal, A.K.; Sharma, D.; Jain, A. Emergence of Orientia tsutsugamushi as an important cause of Acute Encephalitis Syndrome in India. PLoS Negl. Trop. Dis. 2018, 12, e0006346. [Google Scholar] [CrossRef]
- Murhekar, M.; Thangaraj, J.V.; Sadanandane, C.; Mittal, M.; Gupta, N.; Rose, W.; Sahay, S.; Kant, R.; Gupte, M. Investigations of seasonal outbreaks of acute encephalitis syndrome due to Orientia tsutsugamushi in Gorakhpur region, India: A One Health case study. Indian J. Med. Res. 2021, 153, 375–381. [Google Scholar] [CrossRef]
- Kala, D.; Gupta, S.; Nagraik, R.; Verma, V.; Thakur, A.; Kaushal, A. Diagnosis of scrub typhus: Recent advancements and challenges. 3 Biotech 2020, 10, 396. [Google Scholar] [CrossRef]
- Abarca, K.; Martínez-Valdebenito, C.; Angulo, J.; Jiang, J.; Farris, C.M.; Richards, A.L.; Acosta-Jamett, G.; Weitzel, T. Molecular Description of a Novel Orientia Species Causing Scrub Typhus in Chile. Emerg. Infect. Dis. 2020, 26, 2148–2156. [Google Scholar] [CrossRef]
- Kelly, D.J.; Fuerst, P.A.; Ching, W.; Richards, A.L. Scrub Typhus: The Geographic Distribution of Phenotypic and Genotypic Variants of Orientia tsutsugamushi. Clin. Infect. Dis. 2009, 48, S203–S230. [Google Scholar] [CrossRef]
- Bora, T.; Khan, S.A.; Jampa, L.; Laskar, B. Genetic diversity of Orientia tsutsugamushi strains circulating in Northeast India. Trans. R. Soc. Trop. Med. Hyg. 2018, 112, 22–30. [Google Scholar] [CrossRef]
- Chunchanur, S.K.; Venugopal, S.J.; Ambica, R.; Dakshayani, B. Phylogenetic Diversity of Orientia tsutsugamushi Isolates in Patients with Scrub Typhus in Bengaluru, India. Indian J. Med. Microbiol. 2019, 37, 438–441. [Google Scholar] [CrossRef] [PubMed]
- Behera, S.P.; Kumar, N.; Singh, R.; Deval, H.; Zaman, K.; Misra, B.; Pandey, A.; Kant, R.; Kavathekar, A.; Kumar, S.; et al. Molecular Detection and Genetic Characterization of Orientia tsutsugamushi from Hospitalized Acute Encephalitis Syndrome Cases During Two Consecutive Outbreaks in Eastern Uttar Pradesh, India. Vector-Borne Zoonotic Dis. 2021, 21, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Nanaware, N.; Desai, D.; Banerjee, A.; Zaman, K.; Mittal, M.; Kulkarni, S. Genotypic Characterization of Orientia tsutsugamushi Isolated from Acute Encephalitis Syndrome and Acute Febrile Illness Cases in the Gorakhpur Area, Uttar Pradesh, India. Front. Microbiol. 2022, 13, 910757. [Google Scholar] [CrossRef]
- Janardhanan, J.; Prakash, J.A.J.; Abraham, O.C.; Varghese, G.M. Comparison of a conventional and nested PCR for diagnostic confirmation and genotyping of Orientia tsutsugamushi. Diagn. Microbiol. Infect. Dis. 2014, 79, 7–9. [Google Scholar] [CrossRef]
- Bonell, A.; Lubell, Y.; Newton, P.N.; Crump, J.A.; Paris, D.H. Estimating the burden of scrub typhus: A systematic review. PLoS Negl. Trop. Dis. 2017, 11, e0005838. [Google Scholar] [CrossRef]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA Detection Using Recombination Proteins. PLoS Biol. 2006, 4, e204. [Google Scholar] [CrossRef] [PubMed]
- Aman, R.; Mahas, A.; Mahfouz, M. Nucleic Acid Detection Using CRISPR/Cas Biosensing Technologies. ACS Synth. Biol. 2020, 9, 1226–1233. [Google Scholar] [CrossRef]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef]
- Kellner, M.J.; Koob, J.G.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019, 14, 2986–3012. [Google Scholar] [CrossRef]
- Bhardwaj, P.; Kant, R.; Behera, S.P.; Dwivedi, G.R.; Singh, R. Next-Generation Diagnostic with CRISPR/Cas: Beyond Nucleic Acid Detection. Int. J. Mol. Sci. 2022, 23, 6052. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hu, H.; Zanaroli, G.; Xu, P.; Tang, H. A Pseudomonas sp. strain uniquely degrades PAHs and heterocyclic derivatives via lateral dioxygenation pathways. J. Hazard. Mater. 2021, 403, 123956. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Yu, L.; Liu, C.; Ye, S.; Chen, W.; Li, D.; Huang, W. One-tube SARS-CoV-2 detection platform based on RT-RPA and CRISPR/Cas12a. J. Transl. Med. 2021, 19, 74. [Google Scholar] [CrossRef] [PubMed]
- Malcı, K.; Walls, L.E.; Rios-Solis, L. Rational Design of CRISPR/Cas12a-RPA Based One-Pot COVID-19 Detection with Design of Experiments. ACS Synth. Biol. 2022, 11, 1555–1567. [Google Scholar] [CrossRef]
- Zheng, S.Y.; Ma, L.L.; Wang, X.L.; Lu, L.X.; Ma, S.T.; Xu, B.; Ouyang, W. RPA-Cas12aDS: A visual and fast molecular diagnostics platform based on RPA-CRISPR-Cas12a method for infectious bursal disease virus detection. J. Virol. Methods 2022, 304, 114523. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, P.; Behera, S.P.; Nanaware, N.; Zaman, K.; Deval, H.; Kant, R.; Kulkarni, S.; Kumar, R.; Dwivedi, G.R.; Singh, R. Phylogenetic and immunological investigations of complete TSA56 ORF of Orientia tsutsugamushi present in acute encephalitis syndrome cases from eastern Uttar Pradesh, India. Arch. Microbiol. 2023, 205, 178. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef]
- Zhao, D.; Zhang, Y.; Yin, Z.; Zhao, J.; Yang, D.; Zhou, Q. Clinical Predictors of Multiple Organ Dysfunction Syndromes in Pediatric patients with Scrub Typhus. J. Trop. Pediatr. 2016, 63, 167–173. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nguyen, C.T.; Nguyen, U.D.; Le, T.T.; Bui, H.T.; Nguyen, A.N.T.; Thi Nguyen, A.N.; Trieu, N.T.; Trieu, L.P.; Bui, S.T.; Nguyen, C.; et al. Establishment of Recombinase Polymerase Amplification assay for rapid and sensitive detection of Orientia tsutsugamushi in Southeast Asia. Acta Trop. 2020, 210, 105541. [Google Scholar] [CrossRef]
- Kim, D.-M.; Park, G.; Kim, H.S.; Lee, J.Y.; Neupane, G.P.; Graves, S.; Stenos, J. Comparison of Conventional, Nested, and Real-Time Quantitative PCR for Diagnosis of Scrub Typhus. J. Clin. Microbiol. 2011, 49, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Myhrvold, C.; Freije, C.A.; Gootenberg, J.S.; Abudayyeh, O.O.; Metsky, H.C.; Durbin, A.F.; Kellner, M.; Tan, A.; Paul, L.; Parham, L.; et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 2018, 360, 444–448. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.; Guo, J.; Guo, X.; Li, Q.; Li, X.; Sun, Z.; Zhao, Z.; Weng, J.; Wu, J.; Zhang, R.; et al. Fast and visual detection of nucleic acids using a one-step RPA-CRISPR detection (ORCD) system unrestricted by the PAM. Anal. Chim. Acta 2023, 1248, 340938. [Google Scholar] [CrossRef]
- Chao, C.-C.; Belinskaya, T.; Zhang, Z.; Ching, W.-M. Development of Recombinase Polymerase Amplification Assays for Detection of Orientia tsutsugamushi or Rickettsia typhi. PLoS Negl. Trop. Dis. 2015, 9, e0003884. [Google Scholar] [CrossRef]
- Qi, Y.; Yin, Q.; Shao, Y.; Cao, M.; Li, S.; Chen, H.; Shen, W.; Rao, J.; Li, J.; Li, X.; et al. Development of a rapid and visual nucleotide detection method for a Chinese epidemic strain of Orientia tsutsugamushi based on recombinase polymerase amplification assay and lateral flow test. Int. J. Infect. Dis. 2018, 70, 42–50. [Google Scholar] [CrossRef]
- Paris, D.H.; Blacksell, S.D.; Newton, P.N.; Day, N.P. Simple, rapid and sensitive detection of Orientia tsutsugamushi by loop-isothermal DNA amplification. Trans. R. Soc. Trop. Med. Hyg. 2008, 102, 1239–1246. [Google Scholar] [CrossRef]
- Roy, S.; Yadav, S.; Garg, S.; Deshmukh, P.R.; Narang, R. Evaluation of nested PCR and loop mediated isothermal amplification assay (LAMP) targeting 47 kDa gene of Orientia tsutsugamushi for diagnosis of scrub typhus. Indian J. Med. Microbiol. 2021, 39, 475–478. [Google Scholar] [CrossRef]
- Jang, W.S.; Lim, D.H.; Choe, Y.L.; Nam, J.; Moon, K.C.; Kim, C.; Choi, M.; Park, I.; Park, D.W.; Lim, C.S. Developing a multiplex loop-mediated isothermal amplification assay (LAMP) to determine severe fever with thrombocytopenia syndrome (SFTS) and scrub typhus. PLoS ONE 2022, 17, e0262302. [Google Scholar] [CrossRef]
- Paris, D.H.; Blacksell, S.D.; Nawtaisong, P.; Jenjaroen, K.; Teeraratkul, A.; Chierakul, W.; Wuthiekanun, V.; Kantipong, P.; Day, N.P.J. Diagnostic Accuracy of a Loop-Mediated Isothermal PCR Assay for Detection of Orientia tsutsugamushi during Acute Scrub Typhus Infection. PLoS Negl. Trop. Dis. 2011, 5, e1307. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, N.; Koyama, Y.; Urakami, H.; Fukuhara, M.; Tamura, A.; Kawamori, F.; Yamamoto, S.; Kasuya, S.; Yoshimura, K. Demonstration of Antigenic and Genotypic Variation in Orientia tsutsugamushi Which Were Isolated in Japan, and Their Classification into Type and Subtype. Microbiol. Immunol. 1996, 40, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.; Paris, D.H.; Blacksell, S.D.; Laongnualpanich, A.; Kantipong, P.; Chierakul, W.; Wuthiekanun, V.; Day, N.P.; Cooper, B.S.; Limmathurotsakul, D. How to Determine the Accuracy of an Alternative Diagnostic Test when It Is Actually Better than the Reference Tests: A Re-Evaluation of Diagnostic Tests for Scrub Typhus Using Bayesian LCMs. PLoS ONE 2015, 10, e0114930. [Google Scholar] [CrossRef] [PubMed]
- Koo, B.; Kim, D.-E.; Kweon, J.; Jin, C.E.; Kim, S.-H.; Kim, Y.; Shin, Y. CRISPR/dCas9-mediated biosensor for detection of tick-borne diseases. Sens. Actuators B Chem. 2018, 273, 316–321. [Google Scholar] [CrossRef]
- Qiang, W.; Bin, Q.; Qian, C. Fluorescence Isothermal Amplification Primer, Probe, Kit and Detection Method for Orientia Tsutsutsugamushi Nucleic Acid. CN113186304A, 30 July 2021. [Google Scholar]
- Bai, J.; Lin, H.; Li, H.; Zhou, Y.; Liu, J.; Zhong, G.; Wu, L.; Jiang, W.; Du, H.; Yang, J.; et al. Cas12a-Based On-Site and Rapid Nucleic Acid Detection of African Swine Fever. Front. Microbiol. 2019, 10, 2830. [Google Scholar] [CrossRef]
- Xiong, Y.; Cao, G.; Chen, X.; Yang, J.; Shi, M.; Wang, Y.; Nie, F.; Huo, D.; Hou, C. One-pot platform for rapid detecting virus utilizing recombinase polymerase amplification and CRISPR/Cas12a. Appl. Microbiol. Biotechnol. 2022, 106, 4607–4616. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef]
- Chen, P.; Zhou, J.; Wan, Y.; Liu, H.; Li, Y.; Liu, Z.; Wang, H.; Lei, J.; Zhao, K.; Zhang, Y.; et al. A Cas12a ortholog with stringent PAM recognition followed by low off-target editing rates for genome editing. Genome Biol. 2020, 21, 78. [Google Scholar] [CrossRef]
- Wu, Y.; Luo, W.; Weng, Z.; Guo, Y.; Yu, H.; Zhao, R.; Zhang, L.; Zhao, J.; Bai, D.; Zhou, X.; et al. A PAM-free CRISPR/Cas12a ultra-specific activation mode based on toehold-mediated strand displacement and branch migration. Nucleic Acids Res. 2022, 50, 11727–11737. [Google Scholar] [CrossRef]
- Milenia. Milenia Biotec. Available online: https://www.milenia-biotec.com/en/product/hybridetect/ (accessed on 24 July 2023).
- Paul, D.; Lyngdoh, W.V.; Barman, H.; Lyngdoh, C.J.; Lynrah, K.; Durairaj, E. Clinical Performance Analysis and Evaluation of Quantitative Real Time PCR for Diagnosis of Scrub Typhus in North East India. Life Sci. 2020, 2020050440. [Google Scholar] [CrossRef]
- Kang, J.; Li, Y.; Zhao, Y.; Wang, Y.; Ma, C.; Shi, C. Nucleic acid extraction without electrical equipment via magnetic nanoparticles in Pasteur pipettes for pathogen detection. Anal. Biochem. 2021, 635, 114445. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.A.; MacLeod, E.T.; Hide, G.; Welburn, S.C.; Picozzi, K. The best practice for preparation of samples from FTA®cards for diagnosis of blood borne infections using African trypanosomes as a model system. Parasites Vectors 2011, 4, 68. [Google Scholar] [CrossRef] [PubMed]
- Tsou, J.-H.; Leng, Q.; Jiang, F. A CRISPR Test for Detection of Circulating Nuclei Acids. Transl. Oncol. 2019, 12, 1566–1573. [Google Scholar] [CrossRef] [PubMed]
| Feature | Number (n) | Percentage (%) | Total Data Availability (n) |
|---|---|---|---|
| Sex | |||
| Male | 26 | 60.46 | 43 |
| Female | 16 | 39.53 | |
| Age group | |||
| 1.1–5 | 16 | 37.20 | 43 |
| 5.1–10 | 13 | 30.236 | |
| 10.1–15 | 7 | 16.27 | |
| 15.1–20 | 2 | 4.65 | |
| 20.1–30 | 4 | 11.62 | |
| Clinical symptoms | |||
| Fever Grade | |||
| High | 20 | 60.6 | 33 |
| Low | 7 | 21.21 | |
| Vomiting | 18 | 54.54 | |
| Loose motion | 5 | 15.15 | |
| Abdominal pain | 7 | 21.21 | |
| Headache | 9 | 27.27 | |
| Retrorbital | 1 | 3.03 | |
| Cough | 2 | 6.06 | |
| Altered Sensorium | 15 | 45.45 | |
| Seizure | 1 | 3.03 | |
| Body tightening | 19 | 57.57 | |
| Frothing | 12 | 36.36 | |
| Up rolling of eyeball | 16 | 48.48 | |
| Abnormal behavior | 1 | 3.03 | |
| Time interval between symptoms onset and DOA | |||
| ELISA positive | 8.75 | 16 | 40 |
| ELISA negative | 5.8 | 24 | |
| S. No. | Platform | Isothermal Amplification | CRISPR/Cas System | End Point Detection | Gene Target | Sensitivity | Specificity (%) | LOD | Reference |
|---|---|---|---|---|---|---|---|---|---|
| 1 | LAMP assay | LAMP | - | Agarose gel electrophoresis/real-time | groEL | - | - | 3 gene copies | [39] |
| 2 | 47-RPA-nfo and 47-RPA-Exo | RPA | - | Lateral flow/fluorescence | 47 kDa | 80% | 100 | 10–60 gene copies | [37] |
| 3 | dCas9 based SMR | RPA | dCas9 | Resonance-based micro rings | 56 kDa | 1 copy | 100 | 0.54 aM | [45] |
| 4 | RPA-LF | RPA | - | Lateral flow assay | 56 kDa | 100% | 90 | 10 gene copies | [38] |
| 5 | Real time-RPA | RPA | - | Real-time fluorescence | 47 kDa | 80% | 100 | 100 gene copies | [33] |
| 6 | RPA | RPA | - | Fluorescence | 56 kDa | - | - | - | [46] |
| 7 | SFTSV/OT/IC RT-LAMP | LAMP | - | Real-time fluorescence | - | 91.6% | 100 | 25 | [41] |
| 8 | LoCIST | RPA | Cas12a | Lateral flow assay | 56 kDa | 97.6% | 100 | 1 gene copy | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhardwaj, P.; Nanaware, N.S.; Behera, S.P.; Kulkarni, S.; Deval, H.; Kumar, R.; Dwivedi, G.R.; Kant, R.; Singh, R. CRISPR/Cas12a-Based Detection Platform for Early and Rapid Diagnosis of Scrub Typhus. Biosensors 2023, 13, 1021. https://doi.org/10.3390/bios13121021
Bhardwaj P, Nanaware NS, Behera SP, Kulkarni S, Deval H, Kumar R, Dwivedi GR, Kant R, Singh R. CRISPR/Cas12a-Based Detection Platform for Early and Rapid Diagnosis of Scrub Typhus. Biosensors. 2023; 13(12):1021. https://doi.org/10.3390/bios13121021
Chicago/Turabian StyleBhardwaj, Pooja, Nikita Shrikant Nanaware, Sthita Pragnya Behera, Smita Kulkarni, Hirawati Deval, Rajesh Kumar, Gaurav Raj Dwivedi, Rajni Kant, and Rajeev Singh. 2023. "CRISPR/Cas12a-Based Detection Platform for Early and Rapid Diagnosis of Scrub Typhus" Biosensors 13, no. 12: 1021. https://doi.org/10.3390/bios13121021
APA StyleBhardwaj, P., Nanaware, N. S., Behera, S. P., Kulkarni, S., Deval, H., Kumar, R., Dwivedi, G. R., Kant, R., & Singh, R. (2023). CRISPR/Cas12a-Based Detection Platform for Early and Rapid Diagnosis of Scrub Typhus. Biosensors, 13(12), 1021. https://doi.org/10.3390/bios13121021






