Genetic Programming by Nitric Oxide-Sensing Gene Switch System in Tumor-Targeting Bacteria
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
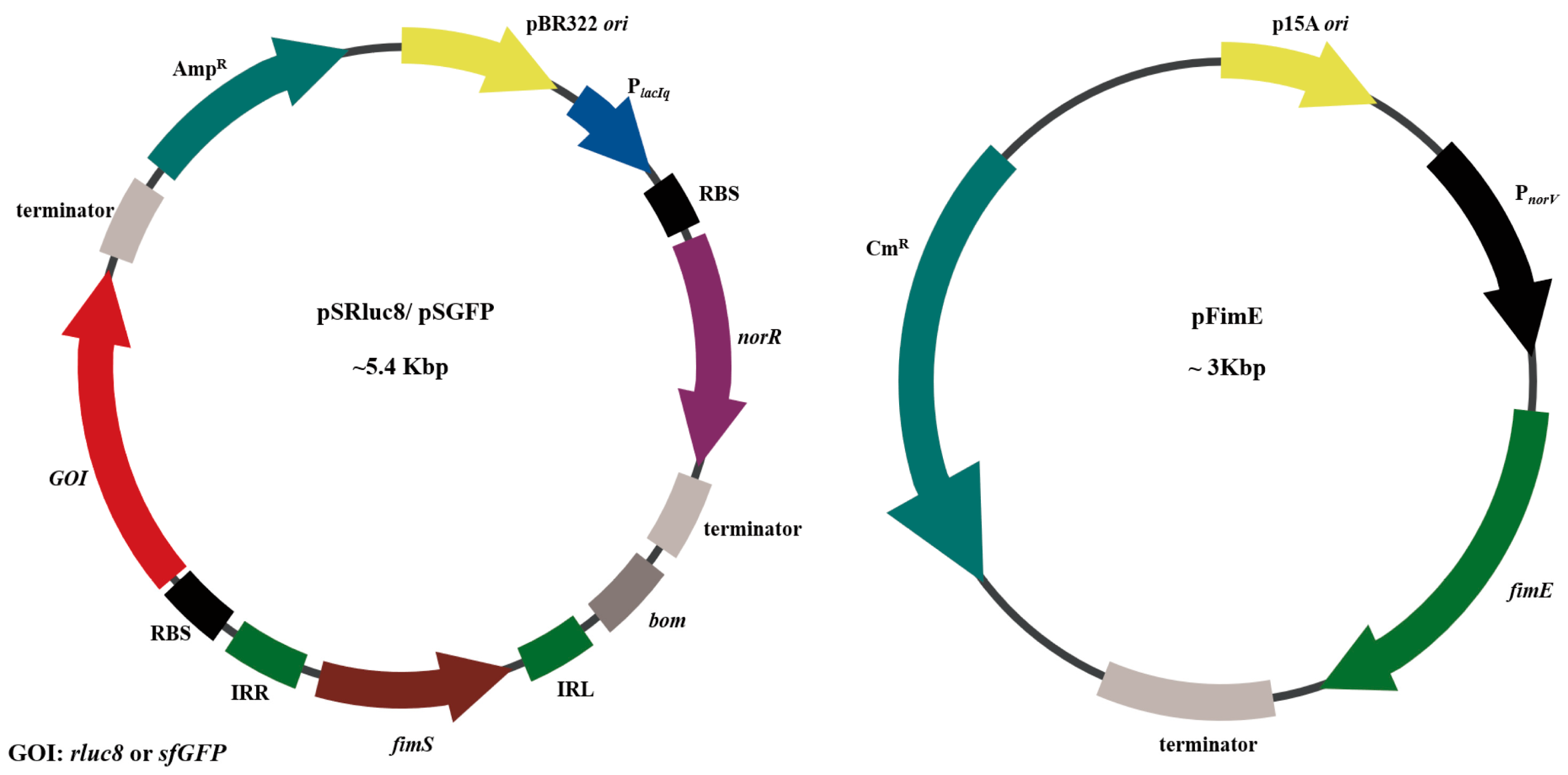
2.2. Construction of Plasmids for NO-Sensing Gene Switch System
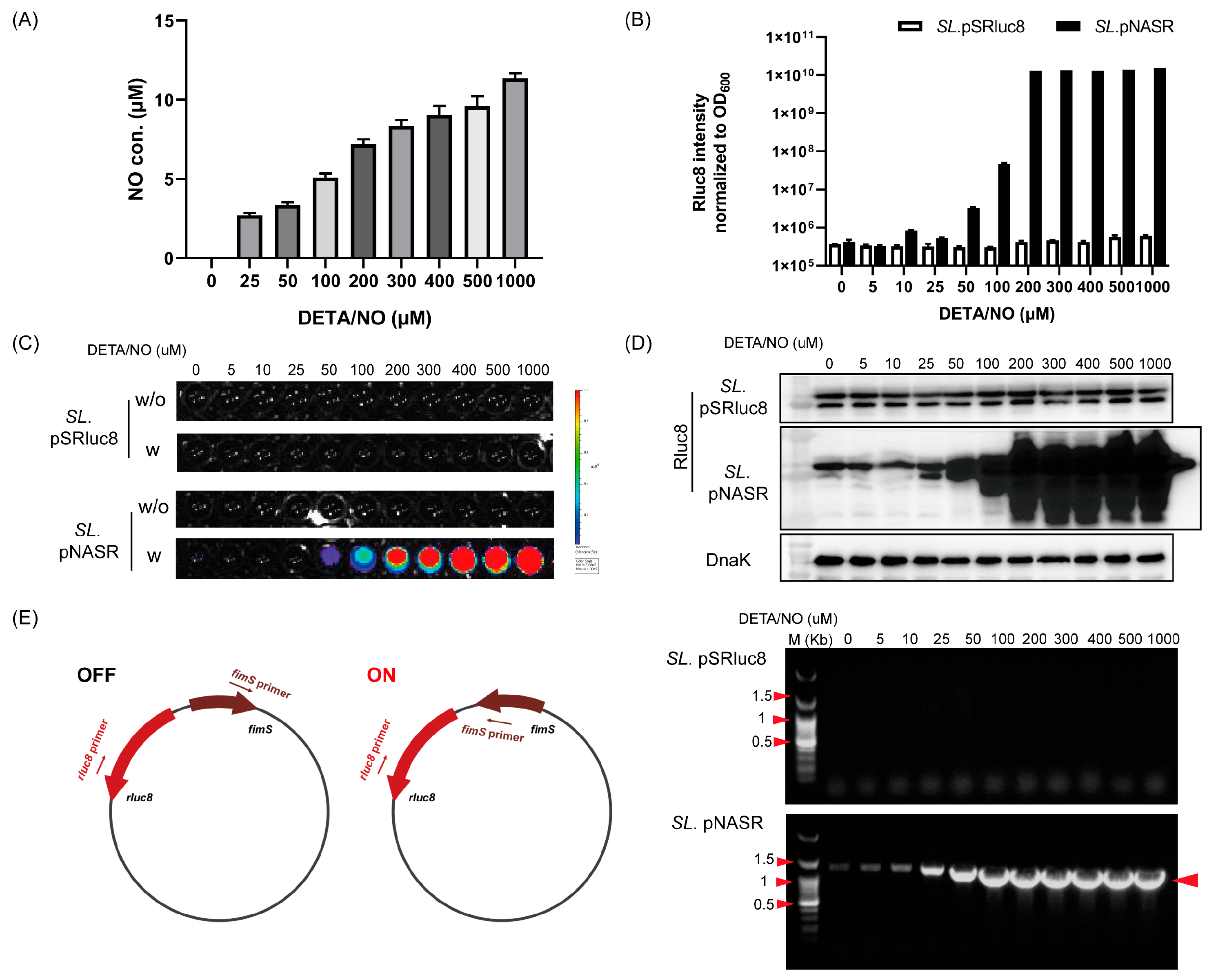
2.3. Analysis of Nitric Oxide (NO)-Sensing Gene Switching Events
2.4. PCR Analysis of Switch Inversion
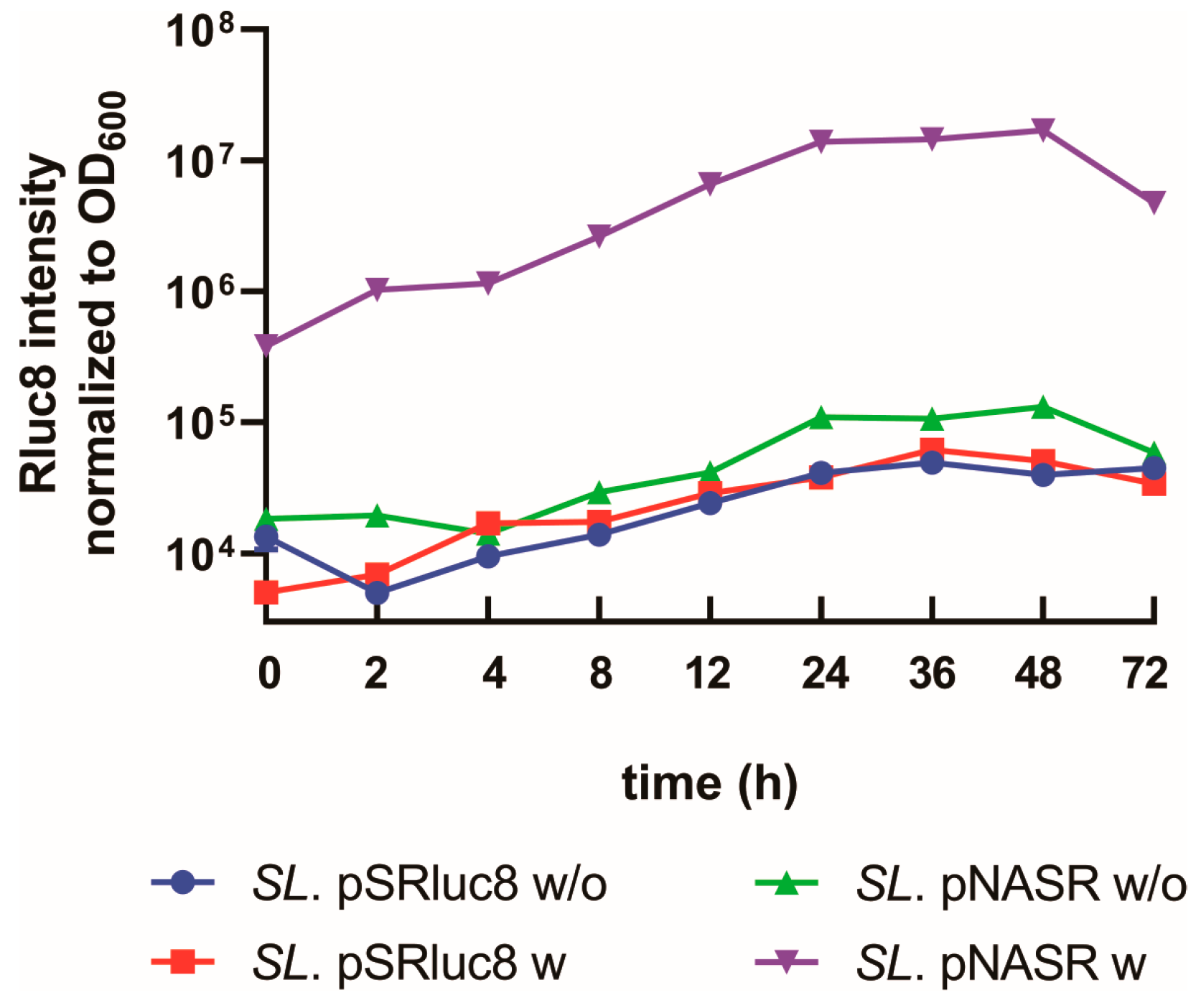
2.5. Measurement of Time-Dependent Switch Activation
2.6. Switch Inheritance Verification
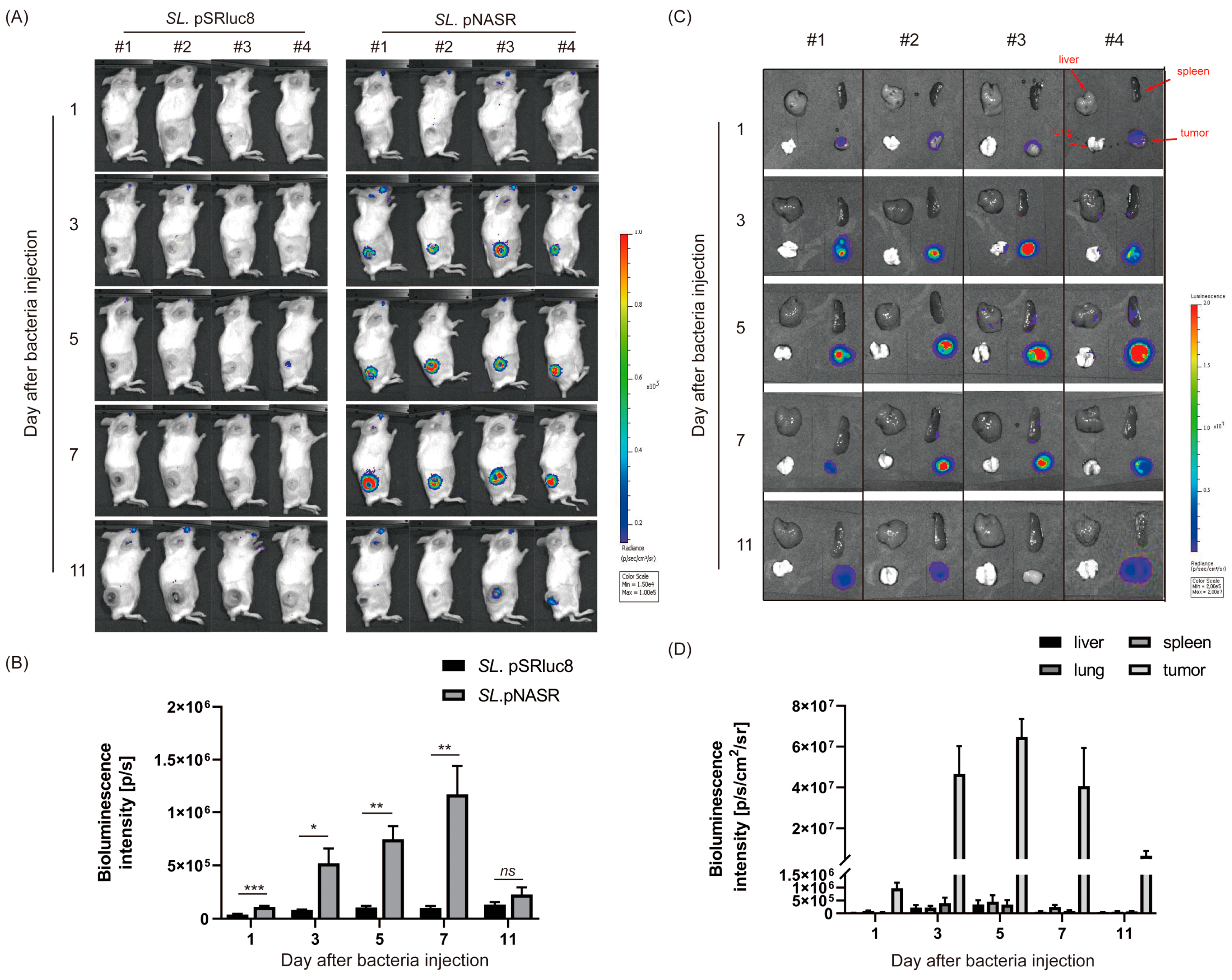
2.7. Analysis of In Vivo Switch Activation via Bioluminescence Imaging of Rluc8 Expression
2.8. In Vivo NO-Specific Switch Activation via Inducible Nitric Oxide Synthase (iNOS) Activity Inhibition
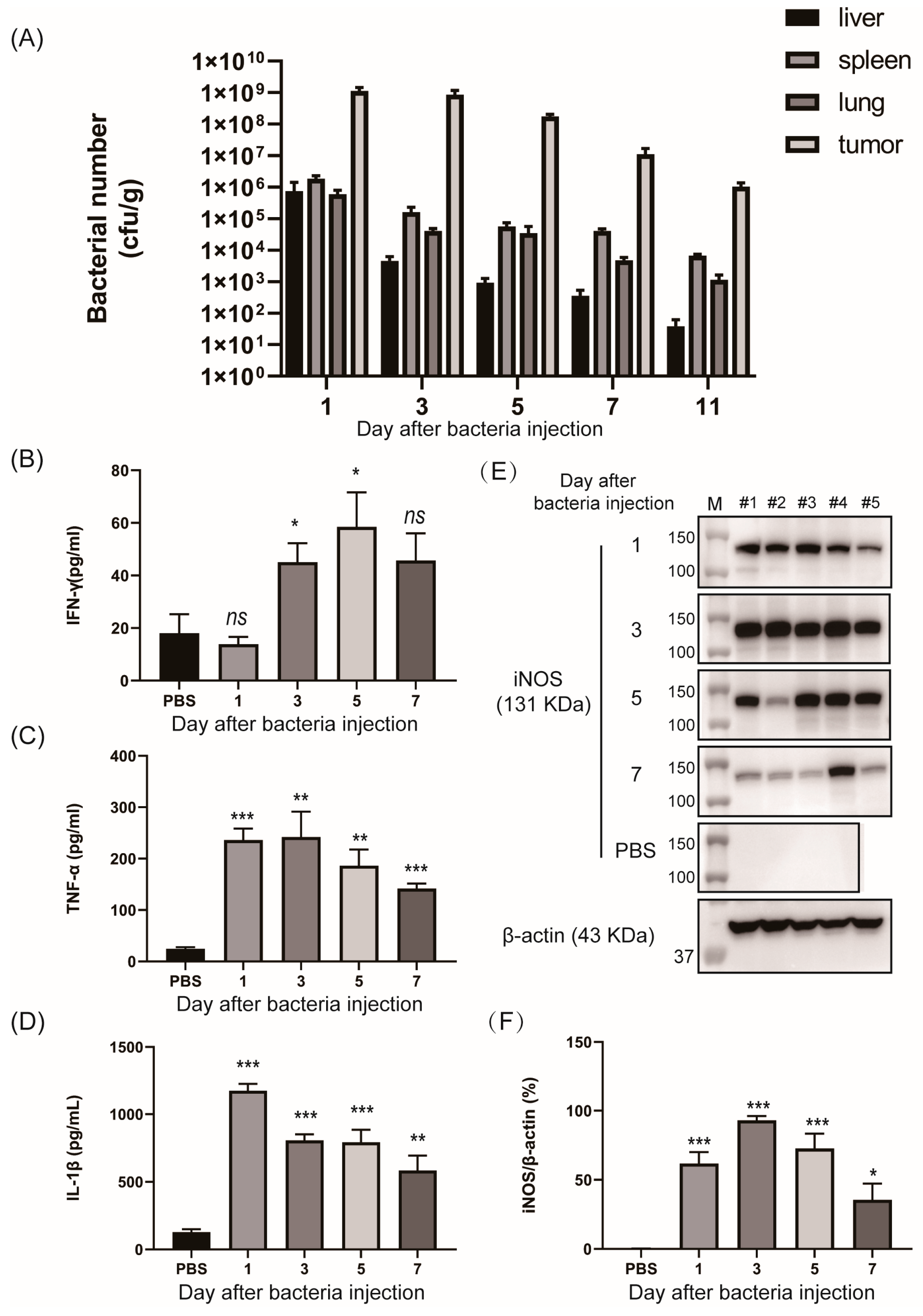
2.9. Measurement of iNOS Expression in Tumors Treated with Bacteria Bearing the NO-Sensing Gene Switch System
2.10. Measurement of Proinflammatory Cytokines by Enzyme-Linked Immunosorbent Assays (ELISA)
2.11. Immunofluorescence Staining of sfGFP in Tumors Treated with Bacteria Carrying the NO-Sensing Gene Switch System
2.12. Statistical Analysis
3. Results
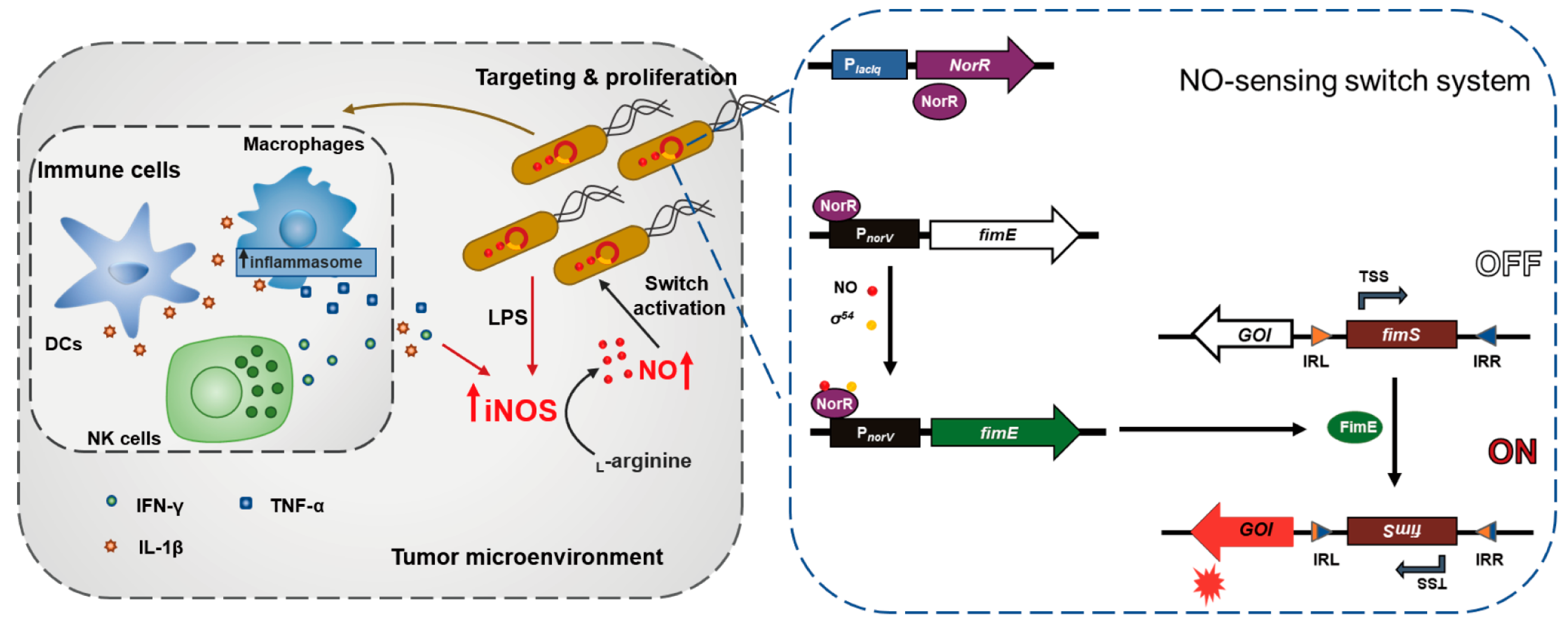
3.1. Study Design and Construction of Nitric Oxide (NO)-Sensing Gene Switch System
3.2. Characterization of Bacteria Transformed with the NO-Sensing Switch System
3.3. In Vivo Tumor-Specific Gene Delivery by Bacteria with NO-Sensing Gene Switch System
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- MacDonald, I.C.; Deans, T.L. Tools and applications in synthetic biology. Adv. Drug Deliv. Rev. 2016, 105 Pt A, 20–34. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; Wei, K.Y.; Smolke, C.D. Synthetic biology: Advancing the design of diverse genetic systems. Annu. Rev. Chem. Biomol. Eng. 2013, 4, 69–102. [Google Scholar] [CrossRef]
- McNerney, M.P.; Doiron, K.E.; Ng, T.L.; Chang, T.Z.; Silver, P.A. Theranostic cells: Emerging clinical applications of synthetic biology. Nat. Rev. Genet. 2021, 22, 730–746. [Google Scholar] [CrossRef]
- Cubillos-Ruiz, A.; Guo, T.; Sokolovska, A.; Miller, P.F.; Collins, J.J.; Lu, T.K.; Lora, J.M. Engineering living therapeutics with synthetic biology. Nat. Rev. Drug Discov. 2021, 20, 941–960. [Google Scholar] [CrossRef]
- Riglar, D.T.; Giessen, T.W.; Baym, M.; Kerns, S.J.; Niederhuber, M.J.; Bronson, R.T.; Kotula, J.W.; Gerber, G.K.; Way, J.C.; Silver, P.A. Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation. Nat. Biotechnol. 2017, 35, 653–658. [Google Scholar] [CrossRef]
- McCarthy, E.F. The Toxins of William B. Coley and the Treatment of Bone and Soft-tissue Sarcomas. Iowa Orthop. J. 2006, 26, 154–158. [Google Scholar]
- Kang, S.R.; Nguyen, D.H.; Yoo, S.W.; Min, J.J. Bacteria and bacterial derivatives as delivery carriers for immunotherapy. Adv. Drug Deliv. Rev. 2022, 181, 114085. [Google Scholar] [CrossRef] [PubMed]
- Forbes, N.S. Engineering the perfect (bacterial) cancer therapy. Nat. Rev. Cancer 2010, 10, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Duong, M.T.; Qin, Y.; You, S.H.; Min, J.J. Bacteria-cancer interactions: Bacteria-based cancer therapy. Exp. Mol. Med. 2019, 51, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.H.; Nguyen, V.H.; Jiang, S.N.; Park, S.H.; Tan, W.; Hong, S.H.; Shin, M.G.; Chung, I.J.; Hong, Y.; Bom, H.S.; et al. Two-step enhanced cancer immunotherapy with engineered Salmonella typhimurium secreting heterologous flagellin. Sci. Transl. Med. 2017, 9, eaak9537. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Phan, T.X.; Nguyen, V.H.; Dinh-Vu, H.V.; Zheng, J.H.; Yun, M.; Park, S.G.; Hong, Y.; Choy, H.E.; Szardenings, M.; et al. Salmonella typhimurium suppresses tumor growth via the pro-inflammatory cytokine interleukin-1β. Theranostics 2015, 5, 1328–1342. [Google Scholar] [CrossRef] [Green Version]
- Phan, T.X.; Nguyen, V.H.; Duong, M.T.Q.; Hong, Y.; Choy, H.E.; Min, J.J. Activation of inflammasome by attenuated Salmonella typhimurium in bacteria-mediated cancer therapy. Microbiol. Immunol. 2015, 59, 664–675. [Google Scholar] [CrossRef]
- Tan, W.; Duong, M.T.Q.; Zuo, C.; Qin, Y.; Zhang, Y.; Guo, Y.; Hong, Y.; Zheng, J.H.; Min, J.J. Targeting of pancreatic cancer cells and stromal cells using engineered oncolytic Salmonella typhimurium. Mol. Ther. 2022, 30, 662–671. [Google Scholar] [CrossRef]
- Hernandez-Luna, M.A.; Luria-Perez, R. Cancer immunotherapy: Priming the host immune response with live attenuated Salmonella enterica. J. Immunol. Res. 2018, 16, 2984247. [Google Scholar] [CrossRef]
- Al-Saafeen, B.H.; Fernandez-Cabezudo, M.J.; Al-Ramadi, B.K. Integration of Salmonella into combination cancer therapy. Cancers 2021, 13, 3228. [Google Scholar] [CrossRef]
- Guo, Y.; Chen, Y.; Liu, X.; Min, J.J.; Tan, W.; Zheng, J.H. Targeted cancer immunotherapy with genetically engineered oncolytic Salmonella typhimurium. Cancer Lett. 2020, 469, 102–110. [Google Scholar] [CrossRef]
- Lundberg, J.O.; Weitzberg, E. Nitric oxide signaling in health and disease. Cell 2022, 185, 2853–5878. [Google Scholar] [CrossRef] [PubMed]
- Bogdan, C. Nitric oxide synthase in innate and adaptive immunity: An update. Trends Immunol. 2015, 36, 161–178. [Google Scholar] [CrossRef]
- Nath, N.; Kashfi, K. Tumor associated macrophages and ‘NO’. Biochem. Pharmacol. 2020, 176, 113899. [Google Scholar] [CrossRef]
- Cinelli, M.A.; Do, H.T.; Miley, G.P.; Silverman, R.B. Inducible nitric oxide synthase: Regulation, structure, and inhibition. Med. Res. Rev. 2020, 40, 158–189. [Google Scholar] [CrossRef]
- Sharma, J.N.; Al-Omran, A.; Parvathy, S.S. Role of nitric oxide in inflammatory diseases. Inflammopharmacology 2007, 15, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Barak, Y.; Schreiber, F.; Thorne, S.H.; Contag, C.H.; Debeer, D.; Matin, A. Role of nitric oxide in Salmonella typhimurium-mediated cancer cell killing. BMC Cancer 2010, 10, 146. [Google Scholar] [CrossRef] [PubMed]
- Tucker, N.P.; D’Autreaux, B.; Spiro, S.; Dixon, R. Mechanism of transcriptional regulation by the Escherichia coli nitric oxide sensor NorR. Biochem. Soc. Trans. 2006, 34 Pt 1, 191–194. [Google Scholar] [CrossRef] [PubMed]
- Bush, M.; Ghosh, T.; Tucker, N.; Zhang, X.; Dixon, R. Transcriptional regulation by the dedicated nitric oxide sensor, NorR: A route towards NO detoxification. Biochem. Soc. Trans. 2011, 39, 289–293. [Google Scholar] [CrossRef]
- Tucker, N.P.; D’Autreaux, B.; Yousafzai, F.K.; Fairhurst, S.A.; Spiro, S.; Dixon, R. Analysis of the Nitric Oxide-sensing Non-heme Iron Center in the NorR Regulatory Protein. J. Biol. Chem. 2008, 283, 908–918. [Google Scholar] [CrossRef]
- Justino, M.C.; Goncalves, V.M.; Saraiva, L.M. Binding of NorR to three DNA sites is essential for promoter activation of the flavorubredoxin gene, the nitric oxide reductase of Escherichia coli. Biochem. Biophys. Res. Commun. 2005, 328, 540–544. [Google Scholar] [CrossRef]
- Trzilova, D.; Tamayo, R. Site-Specific Recombination—How Simple DNA Inversions Produce Complex Phenotypic Heterogeneity in Bacterial Populations. Trends Genet. 2021, 37, 59–72. [Google Scholar] [CrossRef]
- Harbaugh, S.V.; Goodson, M.S.; Dillon, K.; Zabarnick, S.; Kelley-Loughnane, N. Riboswitch-based reversible dual color sensor. ACS Synth. Biol. 2017, 6, 766–781. [Google Scholar] [CrossRef]
- Gally, D.L.; Leathart, J.; Blomfield, I.C. Interaction of FimB and FimE with the fim switch that controls the phase variation of type 1 fimbriae in Escherichia coli K-12. Mol. Microbiol. 1996, 21, 725–738. [Google Scholar] [CrossRef]
- Zhang, H.; Susanto, T.T.; Wan, Y.; Chen, S.L. Comprehensive mutagenesis of the fimS promoter regulatory switch reveals novel regulation of type 1 pili in uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 2016, 113, 4182–4187. [Google Scholar] [CrossRef]
- Blomfield, I.; van der Woude, M. Regulation of fimbrial expression. EcoSal Plus 2007, 2. [Google Scholar] [CrossRef] [PubMed]
- Holden, N.; Blomfield, I.C.; Uhlin, B.E.; Totsika, M.; Kulasekara, D.H.; Gally, D.L. Comparative analysis of FimB and FimE recombinase activity. Microbiology 2007, 153 Pt 12, 4138–4149. [Google Scholar] [CrossRef] [PubMed]
- Archer, E.J.; Robinson, A.B.; Suel, G.M. Engineered E. coli that detect and respond to gut inflammation through nitric oxide sensing. ACS Synth. Biol. 2012, 1, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Kim, H.J.; Kim, E.Y.; Shin, M.; Lee, H.C.; Hong, Y.; Rhee, J.H.; Yoon, H.; Ryu, S.; Lim, S.; et al. ppGpp-dependent stationary phase induction of genes on Salmonella pathogenicity island 1. J. Biol. Chem. 2004, 279, 34183–34190. [Google Scholar] [CrossRef] [PubMed]
- Loening, A.M.; Fenn, T.D.; Wu, A.M.; Gambhir, S.S. Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng. Des. Sel. 2006, 19, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.H.; Kim, H.S.; Ha, J.M.; Hong, Y.; Choy, H.E.; Min, J.J. Genetically engineered Salmonella typhimurium as an imageable therapeutic probe for cancer. Cancer Res. 2010, 70, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Thangam, R.; You, S.H.; Sultonova, R.D.; Venu, A.; Min, J.J.; Hong, Y. Engineering calreticulin-targeting monobodies to detect immunogenic cell death in cancer chemotherapy. Cancers 2021, 13, 2801. [Google Scholar] [CrossRef]
- Prindle, A.; Samayoa, P.; Razinkov, I.; Danino, T.; Tsimring, L.S.; Hasty, J. A sensing array of radically coupled genetic ‘biopixels’. Nature 2011, 481, 39–44. [Google Scholar] [CrossRef]
- Courbet, A.; Endy, D.; Renard, E.; Molina, F.; Bonnet, J. Detection of pathological biomarkers in human clinical samples via amplifying genetic switches and logic gates. Sci. Transl. Med. 2015, 7, 289ra83. [Google Scholar] [CrossRef]
- Moore, S.J.; Lai, H.E.; Kelwick, R.J.; Chee, S.M.; Bell, D.J.; Polizzi, K.M.; Freemont, P.S. EcoFlex: A Multifunctional MoClo Kit for E. coli Synthetic Biology. ACS Synth. Biol. 2016, 5, 1059–1069. [Google Scholar] [CrossRef] [PubMed]
- Garvey, E.P.; Oplinger, J.A.; Furfine, E.S.; Kiff, R.J.; Laszlo, F.; Whittle, B.J.; Knowles, R.G. 1400W is a slow, tight binding, and highly selective inhibitor of inducible nitric-oxide synthase in vitro and in vivo. J. Biol. Chem. 1997, 272, 4959–4963. [Google Scholar] [CrossRef] [Green Version]
- Omer, R.; Mohsin, M.Z.; Mohsin, A.; Mushtaq, B.S.; Huang, X.; Guo, M.; Zhuang, Y.; Huang, J. Engineered bacteria-based living materials for biotherapeutic applications. Front. Bioeng. Biotechnol. 2022, 10, 870675. [Google Scholar] [CrossRef]
- Guzman, L.M.; Belin, D.; Carson, M.J.; Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995, 177, 4121–4130. [Google Scholar] [CrossRef]
- Baron, U.; Freundlieb, S.; Gossen, M.; Bujard, H. Co-regulation of two gene activities by tetracycline via a bidirectional promoter. Nucleic Acids Res. 1995, 23, 3605–3606. [Google Scholar] [CrossRef] [PubMed]
- Turner, P.V.; Brabb, T.; Pekow, C.; Vasbinder, M.A. Administration of substances to laboratory animals: Routes of administration and factors to consider. J. Am. Assoc. Lab. Anim. Sci. 2011, 50, 600–613. [Google Scholar] [PubMed]
- Ryan, R.M.; Green, J.; Williams, P.J.; Tazzyman, S.; Hunt, S.; Harmey, J.H.; Kehoe, S.C.; Lewis, C.E. Bacterial delivery of a novel cytolysin to hypoxic areas of solid tumors. Gene Ther. 2009, 16, 329–339. [Google Scholar] [CrossRef]
- Flentie, K.; Kocher, B.; Gammon, S.T.; Novack, D.V.; McKinney, J.S.; Piwnica-Worms, D. A bioluminescent transposon reporter-trap identifies tumor-specific microenvironment-induced promoters in Salmonella for conditional bacterial-based tumor therapy. Cancer Discov. 2012, 2, 624–637. [Google Scholar] [CrossRef]
- Ozdemir, T.; Fedorec, A.J.H.; Danino, T.; Barnes, C.P. Synthetic biology and engineered live biotherapeutics: Toward increasing system complexity. Cell Syst. 2018, 7, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Riglar, D.T.; Silver, P.A. Engineering bacteria for diagnostic and therapeutic applications. Nat. Rev. Microbiol. 2018, 16, 214–225. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, Y.; You, S.-H.; Zhang, Y.; Venu, A.; Hong, Y.; Min, J.-J. Genetic Programming by Nitric Oxide-Sensing Gene Switch System in Tumor-Targeting Bacteria. Biosensors 2023, 13, 266. https://doi.org/10.3390/bios13020266
Qin Y, You S-H, Zhang Y, Venu A, Hong Y, Min J-J. Genetic Programming by Nitric Oxide-Sensing Gene Switch System in Tumor-Targeting Bacteria. Biosensors. 2023; 13(2):266. https://doi.org/10.3390/bios13020266
Chicago/Turabian StyleQin, Yeshan, Sung-Hwan You, Ying Zhang, Akhil Venu, Yeongjin Hong, and Jung-Joon Min. 2023. "Genetic Programming by Nitric Oxide-Sensing Gene Switch System in Tumor-Targeting Bacteria" Biosensors 13, no. 2: 266. https://doi.org/10.3390/bios13020266
APA StyleQin, Y., You, S.-H., Zhang, Y., Venu, A., Hong, Y., & Min, J.-J. (2023). Genetic Programming by Nitric Oxide-Sensing Gene Switch System in Tumor-Targeting Bacteria. Biosensors, 13(2), 266. https://doi.org/10.3390/bios13020266



