Development of a Novel Phagomagnetic-Assisted Isothermal DNA Amplification System for Endpoint Electrochemical Detection of Listeria monocytogenes
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Solutions
2.2. Microorganisms and Inoculum Preparation
2.2.1. Bacterial Strains and Culture Conditions
2.2.2. Bacteriophage Titration by the Double-Layer Method
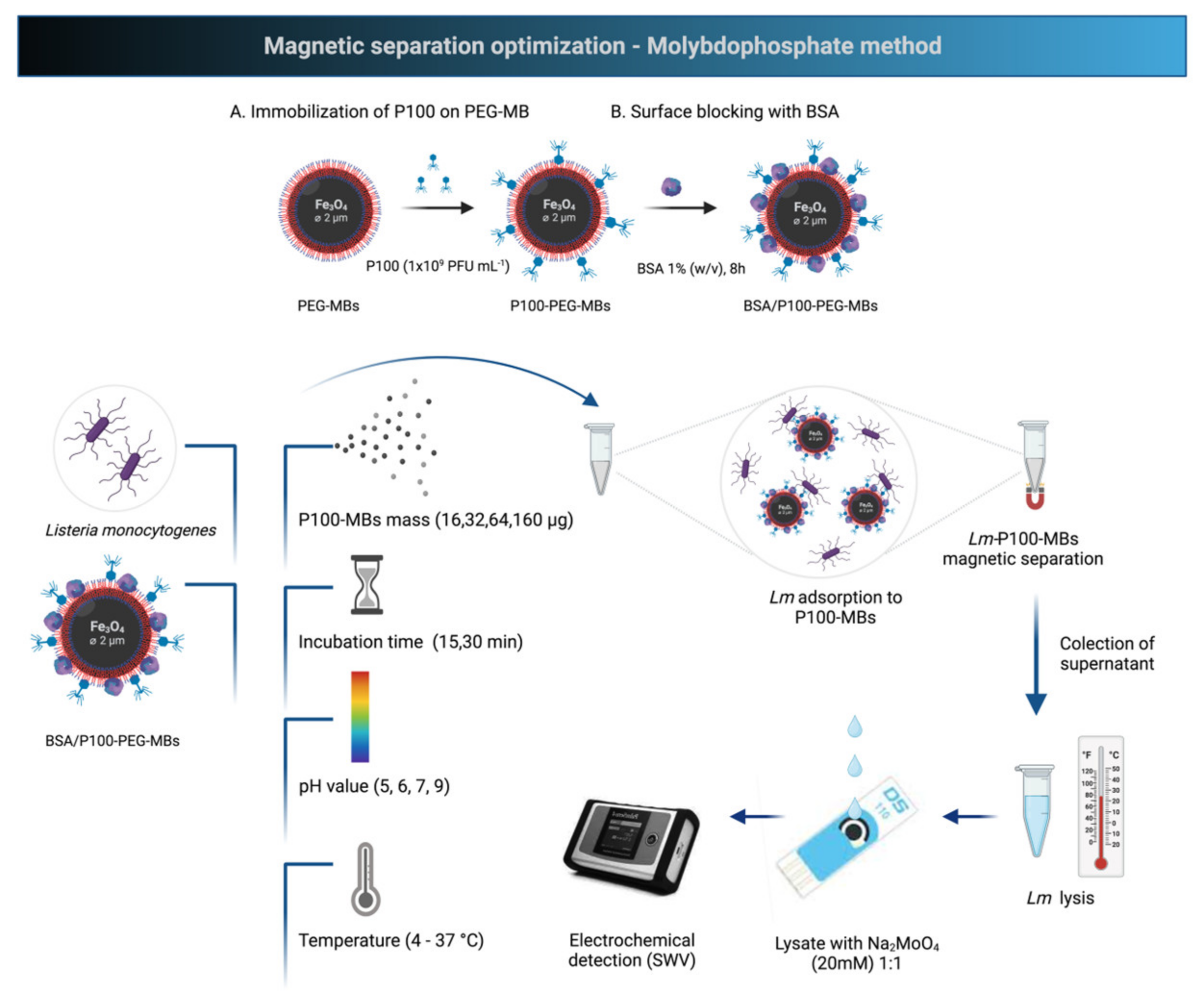
2.3. Preparation of P100 Modified Magnetic Particles
2.4. Phagomagnetic Separation Protocol: Capture Efficiency
2.5. Development of a Novel LAMP Assay Targeting prfA
2.5.1. Preparation of Genomic DNA
2.5.2. Design of the L. monocytogenes-Specific LAMP Primers
2.5.3. Optimization of LAMP Reaction System
2.5.4. Evaluation of LAMP Assay Specificity
2.5.5. Determination of Analytical Sensitivity
2.5.6. PCR Targeting prfA
2.6. Validation of the Applicability of the Developed Method in Pasteurized Milk
2.6.1. Application of Phagomagnetic-Assisted LAMP Method in Milk Samples
2.6.2. Endpoint Electrochemical Detection
2.7. Statistical Analysis
3. Results and Discussion
3.1. Effect of Incubation Solution pH and Immobilization Method
3.2. Influence of Non-Specific Adsorption: Surface Blocking Step Optimization
3.3. Phagomagnetic Separation Protocol Optimization
3.4. LAMP Assay Targeting prfA
3.4.1. Primers Efficiency Evaluation
3.4.2. Assessment of LAMP Specificity—Inclusivity
3.4.3. Evaluation of LAMP Specificity—Exclusivity
3.4.4. Evaluation of LAMP Analytical Performance (LOD95)
3.5. Applicability of the Combined Detection System in Pasteurized Milk
3.5.1. Phagomagnetic Particles Performance in Pasteurized Milk
3.5.2. Phagomagnetic-Assisted LAMP Assay
3.5.3. Detection Limit of the Phagomagnetic-Assisted LAMP Assay in Milk
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union One Health 2021 Zoonoses Report. EFSA J. 2022, 20, e07666. [Google Scholar] [CrossRef]
- Belias, A.; Sullivan, G.; Wiedmann, M.; Ivanek, R. Factors That Contribute to Persistent Listeria in Food Processing Facilities and Relevant Interventions: A Rapid Review. Food Control 2022, 133, 108579. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, S.; Chen, X.; Qu, C. Review Controlling Listeria monocytogenes in Ready-to-Eat Meat and Poultry Products: An Overview of Outbreaks, Current Legislations, Challenges, and Future Prospects. Trends Food Sci. Technol. 2021, 116, 24–35. [Google Scholar] [CrossRef]
- EFSA Panel on Biological Hazards (BIOHAZ); Ricci, A.; Allende, A.; Bolton, D.; Chemaly, M.; Davies, R.; Fernández Escámez, P.S.; Girones, R.; Herman, L.; Koutsoumanis, K.; et al. Listeria monocytogenes Contamination of Ready-to-Eat Foods and the Risk for Human Health in the EU. EFSA J. 2018, 16, e05134. [Google Scholar] [CrossRef]
- Churchill, K.J.; Sargeant, J.M.; Farber, J.M.; O’Connor, A.M. Prevalence of Listeria monocytogenes in Select Ready-to-Eat Foods-Deli Meat, Soft Cheese, and Packaged Salad: A Systematic Review and Meta-Analysis. J. Food Prot. 2019, 82, 344–357. [Google Scholar] [CrossRef]
- Sarno, E.; Pezzutto, D.; Rossi, M.; Liebana, E.; Rizzi, V. A Review of Significant European Foodborne Outbreaks in the Last Decade. J. Food Prot. 2021, 84, 2059–2070. [Google Scholar] [CrossRef] [PubMed]
- Desai, A.N.; Anyoha, A.; Madoff, L.C.; Lassmann, B. Changing Epidemiology of Listeria monocytogenes Outbreaks, Sporadic Cases, and Recalls Globally: A Review of ProMED Reports from 1996 to 2018. Int. J. Infect. Dis. 2019, 84, 48–53. [Google Scholar] [CrossRef]
- Wei, Q.; Wang, X.; Sun, D.-W.; Pu, H. Rapid Detection and Control of Psychrotrophic Microorganisms in Cold Storage Foods: A Review. Trends Food Sci. Technol. 2019, 86, 453–464. [Google Scholar] [CrossRef]
- Ziyaina, M.; Rasco, B.; Sablani, S.S. Rapid Methods of Microbial Detection in Dairy Products. Food Control 2020, 110, 107008. [Google Scholar] [CrossRef]
- Fogaça, M.B.T.; Bhunia, A.K.; Lopes-Luz, L.; de Almeida, E.P.R.P.; Vieira, J.D.G.; Bührer-Sékula, S. Antibody- and Nucleic Acid-Based Lateral Flow Immunoassay for Listeria monocytogenes Detection. Anal. Bioanal. Chem. 2021, 413, 4161–4180. [Google Scholar] [CrossRef]
- Yang, H.; Qu, L.; Wimbrow, A.N.; Jiang, X.; Sun, Y. Rapid Detection of Listeria monocytogenes by Nanoparticle-Based Immunomagnetic Separation and Real-Time PCR. Int. J. Food Microbiol. 2007, 118, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, D.; Kinchla, A.J.; Sela, D.A.; Nugen, S.R. Rapid Screening of Waterborne Pathogens Using Phage-Mediated Separation Coupled with Real-Time PCR Detection. Anal. Bioanal. Chem. 2016, 408, 4169–4178. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Mori, Y.; Tomita, N.; Kanda, H. Loop-Mediated Isothermal Amplification (LAMP): Principle, Features, and Future Prospects. J. Microbiol. 2015, 53, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Dai, Z.; Tian, X.; Jiang, X. Detection of Listeria monocytogenes Based on Combined Aptamers Magnetic Capture and Loop-Mediated Isothermal Amplification. Food Control 2018, 85, 443–452. [Google Scholar] [CrossRef]
- Lee, J.E.; Kim, S.A.; Mun, H.; Kim, S.R.; Ha, K.S.; Shim, W.B. A Rapid and Colorimetric Loop-Mediated Isothermal Amplification (LAMP) Based on HRP-Mimicking Molecular Beacon for the Detection of Major 6 Listeria Species in Enoki Mushroom. Food Control 2022, 133, 108569. [Google Scholar] [CrossRef]
- Wachiralurpan, S.; Phung-On, I.; Chanlek, N.; Areekit, S.; Chansiri, K.; Lieberzeit, P.A. In-Situ Monitoring of Real-Time Loop-Mediated Isothermal Amplification with QCM: Detecting Listeria monocytogenes. Biosensors 2021, 11, 308. [Google Scholar] [CrossRef]
- Wachiralurpan, S.; Sriyapai, T.; Areekit, S.; Kaewphinit, T.; Sriyapai, P.; Santiwatanakul, S.; Chansiri, K. Development of a Rapid Screening Test for Listeria monocytogenes in Raw Chicken Meat Using Loop-Mediated Isothermal Amplification (LAMP) and Lateral Flow Dipstick (LFD). Food Anal. Methods 2017, 10, 3763–3772. [Google Scholar] [CrossRef]
- Kim, J.-H.; Oh, S.-W. Pretreatment Methods for Nucleic Acid-Based Rapid Detection of Pathogens in Food: A Review. Food Control 2021, 121, 107575. [Google Scholar] [CrossRef]
- Shan, S.; Zhong, Z.; Lai, W.; Xiong, Y.; Cui, X.; Liu, D. Immunomagnetic Nanobeads Based on a Streptavidin-Biotin System for the Highly Efficient and Specific Separation of Listeria monocytogenes. Food Control 2014, 45, 138–142. [Google Scholar] [CrossRef]
- Roumani, F.; Azinheiro, S.; Carvalho, J.; Prado, M.; Garrido-Maestu, A. Loop-Mediated Isothermal Amplification Combined with Immunomagnetic Separation and Propidium Monoazide for the Specific Detection of Viable Listeria monocytogenes in Milk Products, with an Internal Amplification Control. Food Control 2021, 125, 107975. [Google Scholar] [CrossRef]
- Jevtuševskaja, J.; Uusna, J.; Andresen, L.; Krõlov, K.; Laanpere, M.; Grellier, T.; Tulp, I.; Langel, Ü. Combination with Antimicrobial Peptide Lyses Improves Loop-Mediated Isothermal Amplification Based Method for Chlamydia trachomatis Detection Directly in Urine Sample. BMC Infect. Dis. 2016, 16, 329. [Google Scholar] [CrossRef]
- Liébana, S.; Spricigo, D.A.; Cortés, M.P.; Barbé, J.; Llagostera, M.; Alegret, S.; Pividori, M.I. Phagomagnetic Separation and Electrochemical Magneto-Genosensing of Pathogenic Bacteria. Anal. Chem. 2013, 85, 3079–3086. [Google Scholar] [CrossRef]
- Fernandes, E.; Martins, V.C.; Nóbrega, C.; Carvalho, C.M.; Cardoso, F.A.; Cardoso, S.; Dias, J.; Deng, D.; Kluskens, L.D.; Freitas, P.P.; et al. A Bacteriophage Detection Tool for Viability Assessment of Salmonella Cells. Biosens. Bioelectron. 2014, 52, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Tlili, C.; Sokullu, E.; Safavieh, M.; Tolba, M.; Ahmed, M.U.; Zourob, M. Bacteria Screening, Viability, And Confirmation Assays Using Bacteriophage-Impedimetric/Loop-Mediated Isothermal Amplification Dual-Response Biosensors. Anal. Chem. 2013, 85, 4893–4901. [Google Scholar] [CrossRef]
- Kropinski, A.M.; Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of Bacteriophages by Double Agar Overlay Plaque Assay. Methods Mol. Biol. 2009, 501, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Ramasamy, R.P. Isolation and Separation of Listeria monocytogenes Using Bacteriophage P100-Modified Magnetic Particles. Colloids Surf. B Biointerfaces 2019, 175, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Liu, S.; Yang, M.; Rasooly, A. Amperometric Genosensor for Culture Independent Bacterial Count. Sens. Actuators B Chem. 2019, 299, 126944. [Google Scholar] [CrossRef]
- Hardinge, P.; Murray, J.A.H. Full Dynamic Range Quantification Using Loop-Mediated Amplification (LAMP) by Combining Analysis of Amplification Timing and Variance between Replicates at Low Copy Number. Sci. Rep. 2020, 10, 916. [Google Scholar] [CrossRef]
- Simon, M.C.; Gray, D.I.; Cook, N. DNA Extraction and PCR Methods for the Detection of Listeria monocytogenes in Cold-Smoked Salmon. Appl. Environ. Microbiol. 1996, 62, 822–824. [Google Scholar] [CrossRef]
- D’Agostino, M.; Wagner, M.; Vazquez-Boland, J.A.; Kuchta, T.; Karpiskova, R.; Hoorfar, J.; Novella, S.; Scortti, M.; Ellison, J.; Murray, A.; et al. A Validated PCR-Based Method to Detect Listeria monocytogenes Using Raw Milk as a Food Model-towards an International Standard. J. Food Prot. 2004, 67, 1646–1655. [Google Scholar] [CrossRef]
- ISO 11290-1:2017; Microbiology of the Food Chain—Horizontal Method for the Detection and Enumeration of Listeria monocytogenes and of Listeria spp.—Part 1: Detection Method. ISO (International Organization for Standardization): Geneva, Switzerland, 2017.
- Bergis, H.; Bonanno, L.; Asséré, A.; Lombard, B. EURL Lm Technical Guidance Document on Challenge Tests and Durability Studies for Assessing Shelf-Life of Ready-to-Eat Foods Related to Listeria monocytogenes. EURL Lm 2021, 4, 60. [Google Scholar]
- O’Connell, L.; Marcoux, P.R.; Roupioz, Y. Strategies for Surface Immobilization of Whole Bacteriophages: A Review. ACS Biomater. Sci. Eng. 2021, 7, 1987–2014. [Google Scholar] [CrossRef] [PubMed]
- Richter, Ł.; Bielec, K.; Leśniewski, A.; Łoś, M.; Paczesny, J.; Hołyst, R. Dense Layer of Bacteriophages Ordered in Alternating Electric Field and Immobilized by Surface Chemical Modification as Sensing Element for Bacteria Detection. ACS Appl. Mater. Interfaces 2017, 9, 19622–19629. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, S. DLS and Zeta Potential—What They Are and What They Are Not? J. Control. Release 2016, 235, 337–351. [Google Scholar] [CrossRef] [PubMed]
- Klumpp, J.; Loessner, M.J. Listeria Phages: Genomes, Evolution, and Application. Bacteriophage 2013, 3, e26861. [Google Scholar] [CrossRef]
- Dika, C.; Duval, J.F.L.; Francius, G.; Perrin, A.; Gantzer, C. Isoelectric Point Is an Inadequate Descriptor of MS2, Phi X 174 and PRD1 Phages Adhesion on Abiotic Surfaces. J. Colloid Interface Sci. 2015, 446, 327–334. [Google Scholar] [CrossRef]
- Sommer, J.; Bromberger, B.; Robben, C.; Kalb, R.; Rossmanith, P.; Mester, P.-J. Liquid-Liquid Extraction of Viral Particles with Ionic Liquids. Sep. Purif. Technol. 2021, 254, 117591. [Google Scholar] [CrossRef]
- Heffron, J.; Mayer, B.K. Virus Isoelectric Point Estimation: Theories and Methods. Appl. Environ. Microbiol. 2021, 87, e02319-20. [Google Scholar] [CrossRef]
- Farooq, U.; Ullah, M.W.; Yang, Q.; Aziz, A.; Xu, J.; Zhou, L.; Wang, S. High-Density Phage Particles Immobilization in Surface-Modified Bacterial Cellulose for Ultra-Sensitive and Selective Electrochemical Detection of Staphylococcus aureus. Biosens. Bioelectron. 2020, 157, 112163. [Google Scholar] [CrossRef]
- Filik, K.; Szermer-Olearnik, B.; Oleksy, S.; Brykała, J.; Brzozowska, E. Bacteriophage Tail Proteins as a Tool for Bacterial Pathogen Recognition-A Literature Review. Antibiotics 2022, 11, 555. [Google Scholar] [CrossRef]
- Xu, J.; Zhao, C.; Chau, Y.; Lee, Y.-K. The Synergy of Chemical Immobilization and Electrical Orientation of T4 Bacteriophage on a Micro Electrochemical Sensor for Low-Level Viable Bacteria Detection via Differential Pulse Voltammetry. Biosens. Bioelectron. 2020, 151, 111914. [Google Scholar] [CrossRef] [PubMed]
- Armanious, A.; Aeppli, M.; Jacak, R.; Refardt, D.; Sigstam, T.; Kohn, T.; Sander, M. Viruses at Solid-Water Interfaces: A Systematic Assessment of Interactions Driving Adsorption. Environ. Sci. Technol. 2016, 50, 732–743. [Google Scholar] [CrossRef] [PubMed]
- Fister, S.; Robben, C.; Witte, A.K.; Schoder, D.; Wagner, M.; Rossmanith, P. Influence of Environmental Factors on Phage-Bacteria Interaction and on the Efficacy and Infectivity of Phage P100. Front. Microbiol. 2016, 7, 1152. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wang, H.; Cao, S.; Xiang, W.; Li, T.; Yang, M. Electrochemical Determination of the Activity and Inhibition of Telomerase Based on the Interaction of DNA with Molybdate. Microchim. Acta 2019, 186, 96. [Google Scholar] [CrossRef]
- Tan, X.; Zhang, L.; Deng, X.; Miao, L.; Li, H.; Zheng, G. Redox Active Molybdophosphate Produced by Cu3(PO4)2 Nanospheres for Enhancing Enzyme-Free Electrochemical Immunoassay of C-Reactive Protein. New J. Chem. 2017, 41, 11867–11871. [Google Scholar] [CrossRef]
- Hou, Y.; Chen, J.; Xie, B.; Li, T.; Yang, M. Electrochemical Assay for Analysis of Circulation Tumor Cells Based on Isolation of the Cell with Magnetic Nanoparticles and Reaction of DNA with Molybdate. Microchim. Acta 2020, 187, 420. [Google Scholar] [CrossRef] [PubMed]
- Cho, A.-R.R.; Dong, H.-J.J.; Seo, K.-H.H.; Cho, S. Development of a Loop-Mediated Isothermal Amplification Assay for Detecting Listeria monocytogenes prfA in Milk. Food Sci. Biotechnol. 2014, 23, 467–474. [Google Scholar] [CrossRef]
- Cooray, K.J.; Nishibori, T.; Xiong, H.; Matsuyama, T.; Fujita, M.; Mitsuyama, M. Detection of Multiple Virulence-Associated Genes of Listeria monocytogenes by PCR in Artificially Contaminated Milk Samples. Appl. Environ. Microbiol. 1994, 60, 3023–3026. [Google Scholar] [CrossRef] [PubMed]
- Orsi, R.H.; Wiedmann, M. Characteristics and Distribution of Listeria spp., Including Listeria Species Newly Described since 2009. Appl. Microbiol. Biotechnol. 2016, 100, 5273–5287. [Google Scholar] [CrossRef]
- Den Bakker, H.C.; Warchocki, S.; Wright, E.M.; Allred, A.F.; Ahlstrom, C.; Manuel, C.S.; Stasiewicz, M.J.; Burrell, A.; Roof, S.; Strawn, L.K.; et al. Listeria floridensis sp. nov., Listeria aquatica sp. nov., Listeria cornellensis sp. nov., Listeria riparia sp. nov. and Listeria grandensis sp. nov., from Agricultural and Natural Environments. Int. J. Syst. Evol. Microbiol. 2014, 64, 1882–1889. [Google Scholar] [CrossRef] [PubMed]
- Rossmanith, P.; Mester, P.; Wagner, M.; Schoder, D. Demonstration of the Effective Performance of a Combined Enrichment/Real-Time PCR Method Targeting the prfA Gene of Listeria monocytogenes by Testing Fresh Naturally Contaminated Acid Curd Cheese. Lett. Appl. Microbiol. 2010, 51, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Rossmanith, P.; Krassnig, M.; Wagner, M.; Hein, I. Detection of Listeria monocytogenes in Food Using a Combined Enrichment/Real-Time PCR Method Targeting the prfA Gene. Res. Microbiol. 2006, 157, 763–771. [Google Scholar] [CrossRef]
- Wernars, K.; Heuvelman, K.; Notermans, S.; Domann, E.; Leimeister-Wächter, M.; Chakraborty, T. Suitability of the prfA Gene, Which Encodes a Regulator of Virulence Genes in Listeria monocytogenes, in the Identification of Pathogenic Listeria spp. Appl. Environ. Microbiol. 1992, 58, 765–768. [Google Scholar] [CrossRef]
- Jung, H.-J.; Park, S.-H.; Ha, S.-D.; Lee, K.-H.; Chung, D.H.; Kim, C.-H.; Kim, M.-G.; Kim, K.-Y.; Kim, K.-S. Species-Specific Detection of Listeria monocytogenes Using Polymerase Chain Reaction Assays Targeting the prfA Virulence Gene Cluster. Biosci. Biotechnol. Biochem. 2009, 73, 1412–1415. [Google Scholar] [CrossRef]
- Wachiralurpan, S.; Chansiri, K.; Lieberzeit, P.A. Direct Detection of Listeria monocytogenes DNA Amplification Products with Quartz Crystal Microbalances at Elevated Temperatures. Sens. Actuators B Chem. 2020, 308, 127678. [Google Scholar] [CrossRef]
- Wachiralurpan, S.; Sriyapai, T.; Areekit, S.; Sriyapai, P.; Thongphueak, D.; Santiwatanakul, S.; Chansiri, K. A One-Step Rapid Screening Test of Listeria monocytogenes in Food Samples Using a Real-Time Loop-Mediated Isothermal Amplification Turbidity Assay. Anal. Methods 2017, 9, 6403–6410. [Google Scholar] [CrossRef]
- Wachiralurpan, S.; Sriyapai, T.; Areekit, S.; Sriyapai, P.; Augkarawaritsawong, S.; Santiwatanakul, S.; Chansiri, K. Rapid Colorimetric Assay for Detection of Listeria monocytogenes in Food Samples Using LAMP Formation of DNA Concatemers and Gold Nanoparticle-DNA Probe Complex. Front. Chem. 2018, 6, 1–9. [Google Scholar] [CrossRef]
- Chen, Q.; Yao, C.; Yang, C.; Liu, Z.; Wan, S. Development of an In-Situ Signal Amplified Electrochemical Assay for Detection of Listeria monocytogenes with Label-Free Strategy. Food Chem. 2021, 358, 129894. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, Y.; Chu, J.; Xu, Z.; Zhong, Q. Development and Application of a Simple Loop-Mediated Isothermal Amplification Method on Rapid Detection of Listeria monocytogenes Strains. Mol. Biol. Rep. 2012, 39, 445–449. [Google Scholar] [CrossRef]
- Hoorfar, J.; Malorny, B.; Abdulmawjood, A.; Cook, N.; Wagner, M.; Fach, P. Practical Considerations in Design of Internal Amplification Controls for Diagnostic PCR Assays. J. Clin. Microbiol. 2004, 42, 1863–1868. [Google Scholar] [CrossRef]
- Rodríguez-Rubio, L.; García, P.; Rodríguez, A.; Billington, C.; Hudson, J.A.; Martínez, B. Listeriaphages and Coagulin C23 Act Synergistically to Kill Listeria monocytogenes in Milk under Refrigeration Conditions. Int. J. Food Microbiol. 2015, 205, 68–72. [Google Scholar] [CrossRef]
- García-Anaya, M.C.; Sepúlveda, D.R.; Rios-Velasco, C.; Zamudio-Flores, P.B.; Romo-Chacón, A.; Acosta-Muñiz, C.H. Stability of Listerial Bacteriophage A511 in Bovine Milk Fat Globules. Int. Dairy J. 2020, 103, 104627. [Google Scholar] [CrossRef]
- Bai, X.; Wang, Z.; Li, W.; Xiao, F.; Huang, J.; Xu, Q.; Xu, H. Rapid and accurate detection of Listeria monocytogenes in milk using ampicillin-mediated magnetic separation coupled with quantitative real-time PCR. Microchem. J. 2022, 183, 108063. [Google Scholar] [CrossRef]
- Swift, B.M.C.C.; Meade, N.; Barron, E.S.; Bennett, M.; Perehenic, T.; Hughes, V.; Stevenson, K.; Rees, C.E.D.D. The Development and Use of Actiphage® to Detect Viable Mycobacteria from Bovine Tuberculosis and Johne’s Disease-Infected Animals. Microb. Biotechnol. 2020, 13, 738–746. [Google Scholar] [CrossRef] [PubMed]
- Lau, H.Y.; Wu, H.; Wee, E.J.H.; Trau, M.; Wang, Y.; Botella, J.R. Specific and Sensitive Isothermal Electrochemical Biosensor for Plant Pathogen DNA Detection with Colloidal Gold Nanoparticles as Probes. Sci. Rep. 2017, 7, 38896. [Google Scholar] [CrossRef]
- Azek, F.; Grossiord, C.; Joannes, M.; Limoges, B.; Brossier, P. Hybridization Assay at a Disposable Electrochemical Biosensor for the Attomole Detection of Amplified Human Cytomegalovirus DNA. Anal. Biochem. 2000, 284, 107–113. [Google Scholar] [CrossRef]
- Azinheiro, S.; Roumani, F.; Prado, M.; Garrido-Maestu, A. Rapid Same-Day Detection of Listeria monocytogenes, Salmonella spp., and Escherichia coli O157 by Colorimetric LAMP in Dairy Products. Food Anal. Methods 2022, 15, 2959–2971. [Google Scholar] [CrossRef]
- Li, W.; Mao, R.; Yue, X.; Wu, J.; Wu, R.; Qiao, Y.; Peng, Q.; Shi, B.; Luo, Y.; Chen, X.; et al. Competitive Annealing Mediated Isothermal Amplification (CAMP) for Rapid and Simple Detection of Listeria monocytogenes in Milk. Food Control 2020, 117, 107347. [Google Scholar] [CrossRef]
- Wang, D.; Huo, G.; Ren, D.; Li, Y. Development and Evaluation of a Loop-Mediated Isothermal Amplification (LAMP) Method for Detecting Listeria monocytogenes in Raw Milk. J. Food Saf. 2010, 30, 251–262. [Google Scholar] [CrossRef]
- Teixeira, A.; Paris, J.L.; Roumani, F.; Diéguez, L.; Prado, M.; Espiña, B.; Abalde-Cela, S.; Garrido-Maestu, A.; Rodriguez-Lorenzo, L. Multifuntional Gold Nanoparticles for the SERS Detection of Pathogens Combined with a LAMP-in-Microdroplets Approach. Materials 2020, 13, 1934. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Ma, A.; Li, D.; Luo, L.; Liu, D.; Hu, S.; Jin, D.; Liu, K.; Ye, C.; et al. The Novel Multiple Inner Primers-Loop-Mediated Isothermal Amplification (MIP-LAMP) for Rapid Detection and Differentiation of Listeria monocytogenes. Molecules 2015, 20, 21515–21531. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, G.; Lu, C.; Deng, R.; Zhi, A.; Guo, J.; Zhao, D.; Xu, Z. Rapid Detection of Listeria monocytogenes in Raw Milk with Loop-Mediated Isothermal Amplification and Chemosensor. J. Food Sci. 2011, 76, M611–M615. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.J.; Zhou, S.; Zhang, X.Y.; Pu, J.H.; Ge, Q.L.; Tang, X.J.; Gao, Y.S. Rapid and Sensitive Detection of Listeria monocytogenes by Loop-Mediated Isothermal Amplification. Curr. Microbiol. 2011, 63, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Wan, C.; Yang, Y.; Xu, H.; Aguilar, Z.P.; Liu, C.; Lai, W.; Xiong, Y.; Xu, F.; Wei, H. Development of a Propidium Monoazide Treatment Combined with Loop-Mediated Isothermal Amplification (PMA-LAMP) Assay for Rapid Detection of Viable Listeria monocytogenes. Int. J. Food Sci. Technol. 2012, 47, 2460–2467. [Google Scholar] [CrossRef]
- Pisamayarom, K.; Suriyasomboon, A.; Chaumpluk, P. Simple Screening of Listeria monocytogenes Based on a Fluorescence Assay via a Laminated Lab-on-Paper Chip. Biosensors 2017, 7, 56. [Google Scholar] [CrossRef]
- Garrido-Maestu, A.; Azinheiro, S.; Carvalho, J.; Fuciños, P.; Prado, M. Development and Evaluation of Loop-Mediated Isothermal Amplification, and Recombinase Polymerase Amplification Methodologies, for the Detection of Listeria monocytogenes in Ready-to-Eat Food Samples. Food Control 2018, 86, 27–34. [Google Scholar] [CrossRef]
- Liu, Z.; Yao, C.; Wang, Y.; Yang, C. A G-Quadruplex DNAzyme-Based LAMP Biosensing Platform for a Novel Colorimetric Detection of Listeria monocytogenes. Anal. Methods 2018, 10, 848–854. [Google Scholar] [CrossRef]
- Nathaniel, B.R.; Ghai, M.; Druce, M.; Maharaj, I.; Olaniran, A.O. Development of a Loop-Mediated Isothermal Amplification Assay Targeting Lmo0753 Gene for Detection of Listeria monocytogenes in Wastewater. Lett. Appl. Microbiol. 2019, 69, 264–270. [Google Scholar] [CrossRef]
- Sharif, S.; Wang, Y.; Ye, Z.; Wang, Z.; Qiu, Q.; Ying, S.; Ying, Y. A Novel Impedimetric Sensor for Detecting LAMP Amplicons of Pathogenic DNA Based on Magnetic Separation. Sens. Actuators B Chem. 2019, 301, 127051. [Google Scholar] [CrossRef]
- Jin, J.; Duan, L.; Fu, J.; Chai, F.; Zhou, Q.; Wang, Y.; Shao, X.; Wang, L.; Yan, M.; Su, X.; et al. A Real-Time LAMP-Based Dual-Sample Microfluidic Chip for Rapid and Simultaneous Detection of Multiple Waterborne Pathogenic Bacteria from Coastal Waters. Anal. Methods 2021, 13, 2710–2721. [Google Scholar] [CrossRef]
- Shi, D.; Shi, H. Combining Loop-Mediated Isothermal Amplification and Nanozyme-Strip for Ultrasensitive and Rapid Detection of Viable Listeria monocytogenes Cells and Biofilms. LWT Food Sci. Technol. 2022, 154, 112641. [Google Scholar] [CrossRef]
- Tirloni, E.; Bernardi, C.; Drago, S.; Stampone, G.; Pomilio, F.; Cattaneo, P.; Stella, S. Evaluation of a Loop-Mediated Isothermal Amplification Method for the Detection of Listeria monocytogenes in Dairy Food. Ital. J. Food Saf. 2017, 6, 179–184. [Google Scholar] [CrossRef] [PubMed]








| Bacterial Species and Serotype | Source | No. of Strains | LAMP Result | PCR Result | |
|---|---|---|---|---|---|
| Listeria spp. | L. monocytogenes | ATCC BAA-679 | 1 | + | + |
| L. monocytogenes (1/2a) | CCESB | 15 | + | + | |
| L. monocytogenes (1/2b) | CCESB | 15 | + | + | |
| L. monocytogenes (1/2c) | CCESB | 15 | + | + | |
| L. monocytogenes (4b) | CCESB | 15 | + | + | |
| L. innocua 2030c | PHLS | 1 | - | - | |
| L. ivanovii | ATCC 19119 | 1 | - | - | |
| L. aquatica | CCESB | 1 | - | - | |
| Enterococcus spp. | E. faecalis | ATCC 29212 | 1 | - | - |
| E. faecalis | CCESB | 3 | - | - | |
| E. faecium | DSMZ 13590 | 1 | - | - | |
| E. faecium | CCESB | 2 | - | - | |
| Staphylococcus spp. | S. aureus | ATCC 29213 | 1 | - | - |
| S. aureus | ATCC 25923 | 1 | - | - | |
| S. aureus | CCESB | 6 | - | - | |
| Lactococcus spp. | L. lactis | DSMZ 4366 | 1 | - | - |
| L. lactis | CCESB | 2 | - | - | |
| Leuconostoc spp. | L. mesenteroides | CCESB | 2 | - | - |
| Pseudomonas spp. | P. aeruginosa | ATCC 27853 | 1 | - | - |
| Escherichia spp. | E. coli | ATCC 25922 | 1 | - | - |
| Salmonella enterica | S. Infantis M2016 | NFCSO | 1 | - | - |
| S. Braenderup | ATCC BAA-664 | 1 | - | - | |
| S. Weltevreden TA 428/97 | EURL | 1 | - | - | |
| S. Senftenberg | ATCC 43845 | 1 | - | - | |
| S. Typhimurium | ATCC 14028 | 1 | - | - | |
| S. Derby | CCESB | 1 | - | - | |
| S. Enteritidis | ATCC 13076 | 1 | - | - | |
| S. Wernigerode | CCESB | 1 | - | - | |
| Acinetobacter spp. | A. baumannii | CCESB | 2 | - | - |
| Campylobacter spp. | C. jejuni | DSMZ 4688 | 1 | - | - |
| C. coli | DSMZ 4689 | 1 | - | - | |
| C. lari | DSMZ 11375 | 1 | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maciel, C.; Silva, N.F.D.; Teixeira, P.; Magalhães, J.M.C.S. Development of a Novel Phagomagnetic-Assisted Isothermal DNA Amplification System for Endpoint Electrochemical Detection of Listeria monocytogenes. Biosensors 2023, 13, 464. https://doi.org/10.3390/bios13040464
Maciel C, Silva NFD, Teixeira P, Magalhães JMCS. Development of a Novel Phagomagnetic-Assisted Isothermal DNA Amplification System for Endpoint Electrochemical Detection of Listeria monocytogenes. Biosensors. 2023; 13(4):464. https://doi.org/10.3390/bios13040464
Chicago/Turabian StyleMaciel, Cláudia, Nádia F. D. Silva, Paula Teixeira, and Júlia M. C. S. Magalhães. 2023. "Development of a Novel Phagomagnetic-Assisted Isothermal DNA Amplification System for Endpoint Electrochemical Detection of Listeria monocytogenes" Biosensors 13, no. 4: 464. https://doi.org/10.3390/bios13040464
APA StyleMaciel, C., Silva, N. F. D., Teixeira, P., & Magalhães, J. M. C. S. (2023). Development of a Novel Phagomagnetic-Assisted Isothermal DNA Amplification System for Endpoint Electrochemical Detection of Listeria monocytogenes. Biosensors, 13(4), 464. https://doi.org/10.3390/bios13040464





