Integration of G-Quadruplex and Pyrene as a Simple and Efficient Ratiometric Fluorescent Platform That Programmed by Contrary Logic Pair for Highly Sensitive and Selective Coralyne (COR) Detection
Abstract
:1. Introduction
2. Experimental Section
2.1. Chemicals
2.2. Apparatus
2.3. COR Detection and Selectivity Tests
2.4. Recovery Tests of COR in Real Samples
3. Results and Discussion
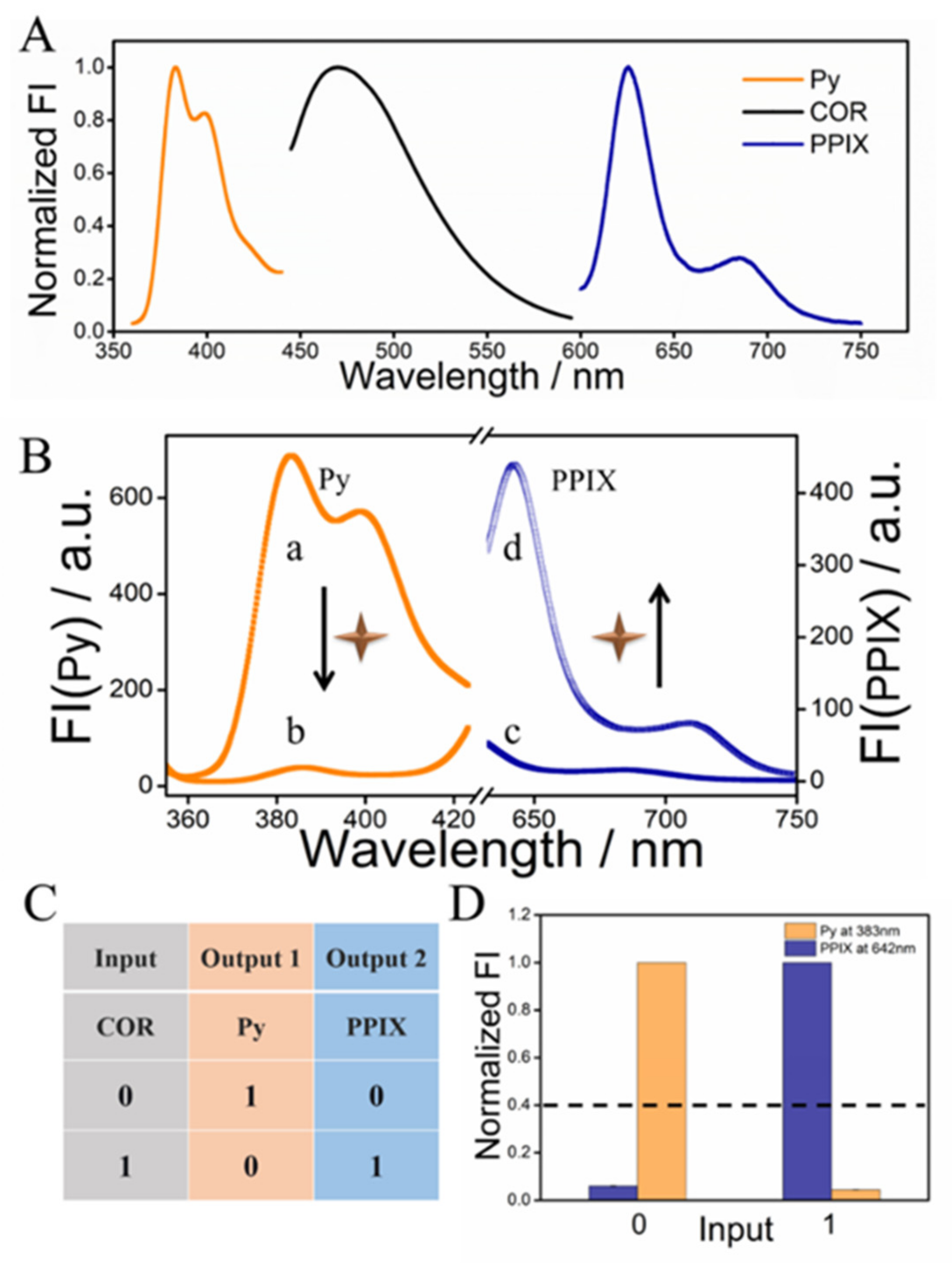
3.1. Sensing Mechanism of the Ratiometric Fluorescent System
3.2. Verification of the Proposed Sensing Mechanism
3.3. Optimization of Reaction Conditions
3.4. Ratiometric Fluorescent Detection of COR
3.5. Selectivity and Real Sample Application of the System
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| COR | Coralyne |
| G4 | G-quadruplex |
| FRET | Fluorescence resonance energy transfer |
| LOD | Limit of detection |
| S/N ratio | Signal-to-noise ratio |
| ss-DNA | Single-stranded DNA |
| Tris | Tris (hydroxymethyl) aminomethane |
| 5-FU | 5-fluorouracil |
| BLM | Bleomycin |
| HPLC-ESI-MS | High-performance liquid chromatography-electrospray ionization-mass spectrometry |
| TMB | 3, 3′, 5, 5′-Tetramethylbenzidine |
| Py | Pyrene |
| G4zyme | G-quadruplex DNAzyme |
| PPIX | Protoporphyrin IX |
| AuNPs | Gold nanoparticles |
| CLP | Contrary logic pair |
| ds-DNA | Double-stranded DNA |
| KCl | Potassium chloride |
| TOB | Tobramycin |
| CD | Circular dichroism |
| FBS | Fetal bovine serum |
| H2O2 | Hydrogen peroxide |
References
- Patro, B.S.; Maity, B.; Chattopadhyay, S. Topoisomerase Inhibitor Coralyne Photosensitizes DNA, Leading to Elicitation of Chk2-Dependent S-phase Checkpoint and p53-Independent Apoptosis in Cancer Cells. Antioxid. Redox Sign. 2010, 12, 945–960. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Badana, A.K.; Mohan, G.M.; Shailender Naik, G.; Malla, R. Synergistic effects of coralyne and paclitaxel on cell migration and proliferation of breast cancer cells lines. Biomed. Pharmacother. 2017, 91, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, R.; Gupta, P.; Bandyopadhyay, S.K.; Patro, B.S.; Chattopadhyay, S. Coralyne, a protoberberine alkaloid, causes robust photosenstization of cancer cells through ATR-p38 MAPK-BAX and JAK2-STAT1-BAX pathways. Chem.-Biol. Interact. 2018, 285, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, R.; Saha, B.; Tyagi, M.; Bandyopadhyay, S.K.; Patro, B.S.; Chattopadhyay, S. Differential modes of photosensitisation in cancer cells by berberine and coralyne. Free Radical Res. 2017, 51, 723–738. [Google Scholar] [CrossRef] [PubMed]
- Wegierek-Ciuk, A.; Arabski, M.; Ciepluch, K.; Brzoska, K.; Lisowska, H.; Czerwinska, M.; Stepkowski, T.; Lis, K.; Lankoff, A. Coralyne Radiosensitizes A549 Cells by Upregulation of CDKN1A Expression to Attenuate Radiation Induced G2/M Block of the Cell Cycle. Int. J. Mol. Sci. 2021, 22, 5791. [Google Scholar] [CrossRef]
- Gatto, B.; Sanders, M.M.; Yu, C.; Wu, H.-Y.; Makhey, D.; LaVoie, E.J.; Liu, L.F. Identification of Topoisomerase I as the Cytotoxic Target of the Protoberberine Alkaloid Coralyne. Cancer Res. 1996, 56, 2795–2800. [Google Scholar]
- Ihmels, H.; Salbach, A. Efficient Photoinduced DNA Damage by Coralyne. Photochem. Photobiol. 2006, 82, 1572–1576. [Google Scholar] [CrossRef]
- Xu, N.; Yang, H.; Cui, M.; Wan, C.; Liu, S. High-performance liquid chromatography-electrospray ionization-mass spectrometry ligand fishing assay: A method for screening triplex DNA binders from natural plant extracts. Anal. Chem. 2012, 84, 2562–2568. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, Y.; Leng, F.; Xiong, Z.H.; Huang, C.Z. Highly selective and sensitive detection of coralyne based on the binding chemistry of aptamer and graphene oxide. Talanta 2013, 112, 117–122. [Google Scholar] [CrossRef]
- Huang, H.; Shi, S.; Zheng, X.; Yao, T. Sensitive detection for coralyne and mercury ions based on homo-A/T DNA by exonuclease signal amplification. Biosens. Bioelectron. 2015, 71, 439–444. [Google Scholar] [CrossRef]
- Chen, M.; Ma, C.; Zhao, H.; Wang, K. Label-free and sensitive detection of coralyne and heparin based on target-induced G-quadruplex formation. Anal. Methods 2019, 11, 1331–1337. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, M.; Ma, C. Sensitive Detection of Coralyne and Heparin Using a Singly Labeled Fluorescent Oligonucleotide Probe. ChemistrySelect 2019, 4, 5686–5690. [Google Scholar] [CrossRef]
- Wang, H.-B.; Li, Y.-H.; Huang, K.-J.; Liu, X.-S.; Yang, Y.-E.; Liu, Y.-M. A label-free and sensitive fluorescence strategy for screening ligands binding to poly(dA) based on exonuclease I-assisted background noise reduction. Anal. Methods 2013, 5, 4852–4858. [Google Scholar] [CrossRef]
- Lv, Z.; Wei, H.; Li, B.; Wang, E. Colorimetric recognition of the coralyne-poly(dA) interaction using unmodified gold nanoparticle probes, and further detection of coralyne based upon this recognition system. Analyst 2009, 134, 1647–1651. [Google Scholar] [CrossRef]
- Tao, Y.; Lin, Y.; Ren, J.; Qu, X. Self-assembled, functionalized graphene and DNA as a universal platform for colorimetric assays. Biomaterials 2013, 34, 4810–4817. [Google Scholar] [CrossRef]
- Hou, T.; Li, C.; Wang, X.; Zhao, C.; Li, F. Label-free colorimetric detection of coralyne utilizing peroxidase-like split G-quadruplex DNAzyme. Anal. Methods 2013, 5, 4671–4674. [Google Scholar] [CrossRef]
- Wang, J.; Yu, J.; Zhou, X.; Miao, P. Exonuclease and Nicking Endonuclease-Assisted Amplified Electrochemical Detection of Coralyne. ChemElectroChem 2017, 4, 1828–1831. [Google Scholar] [CrossRef]
- Lu, Y.; Huang, Q. Label-free selective detection of coralyne due to aptamer-coralyne interaction using DNA modified SiO2@Au core-shell nanoparticles as an effective SERS substrate. Anal. Methods 2013, 5, 3927–3932. [Google Scholar] [CrossRef]
- Chatterjee, T.; Johnson-Buck, A.; Walter, N.G. Highly sensitive protein detection by aptamer-based single-molecule kinetic fingerprinting. Biosens. Bioelectron. 2022, 216, 114639. [Google Scholar] [CrossRef]
- Lee, S.; Godhulayyagari, S.; Nguyen, S.T.; Lu, J.K.; Ebrahimi, S.B.; Samanta, D. Signal Transduction Strategies for Analyte Detection Using DNA-Based Nanostructures. Angew. Chem. Int. Ed. 2022, 61, e202202211. [Google Scholar] [CrossRef]
- Ghotra, G.; Nguyen, B.K.; Chen, J.I.L. DNA-Functionalized Gold Nanoparticles with Toehold-Mediated Strand Displacement for Nucleic Acid Sensors. ACS Appl. Nano Mater. 2020, 3, 10123–10132. [Google Scholar] [CrossRef]
- Pandey, R.; Lu, Y.; McConnell, E.M.; Osman, E.; Scott, A.; Gu, J.; Hoare, T.; Soleymani, L.; Li, Y. Electrochemical DNAzyme-based biosensors for disease diagnosis. Biosens. Bioelectron. 2023, 224, 114983. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Wang, L.L.; Wu, C.Q.; Li, H.; Zhang, Q.L.; Wang, Y.; Xu, L. Controllable DNA hybridization by host-guest complexation-mediated ligand invasion. Nat. Commun. 2022, 13, 5936. [Google Scholar] [CrossRef]
- Deiana, M.; Chand, K.; Jamroskovic, J.; Obi, I.; Chorell, E.; Sabouri, N. A Light-up Logic Platform for Selective Recognition of Parallel G-Quadruplex Structures via Disaggregation-Induced Emission. Angew. Chem. Int. Ed. 2020, 59, 896–902. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.; Wang, K.; Zhu, J.; Xia, Y.; Han, Y.; Liu, Y.; Wang, E. DNA-based visual majority logic gate with one-vote veto function. Chem. Sci. 2015, 6, 1973–1978. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.; Zhu, J.; Zhai, Q.; Wang, E.; Dong, S. Cascade DNA logic device programmed ratiometric DNA analysis and logic devices based on a fluorescent dual-signal probe of a G-quadruplex DNAzyme. Chem. Commun. 2016, 52, 3766–3769. [Google Scholar] [CrossRef]
- Fan, D.; Zhu, J.; Liu, Y.; Wang, E.; Dong, S. Label-free and enzyme-free platform for the construction of advanced DNA logic devices based on the assembly of graphene oxide and DNA-templated AgNCs. Nanoscale 2016, 8, 3834–3840. [Google Scholar] [CrossRef]
- Fan, D.; Wang, E.; Dong, S. Simple, fast, label-free, and nanoquencher-free system for operating multivalued DNA logic gates using polythymine templated CuNPs as signal reporters. Nano Res. 2017, 10, 2560–2569. [Google Scholar] [CrossRef]
- Fan, T.; Du, Y.; Yao, Y.; Wu, J.; Meng, S.; Luo, J.; Zhang, X.; Yang, D.; Wang, C.; Qian, Y.; et al. Rolling circle amplification triggered poly adenine-gold nanoparticles production for label-free electrochemical detection of thrombin. Sens. Actuat. B-Chem. 2018, 266, 9–18. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, H.; Wang, C.; Xu, Y.; Su, J.; Wang, X.; Liu, X.; Feng, D.; Wang, L.; Zuo, X.; et al. Poly-adenine-mediated spherical nucleic acids for strand displacement-based DNA/RNA detection. Biosens. Bioelectron. 2019, 127, 85–91. [Google Scholar] [CrossRef]
- Bhadra, K.; Maiti, M.; Kumar, G.S. DNA-Binding Cytotoxic Alkaloids: Comparative Study of the Energetics of Binding of Berberine, Palmatine, and Coralyne. DNA Cell Biol. 2008, 27, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Xing, F.; Song, G.; Ren, J.; Chaires, J.B.; Qu, X. Molecular recognition of nucleic acids: Coralyne binds strongly to poly(A). FEBS Lett. 2005, 579, 5035–5039. [Google Scholar] [CrossRef] [PubMed]
- Persil, O.; Santai, C.T.; Jain, S.S.; Hud, N.V. Assembly of an Antiparallel Homo-Adenine DNA Duplex by Small-Molecule Binding. J. Am. Chem. Soc. 2004, 126, 8644–8645. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Suresh Kumar, G. Coralyne induced self-structure in polyadenylic acid: Thermodynamics of the structural reorganization. J. Chem. Thermodyn. 2016, 101, 221–226. [Google Scholar] [CrossRef]
- Kim, S.; Choi, J.; Majima, T. Self-assembly of polydeoxyadenylic acid studied at the single-molecule level. J. Phys. Chem. B 2011, 115, 15399–15405. [Google Scholar] [CrossRef]
- Hou, T.; Wang, X.; Liu, X.; Liu, S.; Du, Z.; Li, F. A label-free and colorimetric turn-on assay for coralyne based on coralyne-induced formation of peroxidase-mimicking split DNAzyme. Analyst 2013, 138, 4728–4731. [Google Scholar] [CrossRef]
- Ma, C.; Chen, M.; He, H.; Chen, L. Detection of coralyne and heparin by polymerase extension reaction using SYBR Green I. Mol. Cell. Probe 2019, 46, 101423. [Google Scholar] [CrossRef]
- Fan, D.; Shang, C.; Gu, W.; Wang, E.; Dong, S. Introducing Ratiometric Fluorescence to MnO2 Nanosheet-Based Biosensing: A Simple, Label-Free Ratiometric Fluorescent Sensor Programmed by Cascade Logic Circuit for Ultrasensitive GSH Detection. ACS Appl. Mater. Inter. 2017, 9, 25870–25877. [Google Scholar] [CrossRef]
- Liu, L.; Ga, L.; Ai, J. Ratiometric fluorescence sensing with logical operation: Theory, design and applications. Biosens. Bioelectron. 2022, 213, 114456. [Google Scholar] [CrossRef]
- Fan, D.; Wang, J.; Han, J.; Wang, E.; Dong, S. Engineering DNA logic systems with non-canonical DNA-nanostructures: Basic principles, recent developments and bio-applications. Sci. China Chem. 2021, 65, 284–297. [Google Scholar] [CrossRef]
- Fan, D.; Wang, J.; Wang, E.; Dong, S. Propelling DNA Computing with Materials’ Power: Recent Advancements in Innovative DNA Logic Computing Systems and Smart Bio-Applications. Adv. Sci. 2020, 7, 2001766. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.; Wang, E.; Dong, S. DNA Computing: Versatile Logic Circuits and Innovative Bio-applications. DNA-RNA-Based Comput. Syst. 2021, 12, 231–246. [Google Scholar]
- Orlov, A.V.; Pushkarev, A.V.; Mochalova, E.N.; Nikitin, P.I.; Nikitin, M.P. Development and label-free investigation of logic-gating biolayers for smart biosensing. Sens. Actuat. B-Chem. 2018, 257, 971–979. [Google Scholar] [CrossRef]
- Katz, E.; Poghossian, A.; Schoning, M.J. Enzyme-based logic gates and circuits—Analytical applications and interfacing with electronics. Anal. Bioanal. Chem. 2017, 409, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.; Wang, E.; Dong, S. An intelligent universal system yields double results with half the effort for engineering a DNA “Contrary Logic Pairs” library and various DNA combinatorial logic circuits. Mater. Horiz. 2017, 4, 924–931. [Google Scholar] [CrossRef]
- Fan, D.; Wang, E.; Dong, S. A DNA-based parity generator/checker for error detection through data transmission with visual readout and an output-correction function. Chem. Sci. 2017, 8, 1888–1895. [Google Scholar] [CrossRef]
- Fan, D.; Fan, Y.; Wang, E.; Dong, S. A simple, label-free, electrochemical DNA parity generator/checker for error detection during data transmission based on “aptamer-nanoclaw”-modulated protein steric hindrance. Chem. Sci. 2018, 9, 6981–6987. [Google Scholar] [CrossRef]
- Fan, D.; Wang, J.; Wang, E.; Dong, S. A Janus-inspired amphichromatic system that kills two birds with one stone for operating a “DNA Janus Logic Pair” (DJLP) library. Chem. Sci. 2019, 10, 7290–7298. [Google Scholar] [CrossRef]
- Fan, D.; Wang, E.; Dong, S. Upconversion-chameleon-driven DNA computing: The DNA-unlocked inner-filter-effect (DU-IFE) for operating a multicolor upconversion luminescent DNA logic library and Its biosensing application. Mater. Horiz. 2019, 6, 375–384. [Google Scholar] [CrossRef]
- Han, J.; Wang, J.; Wang, J.; Fan, D.; Dong, S. Recent advancements in coralyne (COR)-based biosensors: Basic principles, various strategies and future perspectives. Biosens. Bioelectron. 2022, 210, 114343. [Google Scholar] [CrossRef]
- Perlikova, P.; Ejlersen, M.; Langkjaer, N.; Wengel, J. Bis-pyrene-modified unlocked nucleic acids: Synthesis, hybridization studies, and fluorescent properties. ChemMedChem 2014, 9, 2120–2127. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Gao, Y.; Li, B.; Jin, Y. Cyclic up-regulation fluorescence of pyrene excimer for studying polynucleotide kinase activity based on dual amplification. Biosens. Bioelectron. 2016, 80, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wang, N.; Li, Y.; Deng, T.; Li, J.; Zhang, K.; Yu, R. Cyclodextrin supramolecular inclusion-enhanced pyrene excimer switching for highly selective detection of RNase H. Anal. Chim. Acta 2019, 1088, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Li, T.; Zhang, J.; Chen, A. A simple FRET-based turn-on fluorescent aptasensor for 17β-estradiol determination in environmental water, urine and milk samples. Sens. Actuat. B-Chem. 2018, 273, 1648–1653. [Google Scholar] [CrossRef]
- Padmapriya, K.; Barthwal, R. Nuclear magnetic resonance studies reveal stabilization of parallel G-quadruplex DNA [d(T2G4T)]4 upon binding to protoberberine alkaloid coralyne. Bioorg. Med. Chem. Lett. 2016, 26, 4915–4918. [Google Scholar] [CrossRef] [PubMed]




| Detection Methods | LOD | Liner Range | Ref. |
|---|---|---|---|
| Colorimetric | 91 nM | 0–728 nM | [14] |
| Colorimetric | 100 nM | 0.1–1 μM | [15] |
| Colorimetric | 19 nM | 0.06–10 μM | [16] |
| Fluorescent | 10 nM | 10–700 nM | [9] |
| Fluorescent | 0.31 nM | 0.2–100 nM | [10] |
| Fluorescent | 5.8 nM | 0.01–5 μM | [11] |
| Fluorescent | 3.1 nM | 1–300 nM | [12] |
| Fluorescent | 3.5 nM | 10–1000 nM | [13] |
| Fluorescent | 0.98 nM | 2–500 nM | [37] |
| Electrochemical | 0.07 nM | 0.1–100 nM | [17] |
| SERS | 100 nM | 0.1–100 μM | [18] |
| Ratiometric fluorescent | 0.63 nM | 0.001–8 μM | This work |
| Sample | Added | Measured | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| COR | 100 nM | 99.8 ± 2.88 nM | 99.8 | 2.9 |
| 4 μM | 4.27 ± 0.24 μM | 106.7 | 5.5 | |
| 8 μM | 8.02 ± 0.37 μM | 100.3 | 4.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, J.; Ding, Y.; Lv, X.; Zhang, Y.; Fan, D. Integration of G-Quadruplex and Pyrene as a Simple and Efficient Ratiometric Fluorescent Platform That Programmed by Contrary Logic Pair for Highly Sensitive and Selective Coralyne (COR) Detection. Biosensors 2023, 13, 489. https://doi.org/10.3390/bios13040489
Han J, Ding Y, Lv X, Zhang Y, Fan D. Integration of G-Quadruplex and Pyrene as a Simple and Efficient Ratiometric Fluorescent Platform That Programmed by Contrary Logic Pair for Highly Sensitive and Selective Coralyne (COR) Detection. Biosensors. 2023; 13(4):489. https://doi.org/10.3390/bios13040489
Chicago/Turabian StyleHan, Jiawen, Yaru Ding, Xujuan Lv, Yuwei Zhang, and Daoqing Fan. 2023. "Integration of G-Quadruplex and Pyrene as a Simple and Efficient Ratiometric Fluorescent Platform That Programmed by Contrary Logic Pair for Highly Sensitive and Selective Coralyne (COR) Detection" Biosensors 13, no. 4: 489. https://doi.org/10.3390/bios13040489
APA StyleHan, J., Ding, Y., Lv, X., Zhang, Y., & Fan, D. (2023). Integration of G-Quadruplex and Pyrene as a Simple and Efficient Ratiometric Fluorescent Platform That Programmed by Contrary Logic Pair for Highly Sensitive and Selective Coralyne (COR) Detection. Biosensors, 13(4), 489. https://doi.org/10.3390/bios13040489





