Oscillatory-Flow PCR Microfluidic Chip Driven by Low Speed Biaxial Centrifugation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biaxial Centrifugation Analysis
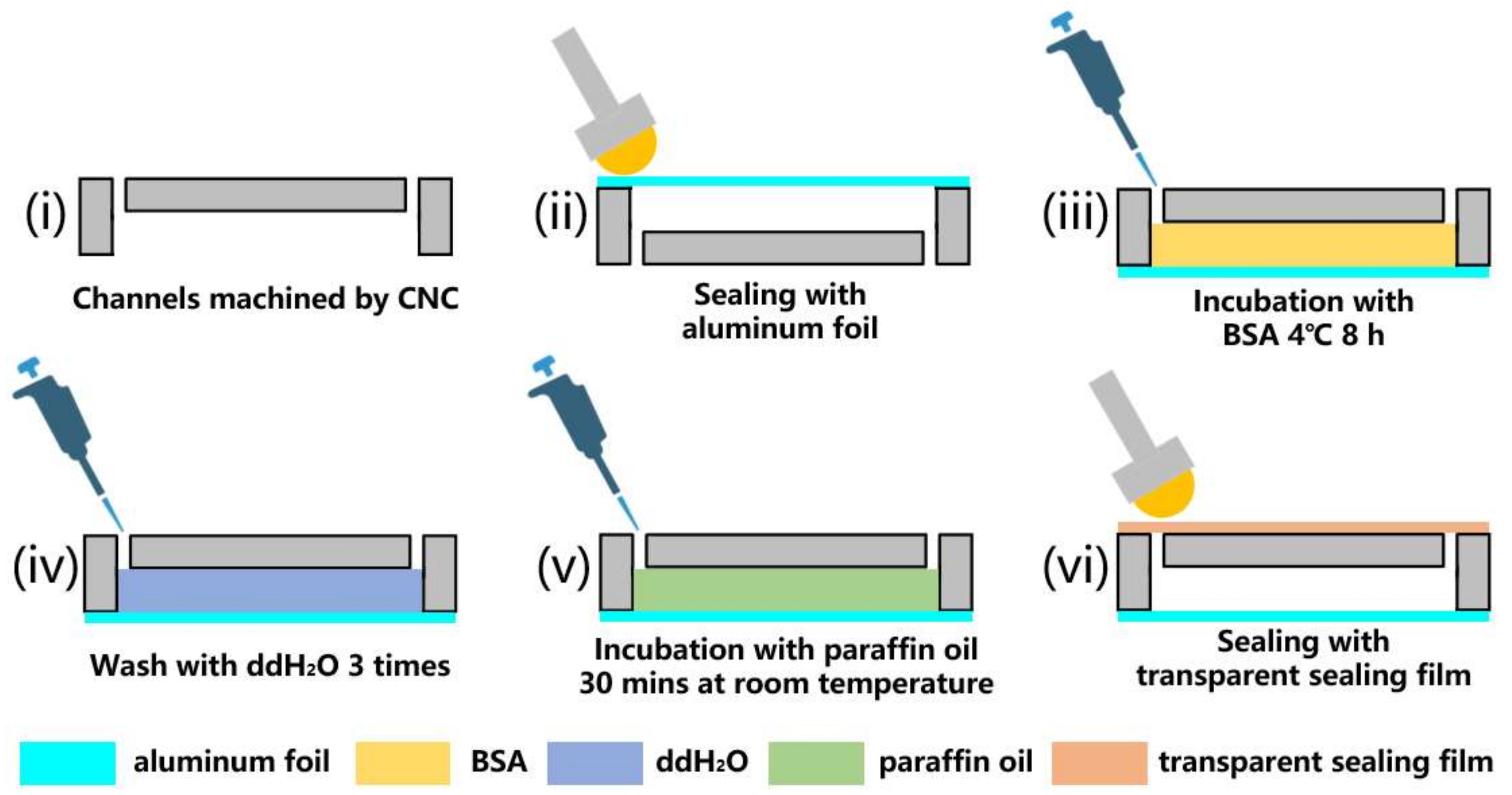
2.1.1. Chip Fabrication
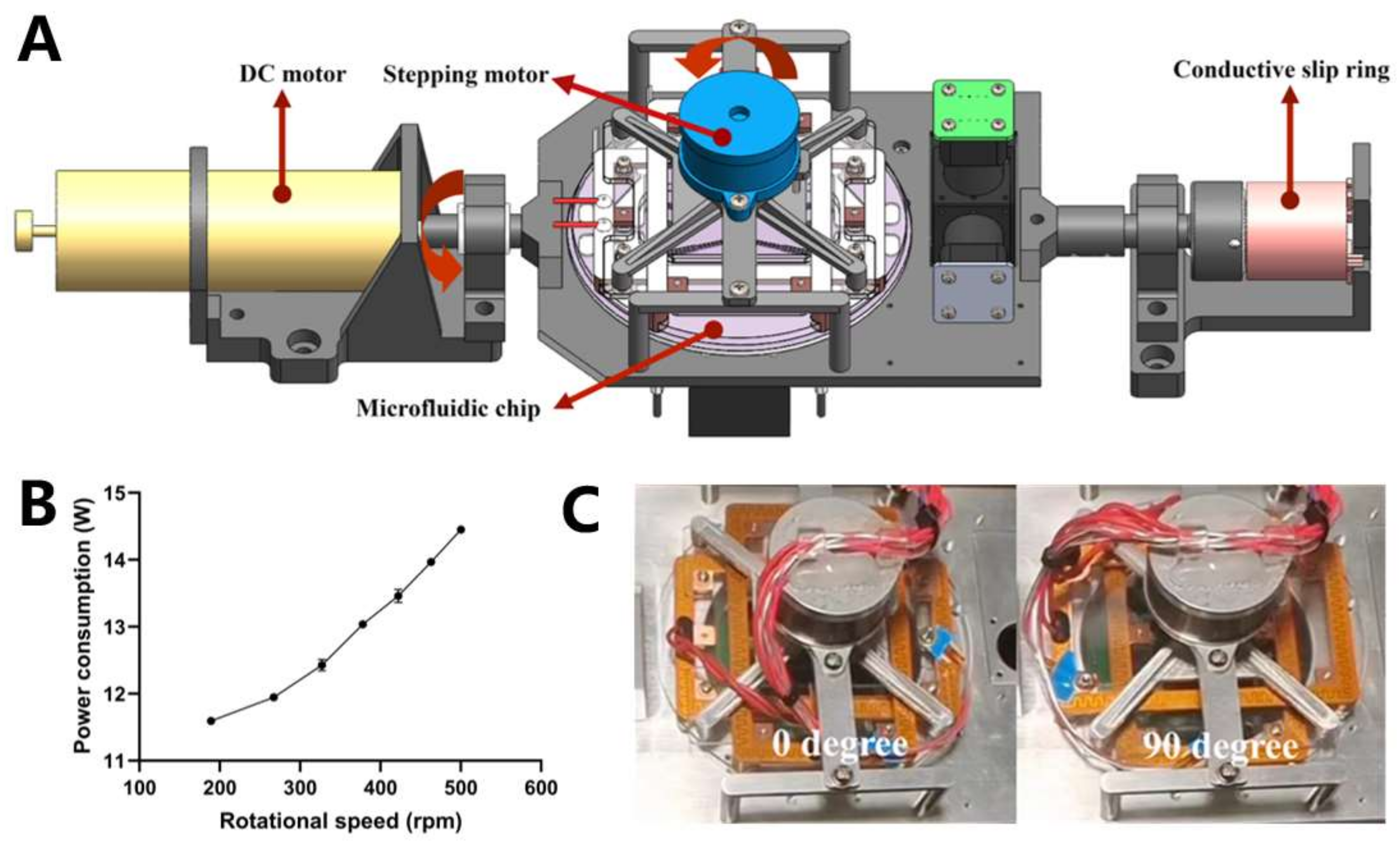
2.1.2. Centrifugal Drive and Measurement
2.2. Device Verification with Chip
2.2.1. Temperature Distribution and Ramp Rate Measurement
2.2.2. Temperature Control Stability and Power Consumption Measurement
2.2.3. Template and Amplification Reagent
- arcC-forward: 5′-TGGTGCAATGTCACAGGGTA-3′;
- arcC-reverse: 5′-CCACGTCCTGCATCTTCTTT-3′;
- yhaI-forward: 5′-ATTTGAGCGGCTGGAAAGAG-3′;
- yhaI-reverse: 5′-AGCGGCCGATATCATGCAT-3′;
- PEX-2-forward: 5′-CGATCAGTCCCCAAGTGAAT-3′;
- PEX-2-reverse: 5′-AGTCTACCGGTGCAACCAAC-3′.
2.2.4. PCR Process and Agarose Gel Electrophoresis Test
3. Results and Discussion
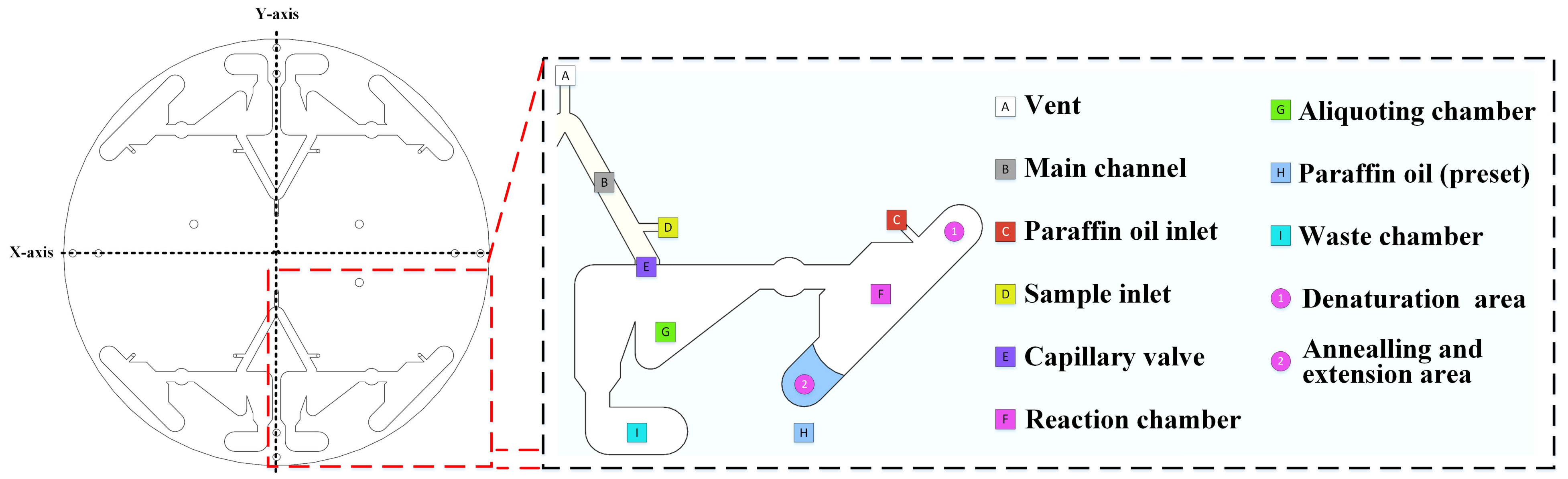
3.1. Chip Design and Centrifugal Drive Validation
3.1.1. Chip Design and Workflow
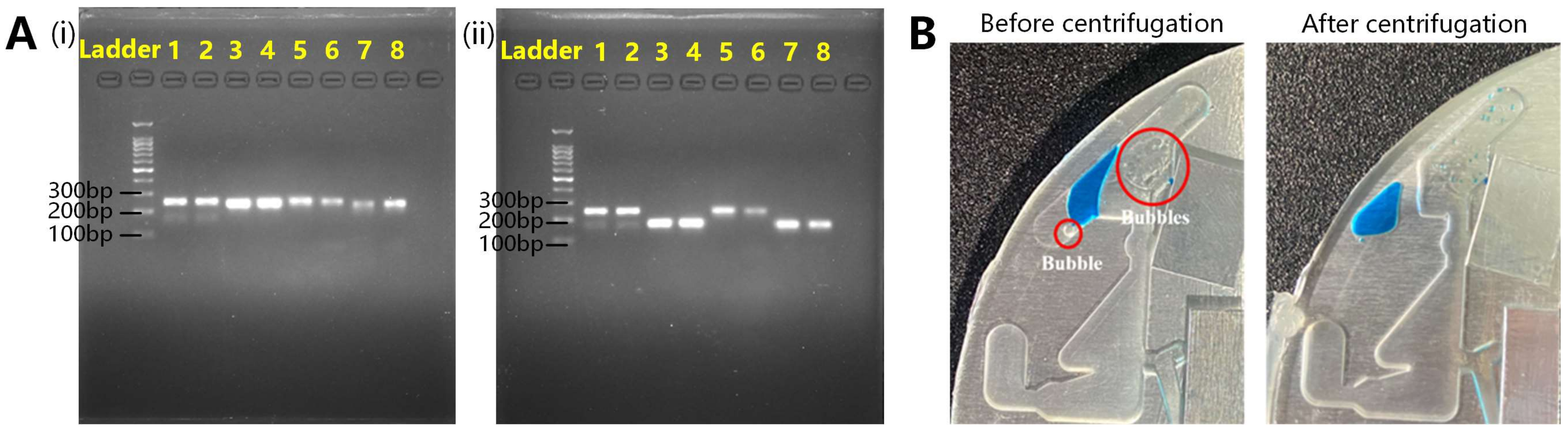
3.1.2. Centrifugal Drive Validation
3.2. Device Design
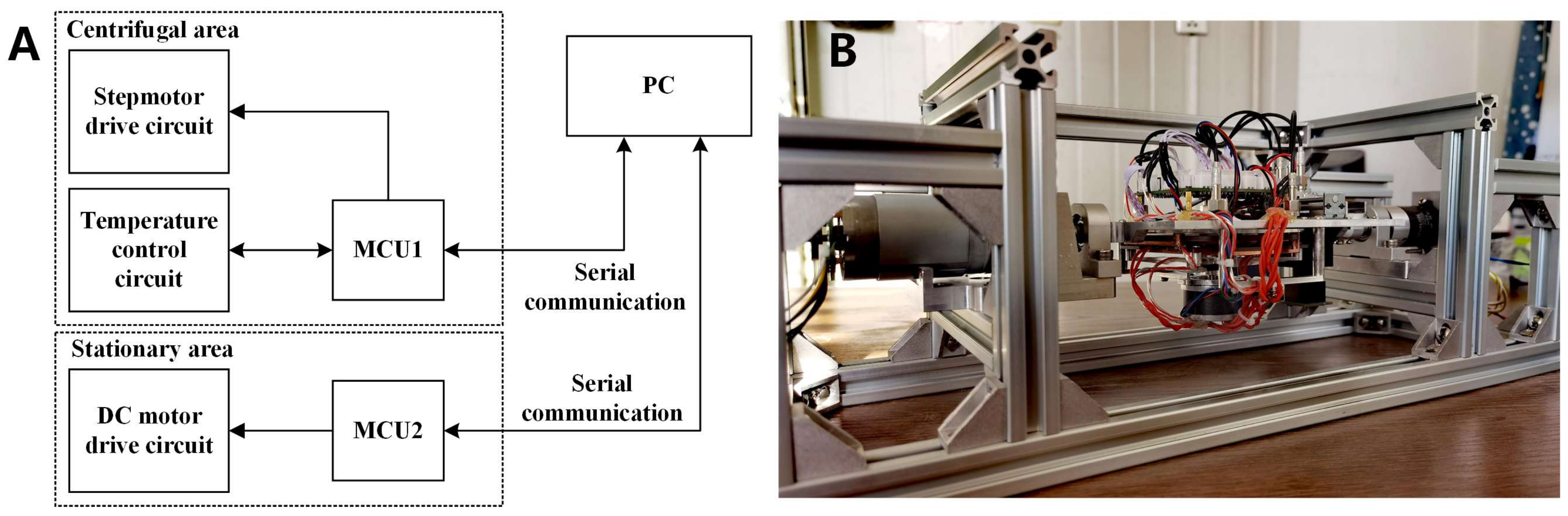
3.2.1. Biaxial Centrifugation Module
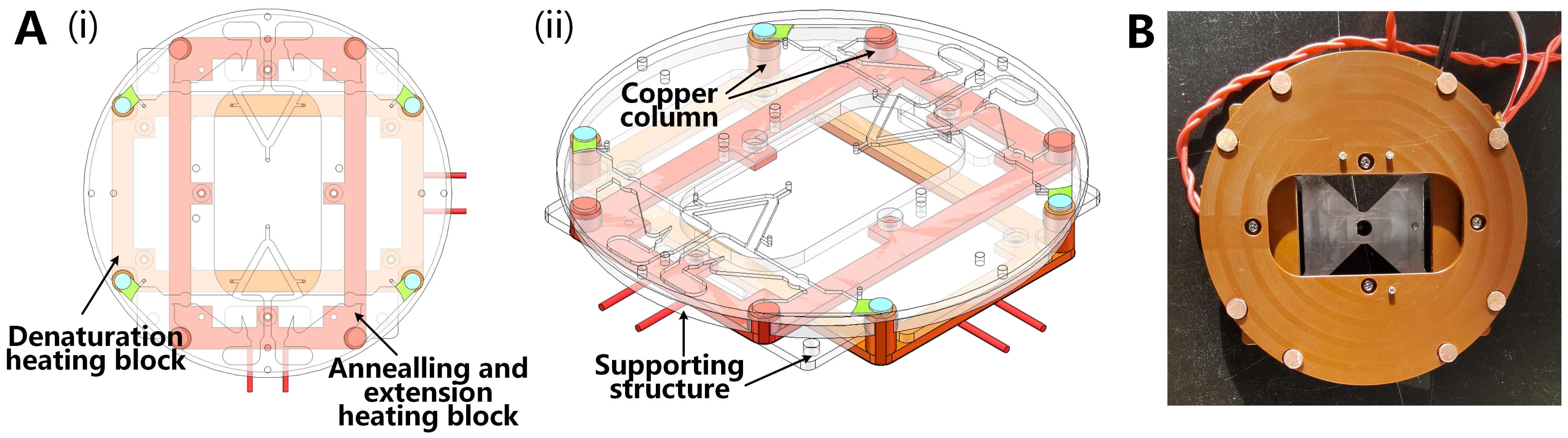
3.2.2. Temperature Control Module
3.2.3. Circuit Module and Device Assembly
3.3. Device Verification with Chip
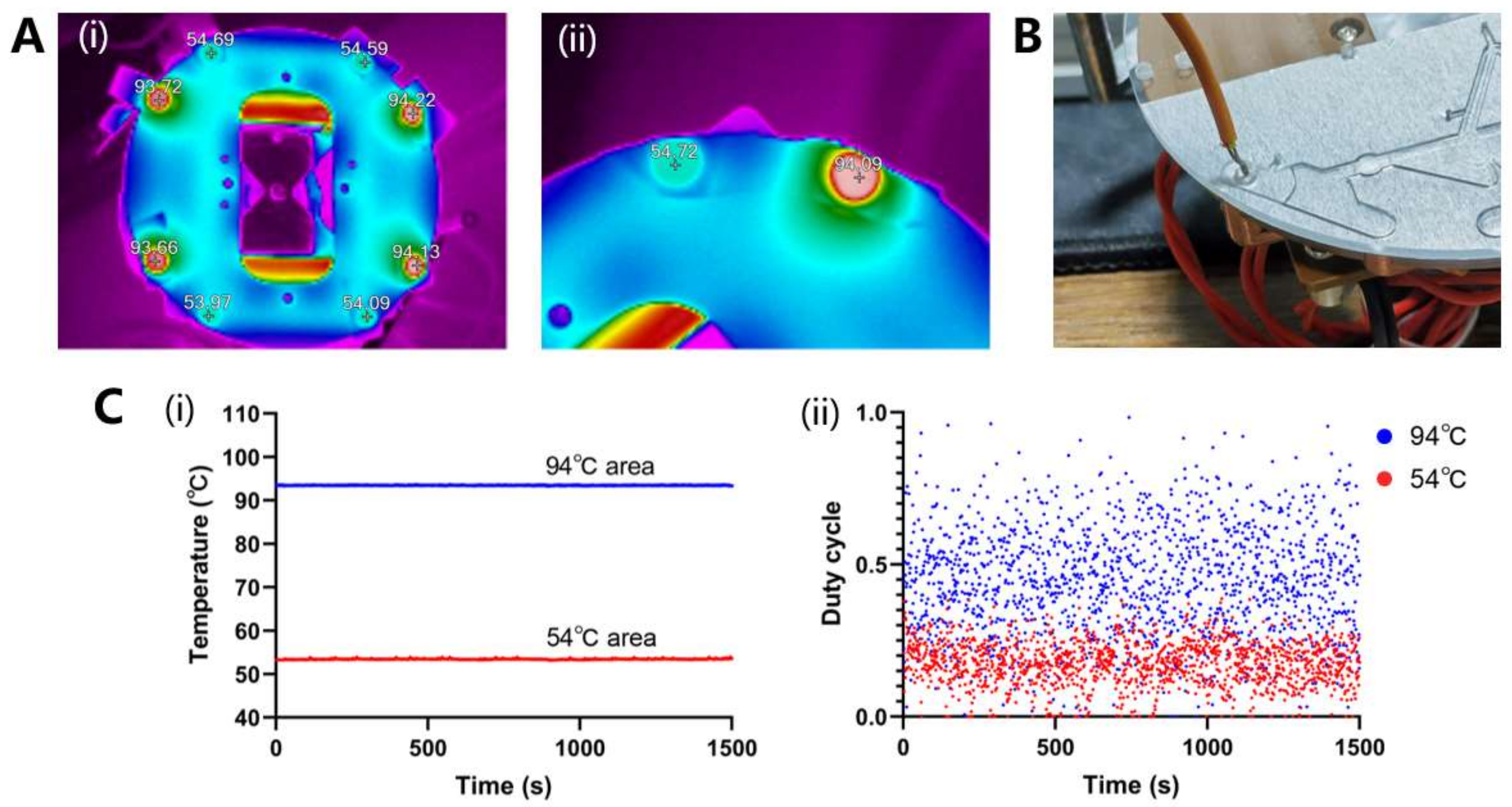
3.3.1. Temperature Distribution and Ramp Rate Measurement
3.3.2. Temperature Control Stability and Power Consumption Measurement
3.3.3. Verification of Oscillatory-Flow PCR
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Wong, S. Diagnostics in space: Will zero gravity add weight to new advances? Expert Rev. Mol. Diagn. 2020, 20, 1–4. [Google Scholar] [CrossRef]
- Robson, D.J.; Cappelletti, C. Biomedical payloads: A maturing application for CubeSats. Acta Astronaut. 2022, 191, 394–403. [Google Scholar] [CrossRef]
- Karouia, F.; Peyvan, K.; Pohorille, A. Toward biotechnology in space: High-throughput instruments for in situ biological research beyond Earth. Biotechnol. Adv. 2017, 35, 905–932. [Google Scholar] [CrossRef]
- Lu, Y.; Yang, L.; Wei, W.; Shi, Q. Microchip-based single-cell functional proteomics for biomedical applications. Lab Chip 2017, 17, 1250–1263. [Google Scholar] [CrossRef]
- Kim, J.; Lim, J.; Lee, C. Quantitative real-time PCR approaches for microbial community studies in wastewater treatment systems: Applications and considerations. Biotechnol. Adv. 2013, 31, 1358–1373. [Google Scholar] [CrossRef]
- Amalfitano, S.; Levantesi, C.; Copetti, D.; Stefani, F.; Locantore, I.; Guarnieri, V.; Lobascio, C.; Bersani, F.; Giacosa, D.; Detsis, E.; et al. Water and microbial monitoring technologies towards the near future space exploration. Water Res. 2020, 177, 115787. [Google Scholar] [CrossRef]
- Adamski, M.G.; Li, Y.; Wagner, E.; Yu, H.; Seales-Bailey, C.; Soper, S.A.; Murphy, M.; Baird, A.E. Next-Generation qPCR for the High-Throughput Measurement of Gene Expression in Multiple Leukocyte Subsets. J. Biomol. Screen 2013, 18, 1008–1017. [Google Scholar] [CrossRef]
- Romsdahl, J.; Blachowicz, A.; Chiang, A.J.; Chiang, Y.-M.; Masonjones, S.; Yaegashi, J.; Countryman, S.; Karouia, F.; Kalkum, M.; Stajich, J.E.; et al. International Space Station conditions alter genomics, proteomics, and metabolomics in Aspergillus nidulans. Appl. Microbiol. Biot. 2019, 103, 1363–1377. [Google Scholar] [CrossRef]
- Mehta, S.K.; Laudenslager, M.L.; Stowe, R.P.; Crucian, B.E.; Feiveson, A.H.; Sams, C.F.; Pierson, D.L. Latent virus reactivation in astronauts on the international space station. NPJ Microgravity 2017, 3, 11. [Google Scholar] [CrossRef]
- Parra, M.; Jung, J.; Boone, T.D.; Tran, L.; Blaber, E.A.; Brown, M.; Chin, M.; Chinn, T.; Cohen, J.; Doebler, R.; et al. Microgravity validation of a novel system for RNA isolation and multiplex quantitative real time PCR analysis of gene expression on the International Space Station. PLoS ONE 2017, 12, e0183480. [Google Scholar] [CrossRef]
- Miao, G.; Zhang, L.; Zhang, J.; Ge, S.; Xia, N.; Qian, S.; Yu, D.; Qiu, X. Free convective PCR: From principle study to commercial applications—A critical review. Anal. Chim. Acta 2020, 1108, 177–197. [Google Scholar] [CrossRef]
- Nestorova, G.G.; Crews, N.; Schramm, A.K.; Aquilina, R.A.; Parra, M.; Chin, M.; Chinn, T.; Hee, L. Spaceflight validation of one-step Gene Sampling tool for genetic analysis on the International Space Station. Acta Astronaut. 2022, 198, 225–232. [Google Scholar] [CrossRef]
- Salman, A.; Carney, H.; Bateson, S.; Ali, Z. Shunting microfluidic PCR device for rapid bacterial detection. Talanta 2020, 207, 120303. [Google Scholar] [CrossRef]
- He, L.; Sang, B.; Wu, W. Battery-Powered Portable Rotary Real-Time Fluorescent qPCR with Low Energy Consumption, Low Cost, and High Throughput. Biosensors 2020, 10, 49. [Google Scholar] [CrossRef]
- Kopparthy, V.L.; Crews, N.D. A versatile oscillating-flow microfluidic PCR system utilizing a thermal gradient for nucleic acid analysis. Biotechnol. Bioeng. 2020, 117, 1525–1532. [Google Scholar] [CrossRef]
- Liu, D.; Liang, G.; Lei, X.; Chen, B.; Wang, W.; Zhou, X. Highly efficient capillary polymerase chain reaction using an oscillation droplet microreactor. Anal. Chim. Acta 2012, 718, 58–63. [Google Scholar] [CrossRef]
- Wang, G.; Ho, H.-P.; Chen, Q.; Yang, A.K.-L.; Kwok, H.-C.; Wu, S.-Y.; Kong, S.-K.; Kwan, Y.-W.; Zhang, X. A lab-in-a-droplet bioassay strategy for centrifugal microfluidics with density difference pumping, power to disc and bidirectional flow control. Lab Chip 2013, 13, 3698–3706. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, H.; Xing, D. Multichannel oscillatory-flow multiplex PCR microfluidics for high-throughput and fast detection of foodborne bacterial pathogens. Biomed. Microdevices 2011, 13, 885–897. [Google Scholar] [CrossRef]
- Brunklaus, S.; Hansen-Hagge, T.E.; Erwes, J.; Höth, J.; Jung, M.; Latta, D.; Strobach, X.; Winkler, C.; Ritzi-Lehnert, M.; Drese, K.S. Fast nucleic acid amplification for integration in point-of-care applications. Electrophoresis 2012, 33, 3222–3228. [Google Scholar] [CrossRef]
- Chen, S.; Sun, Y.; Fan, F.; Chen, S.; Zhang, Y.; Zhang, Y.; Meng, X.; Lin, J.-M. Present status of microfluidic PCR chip in nucleic acid detection and future perspective. Trac-Trend Anal. Chem. 2022, 157, 116737. [Google Scholar] [CrossRef]
- Choi, G.; Jung, J.H.; Park, B.H.; Oh, S.J.; Seo, J.H.; Choi, J.S.; Kim, D.H.; Seo, T.S. A centrifugal direct recombinase polymerase amplification (direct-RPA) microdevice for multiplex and real-time identification of food poisoning bacteria. Lab Chip 2016, 16, 2309–2316. [Google Scholar] [CrossRef]
- Lee, B.S.; Lee, Y.U.; Kim, H.-S.; Kim, T.-H.; Park, J.; Lee, J.-G.; Kim, J.; Kim, H.; Lee, W.G.; Cho, Y.-K. Fully integrated lab-on-a-disc for simultaneous analysis of biochemistry and immunoassay from whole blood. Lab Chip 2011, 11, 70–78. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, J.; Liu, Y.; Lau, W.M.; Yang, J. Modeling of flow burst, flow timing in Lab-on-a-CD systems and its application in digital chemical analysis. Chem. Eng. Technol. 2008, 31, 1328–1335. [Google Scholar] [CrossRef]
- Zhang, L.; Tian, F.; Liu, C.; Feng, Q.; Ma, T.; Zhao, Z.; Li, T.; Jiang, X.; Sun, J. Hand-powered centrifugal microfluidic platform inspired by the spinning top for sample-to-answer diagnostics of nucleic acids. Lab Chip 2018, 18, 610–619. [Google Scholar] [CrossRef]
- Tortajada-Genaro, L.A.; Santiago-Felipe, S.; Amasia, M.; Russom, A.; Maquieira, Á. Isothermal solid-phase recombinase polymerase amplification on microfluidic digital versatile discs (DVDs). Rsc Adv. 2015, 5, 29987–29995. [Google Scholar] [CrossRef]
- Nguyen, H.V.; Nguyen, V.D.; Lee, E.Y.; Seo, T.S. Point-of-care genetic analysis for multiplex pathogenic bacteria on a fully integrated centrifugal microdevice with a large-volume sample. Biosens. Bioelectron. 2019, 136, 132–139. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, Y.; Dai, R.; Lu, S.; Liu, X.; Zhou, T.; Yang, C.; Hu, X.; Lv, X.; Li, X. Oscillatory-Flow PCR Microfluidic Chip Driven by Low Speed Biaxial Centrifugation. Biosensors 2023, 13, 555. https://doi.org/10.3390/bios13050555
Fan Y, Dai R, Lu S, Liu X, Zhou T, Yang C, Hu X, Lv X, Li X. Oscillatory-Flow PCR Microfluidic Chip Driven by Low Speed Biaxial Centrifugation. Biosensors. 2023; 13(5):555. https://doi.org/10.3390/bios13050555
Chicago/Turabian StyleFan, Yunlong, Rongji Dai, Shuyu Lu, Xinyu Liu, Taiyan Zhou, Chunhua Yang, Xiaoming Hu, Xuefei Lv, and Xiaoqiong Li. 2023. "Oscillatory-Flow PCR Microfluidic Chip Driven by Low Speed Biaxial Centrifugation" Biosensors 13, no. 5: 555. https://doi.org/10.3390/bios13050555
APA StyleFan, Y., Dai, R., Lu, S., Liu, X., Zhou, T., Yang, C., Hu, X., Lv, X., & Li, X. (2023). Oscillatory-Flow PCR Microfluidic Chip Driven by Low Speed Biaxial Centrifugation. Biosensors, 13(5), 555. https://doi.org/10.3390/bios13050555




