Biogenesis, Isolation, and Detection of Exosomes and Their Potential in Therapeutics and Diagnostics
Abstract
:1. Introduction
2. Exosomes
2.1. History and Biogenesis of Exosomes
2.2. Key Characteristics
3. Isolation Methods for Exosomes
3.1. Ultracentrifugation
3.2. Precipitation
3.3. Immunoaffinity-Based Capture (IAC)
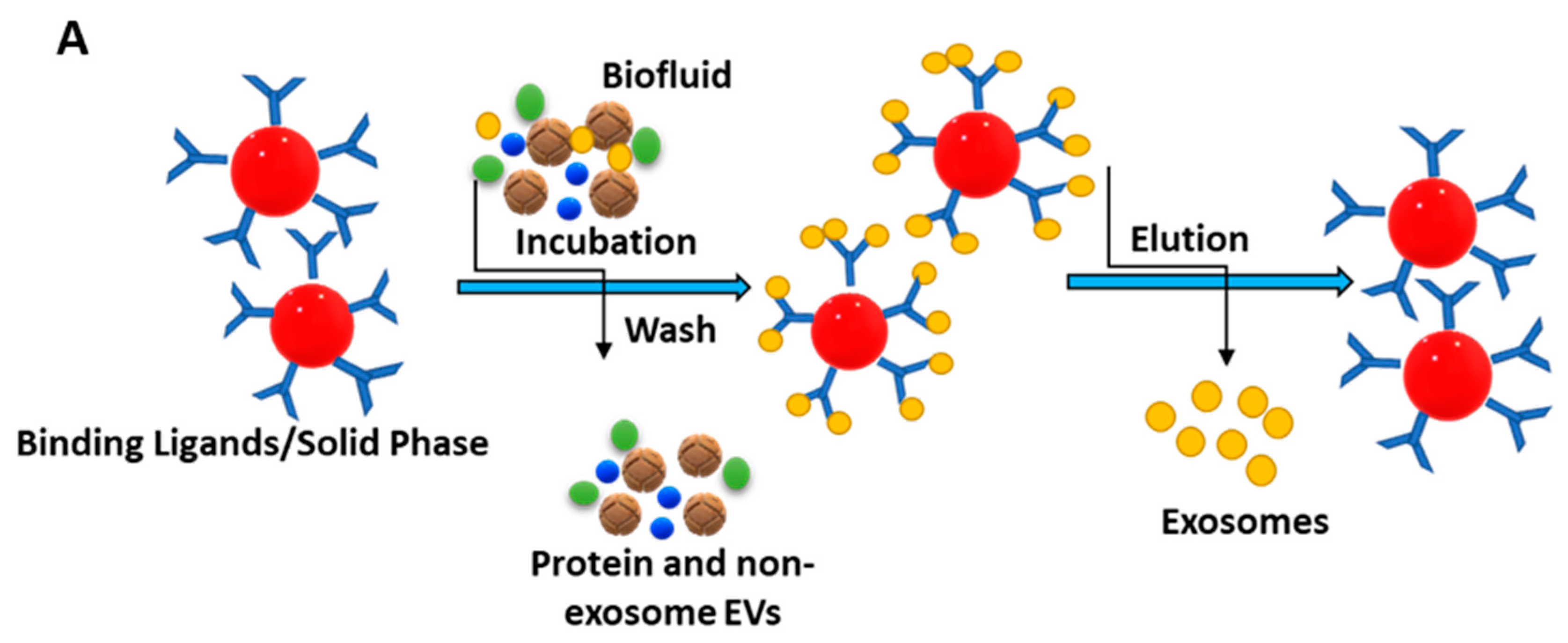
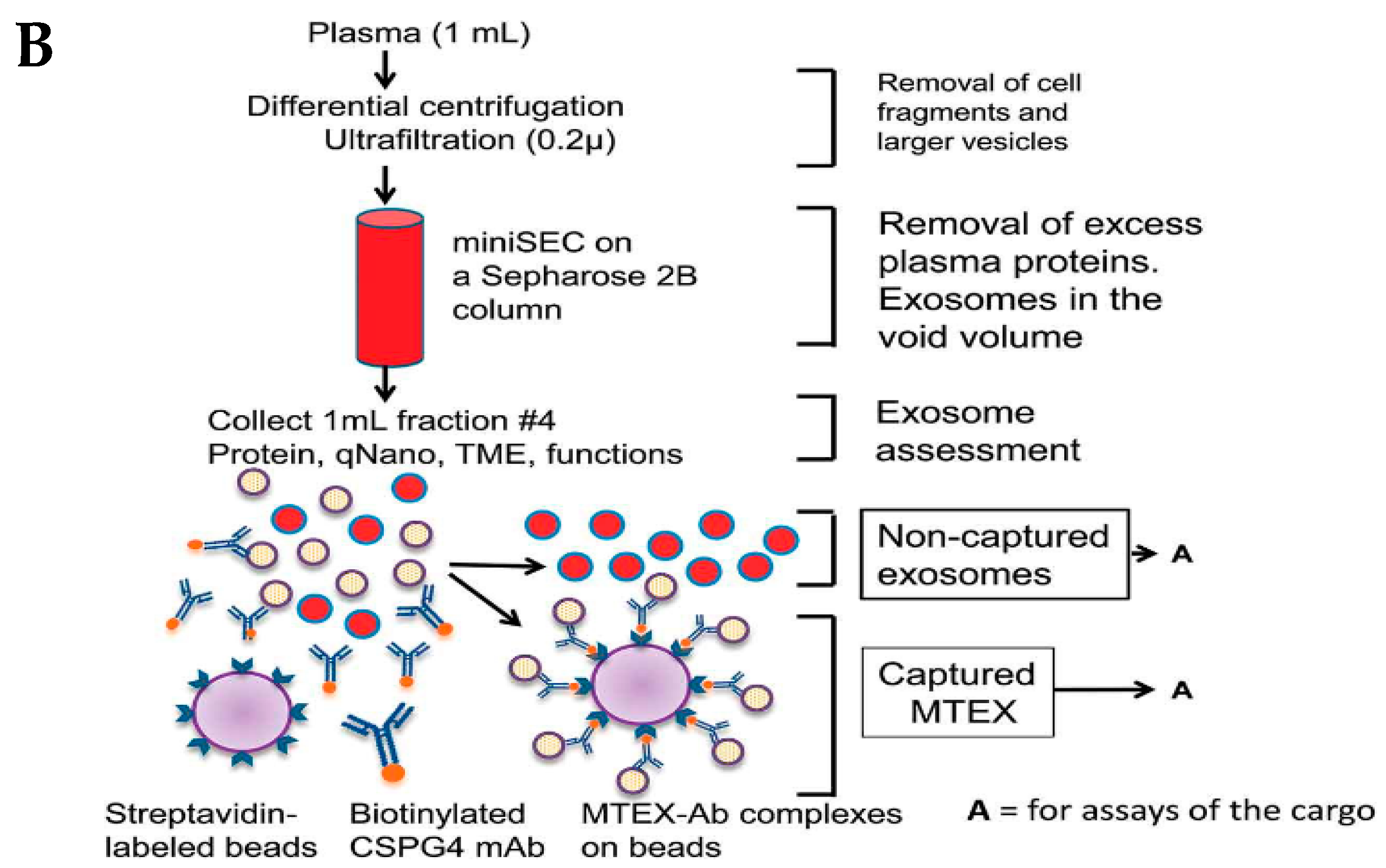
3.4. Size-Based Isolation Method
3.4.1. Ultrafiltration
3.4.2. Size Exclusion Chromatography
3.5. Microfluidic Separation
3.6. Charge-Based Isolation
4. Detection Methods for Exosomes
4.1. Nucleic Acid-Based Detection of Exosomes
4.2. Protein-Based Detection of Exosomes
4.2.1. Aptamer-Based Detection
4.2.2. Immunoreaction-Based Detection
4.2.3. Surface Plasmon Resonance Imaging (SPRi)-Based Detection
4.3. Lipid-Based Detection of Exosomes
4.4. Label-Free Exosome Imaging Methods
4.5. Nanoplatforms and Nano-Biosensors for the Detection of Exosomes
5. Applications of Exosomes
5.1. Exosomes for Early Detection of Diseases
5.1.1. Neurodegenerative Diseases
Parkinson’s Disease
Alzheimer’s Disease
Amyotrophic Lateral Sclerosis
5.1.2. Cancer
Prostate Cancer
Breast Cancer
5.2. Therapeutic Potential of Exosomes
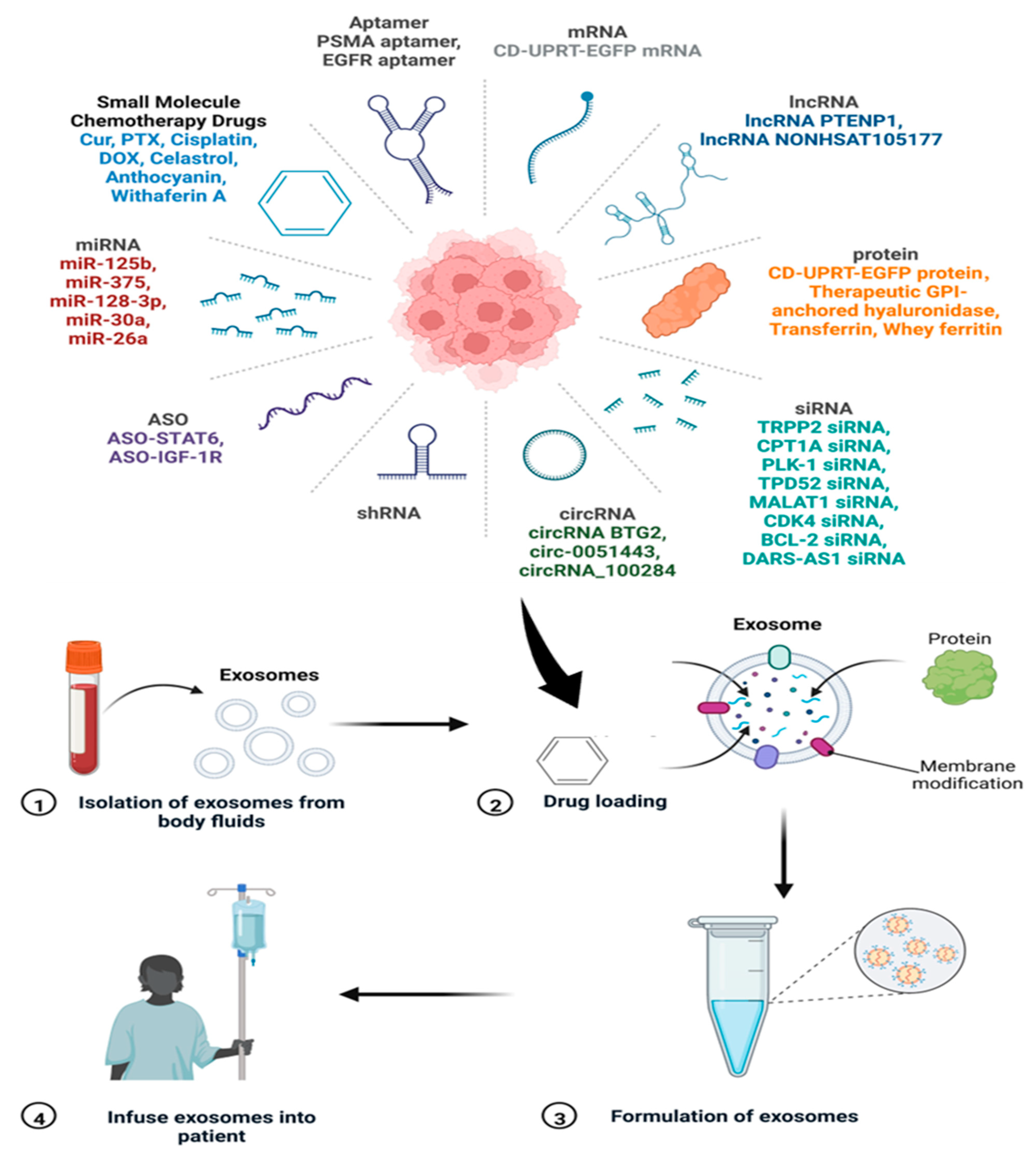
5.3. Exosomes as Drug Delivery Vehicles
6. Conclusions and Future Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, Z.; Yang, J.; Li, H.; Wang, C.; Fletcher, C.; Li, J.; Zhan, Y.; Du, L.; Wang, F.; Jiang, Y. Progress in the research of nanomaterial-based exosome bioanalysis and exosome-based nanomaterials tumor therapy. Biomaterials 2021, 274, 120873. [Google Scholar] [CrossRef] [PubMed]
- Azparren-Angulo, M.; Royo, F.; Gonzalez, E.; Liebana, M.; Brotons, B.; Berganza, J.; Goni-de-Cerio, F.; Manicardi, N.; Abad-Jorda, L.; Gracia-Sancho, J.; et al. Extracellular vesicles in hepatology: Physiological role, involvement in pathogenesis, and therapeutic opportunities. Pharmacol. Ther. 2021, 218, 107683. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Liu, G.; Li, J. Recent Progress on the Isolation and Detection Methods of Exosomes. Chem. Asian J. 2020, 15, 3973–3982. [Google Scholar] [CrossRef]
- Fang, X.; Wang, Y.; Wang, S.; Liu, B. Nanomaterials assisted exosomes isolation and analysis towards liquid biopsy. Mater. Today Bio 2022, 16, 100371. [Google Scholar] [CrossRef]
- Gardiner, C.; Ferreira, Y.J.; Dragovic, R.A.; Redman, C.W.G.; Sargent, I.L. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J. Extracell. Vesicles 2013, 2, 19671. [Google Scholar] [CrossRef] [PubMed]
- Brown, W. Dynamic Light Scattering: The Method and Some Applications; Clarendon Press: Oxford, UK, 1993; Volume 49. [Google Scholar]
- Kurian, T.K.; Banik, S.; Gopal, D.; Chakrabarti, S.; Mazumder, N. Elucidating methods for isolation and quantification of exosomes: A review. Mol. Biotechnol. 2021, 63, 249–266. [Google Scholar] [CrossRef] [PubMed]
- Doldán, X.; Fagúndez, P.; Cayota, A.; Laíz, J.; Tosar, J.P. Electrochemical sandwich immunosensor for determination of exosomes based on surface marker-mediated signal amplification. Anal. Chem. 2016, 88, 10466–10473. [Google Scholar] [CrossRef] [PubMed]
- Ashcroft, B.A.; de Sonneville, J.; Yuana, Y.; Osanto, S.; Bertina, R.; Kuil, M.E.; Oosterkamp, T.H. Determination of the size distribution of blood microparticles directly in plasma using atomic force microscopy and microfluidics. Biomed. Microdevices 2012, 14, 641–649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, M.K.; Mun, J.Y. Sample preparation and imaging of exosomes by transmission electron microscopy. JoVE (J. Vis. Exp.) 2018, 4, e56482. [Google Scholar]
- Jiang, L.; Gu, Y.; Du, Y.; Liu, J. Exosomes: Diagnostic Biomarkers and Therapeutic Delivery Vehicles for Cancer. Mol. Pharm. 2019, 16, 3333–3349. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Pan, Z.; Zhu, X.; Yang, R.; Yang, R.; Yang, T.; Hu, D.; Jing, A.; Liang, G.J.E.R. Mesoporous magnetic nanoparticles conjugated aptamers for exosomes capture and detection of Alzheimer’s disease. Eng. Regen. 2023, 4, 349–356. [Google Scholar] [CrossRef]
- Xin, H.; Li, Y.; Buller, B.; Katakowski, M.; Zhang, Y.; Wang, X.; Shang, X.; Zhang, Z.G.; Chopp, M. Exosome-mediated transfer of miR-133b from multipotent mesenchymal stromal cells to neural cells contributes to neurite outgrowth. Stem Cells 2012, 30, 1556–1564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Wang, M.; Gong, A.H.; Zhang, X.; Wu, X.D.; Zhu, Y.H.; Shi, H.; Wu, L.J.; Zhu, W.; Qian, H.; et al. HucMSC-Exosome Mediated-Wnt4 Signaling Is Required for Cutaneous Wound Healing. Stem Cells 2015, 33, 2158–2168. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.T.; Liu, C.G.; Gao, S.C.; Zhang, Y.; Wang, P.C.J.B.; Sciences, E. The serum exosome derived MicroRNA-135a, -193b, and -384 were potential Alzheimer’s disease biomarkers. Biomed. Environ. Sci. 2018, 31, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Brennan, E.; Wang, B.; McClelland, A.; Mohan, M.; Marai, M.; Beuscart, O.; Derouiche, S.; Gray, S.; Pickering, R.; Tikellis, C.; et al. Protective Effect of let-7 miRNA Family in Regulating Inflammation in Diabetes-Associated Atherosclerosis. Diabetes 2017, 66, 2266–2277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Giau, V.; An, S.S. Emergence of exosomal miRNAs as a diagnostic biomarker for Alzheimer’s disease. J. Neurol. Sci. 2016, 360, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Mosquera-Heredia, M.I.; Morales, L.C.; Vidal, O.M.; Barcelo, E.; Silvera-Redondo, C.; Velez, J.I.; Garavito-Galofre, P. Exosomes: Potential Disease Biomarkers and New Therapeutic Targets. Biomedicines 2021, 9, 1061. [Google Scholar] [CrossRef]
- Lin, Y.; Anderson, J.D.; Rahnama, L.M.A.; Gu, S.V.; Knowlton, A.A. Exosomes in disease and regeneration: Biological functions, diagnostics, and beneficial effects. Am. J. Physiol. Heart Circ. Physiol. 2020, 319, H1162–H1180. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef]
- Cui, S.; Cheng, Z.; Qin, W.; Jiang, L. Exosomes as a liquid biopsy for lung cancer. Lung Cancer 2018, 116, 46–54. [Google Scholar] [CrossRef]
- Pan, B.-T.; Johnstone, R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell 1983, 33, 967–978. [Google Scholar] [CrossRef]
- Enderle, D.; Spiel, A.; Coticchia, C.M.; Berghoff, E.; Mueller, R.; Schlumpberger, M.; Sprenger-Haussels, M.; Shaffer, J.M.; Lader, E.; Skog, J.; et al. Characterization of RNA from exosomes and other extracellular vesicles isolated by a novel spin column-based method. PLoS ONE 2015, 10, e0136133. [Google Scholar] [CrossRef] [Green Version]
- Stoorvogel, W.; Kleijmeer, M.J.; Geuze, H.J.; Raposo, G. The biogenesis and functions of exosomes. Traffic 2002, 3, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Urbanelli, L.; Magini, A.; Buratta, S.; Brozzi, A.; Sagini, K.; Polchi, A.; Tancini, B.; Emiliani, C. Signaling pathways in exosomes biogenesis, secretion and fate. Genes 2013, 4, 152–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Möbius, W.; Ohno-Iwashita, Y.; Donselaar, E.G.V.; Oorschot, V.M.J.; Shimada, Y.; Fujimoto, T.; Heijnen, H.F.G.; Geuze, H.J.; Slot, J.W. Immunoelectron microscopic localization of cholesterol using biotinylated and non-cytolytic perfringolysin O. J. Histochem. Cytochem. 2002, 50, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Kucharzewska, P.; Christianson, H.C.; Welch, J.E.; Svensson, K.J.; Fredlund, E.; Ringnér, M.; Mörgelin, M.; Bourseau-Guilmain, E.; Bengzon, J.; Belting, M. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc. Natl. Acad. Sci. USA 2013, 110, 7312–7317. [Google Scholar] [CrossRef] [PubMed]
- Farooqi, A.A.; Desai, N.N.; Qureshi, M.Z.; Librelotto, D.R.N.; Gasparri, M.L.; Bishayee, A.; Nabavi, S.M.; Curti, V.; Daglia, M. Exosome biogenesis, bioactivities and functions as new delivery systems of natural compounds. Biotechnol. Adv. 2018, 36, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Willms, E.; Johansson, H.J.; Mäger, I.; Lee, Y.; Blomberg, K.E.M.; Sadik, M.; Alaarg, A.; Smith, C.I.E.; Lehtiö, J.; El Andaloussi, S. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci. Rep. 2016, 6, 22519. [Google Scholar] [CrossRef] [Green Version]
- Zeringer, E.; Barta, T.; Li, M.; Vlassov, A.V. Strategies for isolation of exosomes. Cold Spring Harb. Protoc. 2015, 2015, 319–323. [Google Scholar] [CrossRef] [Green Version]
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; Degeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E. Syndecan–syntenin–ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012, 14, 677–685. [Google Scholar] [CrossRef]
- Hornick, N.I.; Huan, J.; Doron, B.; Goloviznina, N.A.; Lapidus, J.; Chang, B.H.; Kurre, P. Serum exosome microRNA as a minimally-invasive early biomarker of AML. Sci. Rep. 2015, 5, 11295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goto, T.; Fujiya, M.; Konishi, H.; Sasajima, J.; Fujibayashi, S.; Hayashi, A.; Utsumi, T.; Sato, H.; Iwama, T.; Ijiri, M. An elevated expression of serum exosomal microRNA-191, -21,-451a of pancreatic neoplasm is considered to be efficient diagnostic marker. BMC Cancer 2018, 18, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kowal, J.; Tkach, M.; Théry, C. Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 2014, 29, 116–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Li, J.; Liu, S.; Wang, T.; Ianni, A.; Bober, E.; Braun, T.; Xiang, R.; Yue, S. Exosomal tetraspanins mediate cancer metastasis by altering host microenvironment. Oncotarget 2017, 8, 62803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronquist, K.G.; Sanchez, C.; Dubois, L.; Chioureas, D.; Fonseca, P.; Larsson, A.; Ullén, A.; Yachnin, J.; Ronquist, G.; Panaretakis, T. Energy-requiring uptake of prostasomes and PC3 cell-derived exosomes into non-malignant and malignant cells. J. Extracell. Vesicles 2016, 5, 29877. [Google Scholar] [CrossRef] [PubMed]
- Doyle, L.M.; Wang, M.Z. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andaloussi, S.E.L.; Lakhal, S.; Mäger, I.; Wood, M.J.A. Exosomes for targeted siRNA delivery across biological barriers. Adv. Drug Deliv. Rev. 2013, 65, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Hergenreder, J.R. Serum Exosome Profile as Related to Early Pregnancy Status in the Mare. Ph.D. Thesis, Colorado State University, Fort Collins, CO, USA, 2011. [Google Scholar]
- Varela-Eirin, M.; Varela-Vazquez, A.; Mateos, M.R.-C.; Vila-Sanjurjo, A.; Fonseca, E.; Mascarenas, J.L.; Vázquez, M.E.; Mayan, M.D. Recruitment of RNA molecules by connexin RNA-binding motifs: Implication in RNA and DNA transport through microvesicles and exosomes. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2017, 1864, 728–736. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.A.; Park, H.; Lim, E.H.; Lee, K.W. Exosomes from breast cancer cells can convert adipose tissue-derived mesenchymal stem cells into myofibroblast-like cells. Int. J. Oncol. 2012, 40, 130–138. [Google Scholar]
- Kojima, R.; Bojar, D.; Rizzi, G.; Hamri, G.C.-E.; El-Baba, M.D.; Saxena, P.; Ausländer, S.; Tan, K.R.; Fussenegger, M. Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson’s disease treatment. Nat. Commun. 2018, 9, 1305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Ren, L.; Li, S.; Li, W.; Zheng, X.; Yang, Y.; Fu, W.; Yi, J.; Wang, J.; Du, G. The biology, function, and applications of exosomes in cancer. Acta Pharm. Sin. B 2021, 11, 2783–2797. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, P.; Zhang, T.; Xu, Z.; Huang, X.; Wang, R.; Du, L. Review on strategies and technologies for exosome isolation and purification. Front. Bioeng. Biotechnol. 2022, 9, 811971. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Kaslan, M.; Lee, S.H.; Yao, J.; Gao, Z. Progress in exosome isolation techniques. Theranostics 2017, 7, 789. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Kang, M.-H.; Jeyaraj, M.; Qasim, M.; Kim, J.-H. Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells 2019, 8, 307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bebelman, M.P.; Smit, M.J.; Pegtel, D.M.; Baglio, S.R. Biogenesis and function of extracellular vesicles in cancer. Pharmacol. Ther. 2018, 188, 1–11. [Google Scholar] [CrossRef]
- Castro, B.M.; Prieto, M.; Silva, L.C. Ceramide: A simple sphingolipid with unique biophysical properties. Prog. Lipid Res. 2014, 54, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef] [Green Version]
- Lobasso, S.; Tanzarella, P.; Mannavola, F.; Tucci, M.; Silvestris, F.; Felici, C.; Ingrosso, C.; Corcelli, A.; Lopalco, P. A lipidomic approach to identify potential biomarkers in exosomes from melanoma cells with different metastatic potential. Front. Physiol. 2021, 12, 748895. [Google Scholar] [CrossRef] [PubMed]
- Admyre, C.; Johansson, S.M.; Qazi, K.R.; Filén, J.-J.; Lahesmaa, R.; Norman, M.; Neve, E.; Scheynius, A.; Gabrielsson, S. Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 2007, 179, 1969–1978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalra, H.; Adda, C.G.; Liem, M.; Ang, C.S.; Mechler, A.; Simpson, R.J.; Hulett, M.D.; Mathivanan, S. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics 2013, 13, 3354–3364. [Google Scholar] [CrossRef]
- Gonzales, P.A.; Pisitkun, T.; Hoffert, J.D.; Tchapyjnikov, D.; Star, R.A.; Kleta, R.; Wang, N.S.; Knepper, M.A. Large-scale proteomics and phosphoproteomics of urinary exosomes. J. Am. Soc. Nephrol. 2009, 20, 363–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokoi, A.; Ochiya, T. Exosomes and extracellular vesicles: Rethinking the essential values in cancer biology. Semin. Cancer Biol. 2021, 74, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Stuffers, S.; Sem Wegner, C.; Stenmark, H.; Brech, A. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 2009, 10, 925–937. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akers, J.C.; Gonda, D.; Kim, R.; Carter, B.S.; Chen, C.C. Biogenesis of extracellular vesicles (EV): Exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J. Neuro-Oncol. 2013, 113, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shu, S.L.; Yang, Y.; Allen, C.L.; Hurley, E.; Tung, K.H.; Minderman, H.; Wu, Y.; Ernstoff, M.S. Purity and yield of melanoma exosomes are dependent on isolation method. J. Extracell. Vesicles 2020, 9, 1692401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Momen-Heravi, F.; Balaj, L.; Alian, S.; Mantel, P.-Y.; Halleck, A.E.; Trachtenberg, A.J.; Soria, C.E.; Oquin, S.; Bonebreak, C.M.; Saracoglu, E. Current methods for the isolation of extracellular vesicles. Biol. Chem. 2013, 394, 1253–1262. [Google Scholar] [CrossRef] [PubMed]
- Konoshenko, M.Y.; Lekchnov, E.A.; Vlassov, A.V.; Laktionov, P.P. Isolation of extracellular vesicles: General methodologies and latest trends. BioMed Res. Int. 2018, 2018, 8545347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, D.; Zhang, W.; Zhang, H.; Zhang, F.; Chen, L.; Ma, L.; Larcher, L.M.; Chen, S.; Liu, N.; Zhao, Q. Progress, opportunity, and perspective on exosome isolation-efforts for efficient exosome-based theranostics. Theranostics 2020, 10, 3684. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Rawat, S.; Arora, V.; Kottarath, S.K.; Dinda, A.K.; Vaishnav, P.K.; Nayak, B.; Mohanty, S. An improvised one-step sucrose cushion ultracentrifugation method for exosome isolation from culture supernatants of mesenchymal stem cells. Stem Cell Res. Ther. 2018, 9, 180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neves, J.S.; Perez, S.A.C.; Spencer, L.A.; Melo, R.C.N.; Weller, P.F. Subcellular fractionation of human eosinophils: Isolation of functional specific granules on isoosmotic density gradients. J. Immunol. Methods 2009, 344, 64–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dettenhofer, M.; Yu, X.-F. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J. Virol. 1999, 73, 1460–1467. [Google Scholar] [CrossRef] [PubMed]
- Ford, T.; Graham, J.; Rickwood, D. Iodixanol: A nonionic iso-osmotic centrifugation medium for the formation of self-generated gradients. Anal. Biochem. 1994, 220, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Skubal, L.R. The Essential Guide to Environmental Chemistry by George Schwedt; John Wiley & Sons Inc.: New York, NY, USA, 2001; Book Review; Volume 1. [Google Scholar] [CrossRef]
- Ritchie, C. Protein Purification; Iowa State University: Ames, IA, USA, 2012; Volume 2, p. 134. [Google Scholar]
- Rider, M.A.; Hurwitz, S.N.; Meckes, D.G. ExtraPEG: A polyethylene glycol-based method for enrichment of extracellular vesicles. Sci. Rep. 2016, 6, 23978. [Google Scholar] [CrossRef]
- Brownlee, Z.; Lynn, K.D.; Thorpe, P.E.; Schroit, A.J. A novel “salting-out” procedure for the isolation of tumor-derived exosomes. J. Immunol. Methods 2014, 407, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.L.; Khosroheidari, M.; Ravi, R.K.; DiStefano, J.K. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 2012, 82, 1024–1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertokova, A.; Svecova, N.; Kozics, K.; Gabelova, A.; Vikartovska, A.; Jane, E.; Hires, M.; Bertok, T.; Tkac, J. Exosomes from prostate cancer cell lines: Isolation optimisation and characterisation. Biomed. Pharmacother. 2022, 151, 113093. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Yang, H.C.; Rhee, W.J. Development and comparative analysis of human urine exosome isolation strategies. Process Biochem. 2020, 88, 197–203. [Google Scholar] [CrossRef]
- Weng, Y.; Sui, Z.; Shan, Y.; Hu, Y.; Chen, Y.; Zhang, L.; Zhang, Y. Effective isolation of exosomes with polyethylene glycol from cell culture supernatant for in-depth proteome profiling. Analyst 2016, 141, 4640–4646. [Google Scholar] [CrossRef] [PubMed]
- Gallart-Palau, X.; Serra, A.; Sze, S.K. Enrichment of extracellular vesicles from tissues of the central nervous system by PROSPR. Mol. Neurodegener. 2016, 11, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [PubMed]
- Zarovni, N.; Corrado, A.; Guazzi, P.; Zocco, D.; Lari, E.; Radano, G.; Muhhina, J.; Fondelli, C.; Gavrilova, J.; Chiesi, A. Integrated isolation and quantitative analysis of exosome shuttled proteins and nucleic acids using immunocapture approaches. Methods 2015, 87, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Lane, R.E.; Korbie, D.; Trau, M.; Hill, M.M. Purification protocols for extracellular vesicles. Extracell. Vesicles Methods Protoc. 2017, 1660, 111–130. [Google Scholar]
- Sharma, P.; Ludwig, S.; Muller, L.; Hong, C.S.; Kirkwood, J.M.; Ferrone, S.; Whiteside, T.L. Immunoaffinity-based isolation of melanoma cell-derived exosomes from plasma of patients with melanoma. J. Extracell. Vesicles 2018, 7, 1435138. [Google Scholar] [CrossRef] [PubMed]
- Saad, M.H.; Badierah, R.; Redwan, E.M.; El-Fakharany, E.M. A comprehensive insight into the role of exosomes in viral infection: Dual faces bearing different functions. Pharmaceutics 2021, 13, 1405. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Huang, W.; Wani, M.; Yu, X.; Ashraf, M. Ischemic preconditioning potentiates the protective effect of stem cells through secretion of exosomes by targeting Mecp2 via miR-22. PLoS ONE 2014, 9, e88685. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.-Y.; Sung, C.W.-H.; Chen, C.; Cheng, C.-M.; Lin, D.P.-C.; Huang, C.-T.; Hsu, M.-Y. Advances in exosomes technology. Clin. Chim. Acta 2019, 493, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Boussac, M.; Véron, P.; Ricciardi-Castagnoli, P.; Raposo, G.; Garin, J.; Amigorena, S. Proteomic analysis of dendritic cell-derived exosomes: A secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol. 2001, 166, 7309–7318. [Google Scholar] [CrossRef]
- Chahar, H.S.; Bao, X.; Casola, A. Exosomes and their role in the life cycle and pathogenesis of RNA viruses. Viruses 2015, 7, 3204–3225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witwer, K.W.; Buzás, E.I.; Bemis, L.T.; Bora, A.; Lässer, C.; Lötvall, J.; Nolte-‘t Hoen, E.N.; Piper, M.G.; Sivaraman, S.; Skog, J. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2013, 2, 20360. [Google Scholar] [CrossRef] [PubMed]
- Kosanović, M.; Milutinović, B.; Goč, S.; Mitić, N.; Janković, M. Ion-exchange chromatography purification of extracellular vesicles. Biotechniques 2017, 63, 65–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busatto, S.; Vilanilam, G.; Ticer, T.; Lin, W.-L.; Dickson, D.W.; Shapiro, S.; Bergese, P.; Wolfram, J. Tangential flow filtration for highly efficient concentration of extracellular vesicles from large volumes of fluid. Cells 2018, 7, 273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.Y.; Rhim, W.-K.; Yoo, Y.-I.; Kim, D.-S.; Ko, K.-W.; Heo, Y.; Park, C.G.; Han, D.K. Defined MSC exosome with high yield and purity to improve regenerative activity. J. Tissue Eng. 2021, 12, 20417314211008626. [Google Scholar] [CrossRef] [PubMed]
- Mulder, M. Basic Principles of Membrane Technology; Springer Science & Business Media: New York, NY, USA, 1996. [Google Scholar]
- Parimon, T.; Garrett Iii, N.E.; Chen, P.; Antes, T.J. Isolation of extracellular vesicles from murine bronchoalveolar lavage fluid using an ultrafiltration centrifugation technique. J. Vis. Exp. JoVE 2018, 9, e58310. [Google Scholar]
- Xu, R.; Simpson, R.J.; Greening, D.W. A protocol for isolation and proteomic characterization of distinct extracellular vesicle subtypes by sequential centrifugal ultrafiltration. Exosomes Microvesicles Methods Protoc. 2017, 1545, 91–116. [Google Scholar]
- Guerreiro, E.M.; Vestad, B.; Steffensen, L.A.; Aass, H.C.D.; Saeed, M.; Øvstebø, R.; Costea, D.E.; Galtung, H.K.; Søland, T.M. Efficient extracellular vesicle isolation by combining cell media modifications, ultrafiltration, and size-exclusion chromatography. PLoS ONE 2018, 13, e0204276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klymiuk, M.C.; Balz, N.; Elashry, M.I.; Heimann, M.; Wenisch, S.; Arnhold, S. Exosomes isolation and identification from equine mesenchymal stem cells. BMC Vet. Res. 2019, 15, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, C.-S.; Funk, S.; Muller, L.; Boyiadzis, M.; Whiteside, T.L. Isolation of biologically active and morphologically intact exosomes from plasma of patients with cancer. J. Extracell. Vesicles 2016, 5, 29289. [Google Scholar] [CrossRef] [PubMed]
- Böing, A.N.; Van Der Pol, E.; Grootemaat, A.E.; Coumans, F.A.W.; Sturk, A.; Nieuwland, R. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J. Extracell. Vesicles 2014, 3, 23430. [Google Scholar] [CrossRef] [PubMed]
- Koh, Y.Q.; Almughlliq, F.B.; Vaswani, K.; Peiris, H.N.; Mitchell, M.D. Exosome enrichment by ultracentrifugation and size exclusion chromatography. Front. Biosci. 2018, 23, 865–874. [Google Scholar]
- Contreras-Naranjo, J.C.; Wu, H.-J.; Ugaz, V.M. Microfluidics for exosome isolation and analysis: Enabling liquid biopsy for personalized medicine. Lab. A Chip 2017, 17, 3558–3577. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, A.C.; Gernaey, K.V.; Krühne, U. Connecting worlds–a view on microfluidics for a wider application. Biotechnol. Adv. 2018, 36, 1341–1366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, J.; Wei, X.; Zhang, X.; Wu, C.; Wang, Z.; Chen, M.; Wang, J. Microfluidic strategies for the isolation and profiling of exosomes. TrAC Trends Anal. Chem. 2022, 158, 116834. [Google Scholar] [CrossRef]
- Shirejini, S.Z.; Inci, F. The Yin and Yang of exosome isolation methods: Conventional practice, microfluidics, and commercial kits. Biotechnol. Adv. 2022, 54, 107814. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Ouyang, Y.; Wang, Z.; Zhang, R.; Huang, P.-H.; Chen, C.; Li, H.; Li, P.; Quinn, D.; Dao, M. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc. Natl. Acad. Sci. USA 2017, 114, 10584–10589. [Google Scholar] [CrossRef]
- Kang, Y.T.; Purcell, E.; Palacios-Rolston, C.; Lo, T.W.; Ramnath, N.; Jolly, S.; Nagrath, S. Isolation and profiling of circulating tumor-associated exosomes using extracellular vesicular lipid–protein binding affinity based microfluidic device. Small 2019, 15, 1903600. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Liao, C.; Zuo, P.; Liu, Z.; Ye, B.-C. Magnetic-based microfluidic device for on-chip isolation and detection of tumor-derived exosomes. Anal. Chem. 2018, 90, 13451–13458. [Google Scholar] [CrossRef]
- Liu, C.; Feng, Q.; Sun, J. Lipid nanovesicles by microfluidics: Manipulation, synthesis, and drug delivery. Adv. Mater. 2019, 31, 1804788. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.; Bae, J.; Jeon, H.; Lee, S.; Kim, T. Pervaporation-assisted in situ formation of nanoporous microchannels with various material and structural properties. Lab. A Chip 2022, 22, 1474–1485. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Liu, X.; Zhang, Z.; Liu, L.-E.; He, S.; Wu, Y.; Effah, C.Y.; Yang, R.; Zhang, A.; Chen, W.; et al. Magnetic-nanowaxberry-based microfluidic ExoSIC for affinity and continuous separation of circulating exosomes towards cancer diagnosis. R. Soc. Chem. 2023, 23, 1694–1702. [Google Scholar] [CrossRef]
- Yang, F.; Liao, X.; Tian, Y.; Li, G. Exosome separation using microfluidic systems: Size-based, immunoaffinity-based and dynamic methodologies. Biotechnol. J. 2017, 12, 1600699. [Google Scholar] [CrossRef] [PubMed]
- Morani, M.; Mai, T.D.; Krupova, Z.; Defrenaix, P.; Multia, E.; Riekkola, M.-L.; Taverna, M. Electrokinetic characterization of extracellular vesicles with capillary electrophoresis: A new tool for their identification and quantification. Anal. Chim. Acta 2020, 1128, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Deregibus, M.C.; Figliolini, F.; D’Antico, S.; Manzini, P.M.; Pasquino, C.; De Lena, M.; Tetta, C.; Brizzi, M.F.; Camussi, G. Charge-based precipitation of extracellular vesicles. Int. J. Mol. Med. 2016, 38, 1359–1366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.-K.; Nishida, H.; An, S.Y.; Shetty, A.K.; Bartosh, T.J.; Prockop, D.J. Chromatographically isolated CD63+ CD81+ extracellular vesicles from mesenchymal stromal cells rescue cognitive impairments after TBI. Proc. Natl. Acad. Sci. USA 2016, 113, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Heath, N.; Grant, L.; De Oliveira, T.M.; Rowlinson, R.; Osteikoetxea, X.; Dekker, N.; Overman, R. Rapid isolation and enrichment of extracellular vesicle preparations using anion exchange chromatography. Sci. Rep. 2018, 8, 5730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martins, T.S.; Vaz, M.; Henriques, A.G. A review on comparative studies addressing exosome isolation methods from body fluids. Anal. Bioanal. Chem. 2023, 415, 1239–1263. [Google Scholar] [CrossRef] [PubMed]
- Hasan, A.; Pattanayek, S.K.; Pandey, L.M. Effect of Functional Groups of Self-Assembled Monolayers on Protein Adsorption and Initial Cell Adhesion. ACS Biomater. Sci. Eng. 2018, 4, 3224–3233. [Google Scholar] [CrossRef]
- Saad, M.G.; Beyenal, H.; Dong, W.-J. Exosomes as powerful engines in cancer: Isolation, characterization and detection techniques. Biosensors 2021, 11, 518. [Google Scholar] [CrossRef]
- Zhou, S.; Yang, Y.; Wu, Y.; Liu, S. Multiplexed profiling of biomarkers in extracellular vesicles for cancer diagnosis and therapy monitoring. Anal. Chim. Acta 2021, 1175, 338633. [Google Scholar] [CrossRef] [PubMed]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajput, A.; Varshney, A.; Bajaj, R.; Pokharkar, V. Exosomes as New Generation Vehicles for Drug Delivery: Biomedical Applications and Future Perspectives. Molecules 2022, 27, 7289. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; He, L.; Xia, Y.; Xu, H.; Liu, C.; Xie, H.; Wang, S.; Peng, L.; Liu, Y.; Liu, Y. A sensitive aptasensor based on a hemin/G-Quadruplex-assisted signal amplification strategy for electrochemical detection of gastric cancer exosomes. Small 2019, 15, 1900735. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Li, Y.-T.; Jin, D.; Yang, F.; Wu, D.; Xiao, M.-M.; Zhang, H.; Zhang, Z.-Y.; Zhang, G.-J. Electrical and label-free quantification of exosomes with a reduced graphene oxide field effect transistor biosensor. Anal. Chem. 2019, 91, 10679–10686. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, Z.; Wang, F.; Zhang, Y.; Wang, H.; Liu, Y. In situ formation of gold nanoparticles decorated Ti3C2 MXenes nanoprobe for highly sensitive electrogenerated chemiluminescence detection of exosomes and their surface proteins. Anal. Chem. 2020, 92, 5546–5553. [Google Scholar] [CrossRef]
- Xu, L.; Shoaie, N.; Jahanpeyma, F.; Zhao, J.; Azimzadeh, M.; Al Jamal, K.T. Optical, electrochemical and electrical (nano)biosensors for detection of exosomes: A comprehensive overview. Biosens. Bioelectron. 2020, 161, 112222. [Google Scholar] [CrossRef] [PubMed]
- Zong, S.F.; Wang, L.; Chen, C.; Lu, J.; Zhu, D.; Zhang, Y.Z.; Wang, Z.Y.; Cui, Y.P. Facile detection of tumor-derived exosomes using magnetic nanobeads and SERS nanoprobes. Anal. Methods 2016, 8, 5001–5008. [Google Scholar] [CrossRef]
- He, F.; Liu, H.; Guo, X.; Yin, B.-C.; Ye, B.-C. Direct exosome quantification via bivalent-cholesterol-labeled DNA anchor for signal amplification. Anal. Chem. 2017, 89, 12968–12975. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.-T.; Kim, Y.J.; Bu, J.; Cho, Y.-H.; Han, S.-W.; Moon, B.-I. High-purity capture and release of circulating exosomes using an exosome-specific dual-patterned immunofiltration (ExoDIF) device. Nanoscale 2017, 9, 13495–13505. [Google Scholar] [CrossRef]
- Nie, P.; Bai, Y.; Mei, H. Synthetic life with alternative nucleic acids as genetic materials. Molecules 2020, 25, 3483. [Google Scholar] [CrossRef]
- Huang, L.; Gu, N.; Zhang, X.E.; Wang, D.B. Light-Inducible Exosome-Based Vehicle for Endogenous RNA Loading and Delivery to Leukemia Cells. Adv. Funct. Mater. 2019, 29, 1807189. [Google Scholar] [CrossRef]
- Dong, H.; Chen, H.; Jiang, J.; Zhang, H.; Cai, C.; Shen, Q. Highly sensitive electrochemical detection of tumor exosomes based on aptamer recognition-induced multi-DNA release and cyclic enzymatic amplification. Anal. Chem. 2018, 90, 4507–4513. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, L.; Wan, S.; Cansiz, S.; Cui, C.; Liu, Y.; Cai, R.; Hong, C.; Teng, I.T.; Shi, M. Aptasensor with expanded nucleotide using DNA nanotetrahedra for electrochemical detection of cancerous exosomes. ACS Nano 2017, 11, 3943–3949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; Aswath, K.; Schroeder, S.G.; Lippolis, J.D.; Reinhardt, T.A.; Sonstegard, T.S. MicroRNA expression profiles of bovine milk exosomes in response to Staphylococcus aureus infection. BMC Genom. 2015, 16, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Li, J.; Liu, Y.; Yuan, Y.; Xu, G. Construction of a Concanavalin A electrochemical sensor base on a novel sandwich capture mode. Sens. Actuators B Chem. 2017, 248, 201–206. [Google Scholar] [CrossRef]
- Zhou, H.; Pisitkun, T.; Aponte, A.; Yuen, P.S.T.; Hoffert, J.D.; Yasuda, H.; Hu, X.; Chawla, L.; Shen, R.F.; Knepper, M.A. Exosomal Fetuin-A identified by proteomics: A novel urinary biomarker for detecting acute kidney injury. Kidney Int. 2006, 70, 1847–1857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-Sánchez, A.; Madrigal-Santillán, E.; Bautista, M.; Esquivel-Soto, J.; Morales-González, Á.; Esquivel-Chirino, C.; Durante-Montiel, I.; Sánchez-Rivera, G.; Valadez-Vega, C.; Morales-González, J.A. Inflammation, oxidative stress, and obesity. Int. J. Mol. Sci. 2011, 12, 3117–3132. [Google Scholar] [CrossRef] [Green Version]
- An, Y.; Jin, T.; Zhu, Y.; Zhang, F.; He, P. An ultrasensitive electrochemical aptasensor for the determination of tumor exosomes based on click chemistry. Biosens. Bioelectron. 2019, 142, 111503. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Liu, M.; Wang, L.; Yan, A.; He, W.; Chen, M.; Lan, J.; Xu, J.; Guan, L.; Chen, J. A visible and colorimetric aptasensor based on DNA-capped single-walled carbon nanotubes for detection of exosomes. Biosens. Bioelectron. 2017, 92, 8–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, L.; Ba, L.; Xiong, Y.; Peng, G. A hybridization chain reaction based assay for fluorometric determination of exosomes using magnetic nanoparticles and both aptamers and antibody as recognition elements. Microchim. Acta 2019, 186, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Y.; Cui, D.; Huang, J.; Fan, W.; Miao, Y.; Pu, K. Near-infrared afterglow semiconducting nano-polycomplexes for the multiplex differentiation of cancer exosomes. Angew. Chem. Int. Ed. 2019, 58, 4983–4987. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhao, J.; Tian, F.; Cai, L.; Zhang, W.; Feng, Q.; Chang, J.; Wan, F.; Yang, Y.; Dai, B. Low-cost thermophoretic profiling of extracellular-vesicle surface proteins for the early detection and classification of cancers. Nat. Biomed. Eng. 2019, 3, 183–193. [Google Scholar] [CrossRef]
- Zhang, Z.; Tang, C.; Zhao, L.; Xu, L.; Zhou, W.; Dong, Z.; Yang, Y.; Xie, Q.; Fang, X. Aptamer-based fluorescence polarization assay for separation-free exosome quantification. Nanoscale 2019, 11, 10106–10113. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Shi, M.; Liu, Y.; Wan, S.; Cui, C.; Zhang, L.; Tan, W. Aptamer/AuNP biosensor for colorimetric profiling of exosomal proteins. Angew. Chem. Int. Ed. 2017, 56, 11916–11920. [Google Scholar] [CrossRef] [PubMed]
- Fang, D.; Zhao, D.; Zhang, S.; Huang, Y.; Dai, H.; Lin, Y. Black phosphorus quantum dots functionalized MXenes as the enhanced dual-mode probe for exosomes sensing. Sens. Actuators B Chem. 2020, 305, 127544. [Google Scholar] [CrossRef]
- Wang, L.; Yang, Y.; Liu, Y.; Ning, L.; Xiang, Y.; Li, G. Bridging exosome and liposome through zirconium–phosphate coordination chemistry: A new method for exosome detection. Chem. Commun. 2019, 55, 2708–2711. [Google Scholar] [CrossRef] [PubMed]
- Pandey, L.M. Design of engineered surfaces for prospective detection of SARS-CoV-2 using quartz crystal microbalance-based techniques. Expert Rev. Proteom. 2020, 17, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Fopase, R.; Panda, C.; Rajendran, A.P.; Uludag, H.; Pandey, L.M. Potential of siRNA in COVID-19 therapy: Emphasis on in silico design and nanoparticles based delivery. Front. Bioeng. Biotechnol. 2023, 11, 1112755. [Google Scholar] [CrossRef] [PubMed]
- Moura, S.L.; Martín, C.G.; Martí, M.; Pividori, M.I. Multiplex detection and characterization of breast cancer exosomes by magneto-actuated immunoassay. Talanta 2020, 211, 120657. [Google Scholar] [CrossRef]
- Wang, C.; Wang, C.; Jin, D.; Yu, Y.; Yang, F.; Zhang, Y.; Yao, Q.; Zhang, G.-J. AuNP-amplified surface acoustic wave sensor for the quantification of exosomes. ACS Sens. 2020, 5, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Duan, X.; Zhao, M.; Wei, X.; Wu, J.; Chen, W.; Liu, P.; Cheng, W.; Cheng, Q.; Ding, S. High-sensitive and multiplex biosensing assay of NSCLC-derived exosomes via different recognition sites based on SPRi array. Biosens. Bioelectron. 2020, 154, 112066. [Google Scholar] [CrossRef] [PubMed]
- Picciolini, S.; Gualerzi, A.; Vanna, R.; Sguassero, A.; Gramatica, F.; Bedoni, M.; Masserini, M.; Morasso, C. Detection and characterization of different brain-derived subpopulations of plasma exosomes by surface plasmon resonance imaging. Anal. Chem. 2018, 90, 8873–8880. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Luo, C.; Mei, Q.; Zhang, H.; Zhang, W.; Su, D.; Fu, W.; Luo, Y. Aptamer-cholesterol-mediated proximity ligation assay for accurate identification of exosomes. Anal. Chem. 2020, 92, 5411–5418. [Google Scholar] [CrossRef]
- Skotland, T.; Ekroos, K.; Kauhanen, D.; Simolin, H.; Seierstad, T.; Berge, V.; Sandvig, K.; Llorente, A. Molecular lipid species in urinary exosomes as potential prostate cancer biomarkers. Eur. J. Cancer 2017, 70, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, D.; Yue, S.; Lu, Y.; Yang, C.; Fang, J.; Xu, Z. Sensitive multicolor visual detection of exosomes via dual signal amplification strategy of enzyme-catalyzed metallization of Au nanorods and hybridization chain reaction. ACS Sens. 2019, 4, 3210–3218. [Google Scholar] [CrossRef]
- Zhang, P.; Jiang, J.; Zhou, X.; Kolay, J.; Wang, R.; Wan, Z.; Wang, S. Label-free imaging and biomarker analysis of exosomes with plasmonic scattering microscopy. Chem. Sci. 2022, 13, 12760–12768. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Huang, Z.; Liu, C.; Yang, S.; Zhang, Y.; Sun, J.; Chen, J.; Chen, J.; Xu, F.; Chen, Y. Label free imaging and deep tracking of single biological nanoparticles in free solution by reflection enhanced dark field scattering microscopy. Sens. Actuators B Chem. 2022, 355, 131317. [Google Scholar] [CrossRef]
- Cheng, J.-X.; Xie, X.S. Vibrational spectroscopic imaging of living systems: An emerging platform for biology and medicine. Science 2015, 350, aaa8870. [Google Scholar] [CrossRef] [PubMed]
- Milane, L.; Singh, A.; Mattheolabakis, G.; Suresh, M.; Amiji, M.M. Exosome mediated communication within the tumor microenvironment. J. Control. Release 2015, 219, 278–294. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Shan, X.; Wang, S.; Chen, H.; Tao, N. Molecular scale origin of surface plasmon resonance biosensors. Anal. Chem. 2014, 86, 8992–8997. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Shen, G.; Wang, H.; Li, H.; Zhang, T.; Tao, N.; Ding, X.; Yu, H. Interferometric plasmonic imaging and detection of single exosomes. Proc. Natl. Acad. Sci. USA 2018, 115, 10275–10280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehak; Thummer, R.P.; Pandey, L.M. Surface modified iron-oxide based engineered nanomaterials for hyperthermia therapy of cancer cells. Biotechnol. Genet. Eng. Rev. 2023, 29, 1–47. [Google Scholar] [CrossRef]
- Jawed, A.; Saxena, V.; Pandey, L.M. Engineered nanomaterials and their surface functionalization for the removal of heavy metals: A review. J. Water Process Eng. 2020, 33, 101009. [Google Scholar] [CrossRef]
- Pandey, L.M. Surface engineering of nano-sorbents for the removal of heavy metals: Interfacial aspects. J. Environ. Chem. Eng. 2021, 9, 104586. [Google Scholar] [CrossRef]
- Pandey, L.M. Surface engineering of personal protective equipments (PPEs) to prevent the contagious infections of SARS-CoV-2. Surf. Eng. 2020, 36, 901–907. [Google Scholar] [CrossRef]
- Chandra, P.; Pandey, L.M. Biointerface Engineering: Prospects in Medical Diagnostics and Drug Delivery; Springer Nature: Singapore, 2020. [Google Scholar]
- Pandey, L.M.; Hasan, A. Nanoscale Engineering of Biomaterials: Properties and Applications; Springer Nature: Singapore, 2022. [Google Scholar]
- Hasan, A.; Saxena, V.; Pandey, L.M. Surface Functionalization of Ti6Al4V via Self-assembled Monolayers for Improved Protein Adsorption and Fibroblast Adhesion. Langmuir 2018, 34, 3494–3506. [Google Scholar] [CrossRef] [PubMed]
- Hasan, A.; Pandey, L.M. Kinetic studies of attachment and re-orientation of octyltriethoxysilane for formation of self-assembled monolayer on a silica substrate. Mater. Sci. Eng. C 2016, 68, 423–429. [Google Scholar] [CrossRef]
- Pandey, L.M. Design of biocompatible and self-antibacterial titanium surfaces for biomedical applications. Curr. Opin. Biomed. Eng. 2023, 25, 100423. [Google Scholar] [CrossRef]
- Back, S.J.; Kim, W.; Kim, D.Y.; Kim, S.J.; Hwang, S.R.; Jung, G.B. Rapid and simple isolation and detection of exosomes using CaTiO3:Eu3+@Fe3O4 multifunctional nanocomposites. Anal. Biochem. 2023, 673, 115161. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wang, B.C.; Feng, Q.S.; Fang, X.; Dai, X.H.; Yan, Y.H.; Ding, C.F. Magnetic guanidyl-functionalized covalent organic framework composite: A platform for specific capture and isolation of phosphopeptides and exosomes. Microchim. Acta 2022, 189, 330. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.O.; Ahn, H.S.; Dao, T.N.T.; Hong, J.Y.; Shin, W.; Lim, Y.M.; Chung, S.J.; Lee, J.H.; Liu, H.F.; Koo, B.; et al. Magnetic transferrin nanoparticles (MTNs) assay as a novel isolation approach for exosomal biomarkers in neurological diseases. Biomater. Res. 2023, 27, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Farsani, A.M.; Rahimi, F.; Taebnia, N.; Salimi, M.; Arpanaei, A. Tailored design and preparation of magnetic nanocomposite particles for the isolation of exosomes. Nanotechnology 2023, 34, 155603. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Pathak, A.; Kumar, S.; Malik, P.S.; Elangovan, R. Rapid immunomagnetic co-capture assay for quantification of lung cancer associated exosomes. J. Immunol. Methods 2022, 508, 113324. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.J.; Yu, Y.R.; Shou, X.; Zhang, D.G.; Liang, G.F.; Zhao, Y.J. Hedgehog-inspired magnetic nanoparticles for effectively capturing and detecting exosomes. NPG Asia Mater. 2021, 13, 78. [Google Scholar] [CrossRef]
- Pammi Guru, K.T.; Praween, N.; Basu, P.K. Isolation of Exosomes from Human Serum Using Gold-Nanoparticle-Coated Silicon Surface. Nanomaterials 2023, 13, 387. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, P.; Biswas, A.; Biswas, S.C. Brain-enriched miR-128: Reduced in exosomes from Parkinson’s patient plasma, improves synaptic integrity, and prevents 6-OHDA mediated neuronal apoptosis. Front. Cell Neurosci. 2023, 16, 680. [Google Scholar] [CrossRef]
- Gui, Y.; Liu, H.; Zhang, L.; Lv, W.; Hu, X. Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget 2015, 6, 37043–37053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, X.Y.; Lu, J.M.; Zhao, Z.Q.; Li, M.C.; Lu, T.; An, X.S.; Xue, L.J. MicroRNA biomarkers of Parkinson’s disease in serum exosome-like microvesicles. Neurosci. Lett. 2017, 644, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.F.; Qu, M.W.; Li, G.C.; Zhang, F.B.; Rui, H.C. Circulating exosomal miRNAs as diagnostic biomarkers in Parkinson’s disease. Eur. Rev. Med. Pharm. 2018, 22, 5278–5283. [Google Scholar] [CrossRef]
- Manna, I.; Quattrone, A.; De Benedittis, S.; Vescio, B.; Iaccino, E.; Quattrone, A. Exosomal miRNA as peripheral biomarkers in Parkinson’s disease and progressive supranuclear palsy: A pilot study. Park. Relat. D 2021, 93, 77–84. [Google Scholar] [CrossRef]
- Tomlinson, P.R.; Zheng, Y.; Fischer, R.; Heidasch, R.; Gardiner, C.; Evetts, S.; Hu, M.; Wade-Martins, R.; Turner, M.R.; Morris, J.; et al. Identification of distinct circulating exosomes in Parkinson’s disease. Ann. Clin. Transl. Neurol. 2015, 2, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Sharma, L.G.; Pandey, L.M. Shear-induced aggregation of amyloid β (1–40) in a parallel plate geometry. J. Biomol. Struct. Dyn. 2021, 39, 6415–6423. [Google Scholar] [CrossRef]
- Panda, C.; Voelz, C.; Habib, P.; Mevissen, C.; Pufe, T.; Beyer, C.; Gupta, S.; Slowik, A. Aggregated Tau-PHF6 (VQIVYK) Potentiates NLRP3 Inflammasome Expression and Autophagy in Human Microglial Cells. Cells 2021, 10, 1652. [Google Scholar] [CrossRef]
- Pandey, L.M. Physicochemical factors of bioprocessing impact the stability of therapeutic proteins. Biotechnol. Adv. 2022, 55, 107909. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Zhao, Y.; Zhou, X.; Luan, J.; Cui, Y.; Han, J. Comparison of the extraction and determination of serum exosome and miRNA in serum and the detection of miR-27a-3p in serum exosome of ALS patients. Intractable Rare Dis. Res. 2018, 7, 13–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katsu, M.; Hama, Y.; Utsumi, J.; Takashina, K.; Yasumatsu, H.; Mori, F.; Wakabayashi, K.; Shoji, M.; Sasaki, H. MicroRNA expression profiles of neuron-derived extracellular vesicles in plasma from patients with amyotrophic lateral sclerosis. Neurosci. Lett. 2019, 708, 134176. [Google Scholar] [CrossRef]
- Lo, T.W.; Figueroa-Romero, C.; Hur, J.; Pacut, C.; Stoll, E.; Spring, C.; Lewis, R.; Nair, A.; Goutman, S.A.; Sakowski, S.A.; et al. Extracellular Vesicles in Serum and Central Nervous System Tissues Contain microRNA Signatures in Sporadic Amyotrophic Lateral Sclerosis. Front. Mol. Neurosci. 2021, 14, 739016. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.A.; Park, C.; Sung, J.J.; Seo, D.J.; Choi, S.J.; Hong, Y.H. Small RNA sequencing of circulating small extracellular vesicles microRNAs in patients with amyotrophic lateral sclerosis. Sci. Rep. 2023, 13, 5528. [Google Scholar] [CrossRef] [PubMed]
- Saucier, D.; Wajnberg, G.; Roy, J.; Beauregard, A.P.; Chacko, S.; Crapoulet, N.; Fournier, S.; Ghosh, A.; Lewis, S.M.; Marrero, A.; et al. Identification of a circulating miRNA signature in extracellular vesicles collected from amyotrophic lateral sclerosis patients. Brain Res. 2019, 1708, 100–108. [Google Scholar] [CrossRef]
- Cai, K.; Li, T.; Guo, L.; Guo, H.; Zhu, W.; Yan, L.; Li, F. Long non-coding RNA LINC00467 regulates hepatocellular carcinoma progression by modulating miR-9-5p/PPARA expression. Open Biol. 2019, 9, 190074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, Y.; Zhao, H. Liquid biopsy in tumors: Opportunities and challenges. Ann. Transl. Med. 2018, 6, S89. [Google Scholar] [CrossRef] [PubMed]
- Ignatiadis, M.; Sledge, G.W.; Jeffrey, S.S. Liquid biopsy enters the clinic—Implementation issues and future challenges. Nat. Rev. Clin. Oncol. 2021, 18, 297–312. [Google Scholar] [CrossRef]
- Li, Q.; Wang, Y.; Ling, L.; Qiao, L.; Chen, H.; Ding, C.; Yu, S. Rapid and specific detection nanoplatform of serum exosomes for prostate cancer diagnosis. Mikrochim. Acta 2021, 188, 283. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, J.H.; Mondal, J.; Hwang, J.; Kim, H.S.; Kumar, V.; Raj, A.; Hwang, S.R.; Lee, Y.K. Magnetofluoro-Immunosensing Platform Based on Binary Nanoparticle-Decorated Graphene for Detection of Cancer Cell-Derived Exosomes. Int. J. Mol. Sci. 2022, 23, 9619. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zhu, N.; Zhou, J.; Kang, K.; Zhou, X.; Ying, B.; Yi, Q.; Wu, Y. A magnetic surface-enhanced Raman scattering platform for performing successive breast cancer exosome isolation and analysis. J. Mater. Chem. B 2021, 9, 2709–2716. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Xiang, Y.; Li, N.; Wei, B.; Chen, Y.; Fang, D.; Fang, M.; Li, Q.; Liu, J.; Tang, Y.; et al. Simultaneous detection of cancerous exosomal miRNA-21 and PD-L1 with a sensitive dual-cycling nanoprobe. Biosens. Bioelectron. 2022, 216, 114636. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Li, Y.; Huang, L.; He, W.; Wang, S.; Wang, C.; Zhou, G.; Chen, Y.; Zhou, X.; Huang, Y.; et al. Label-free diagnosis for colorectal cancer through coffee ring-assisted surface-enhanced Raman spectroscopy on blood serum. J. Biophotonics 2020, 13, e201960176. [Google Scholar] [CrossRef]
- Chinnappan, R.; Ramadan, Q.; Zourob, M. An integrated lab-on-a-chip platform for pre-concentration and detection of colorectal cancer exosomes using anti-CD63 aptamer as a recognition element. Biosens. Bioelectron. 2023, 220, 114856. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.H.; Pulin, A.A.; Seo, M.J.; Kota, D.J.; Ylostalo, J.; Larson, B.L.; Semprun-Prieto, L.; Delafontaine, P.; Prockop, D.J. Intravenous hMSCs Improve Myocardial Infarction in Mice because Cells Embolized in Lung Are Activated to Secrete the Anti-inflammatory Protein TSG-6. Cell Stem Cell 2009, 5, 54–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, R.C.; Arslan, F.; Lee, M.M.; Sze, N.S.; Choo, A.; Chen, T.S.; Salto-Tellez, M.; Timmers, L.; Lee, C.N.; El Oakley, R.M.; et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010, 4, 214–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, Z.C.; Lu, J.; Wang, S.Z.; Wu, H.; Zhang, Y.T.; Xu, S.G. Stem cell-derived exosomes: A promising strategy for fracture healing. Cell Prolif. 2017, 50, e12359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, X.J.; Sun, X.Y.; Huang, K.M.; Zhang, L.; Yang, Z.S.; Zou, D.D.; Wang, B.; Warnock, G.L.; Dai, L.J.; Luo, J. Therapeutic potential of CAR-T cell-derived exosomes: A cell-free modality for targeted cancer therapy. Oncotarget 2015, 6, 44179–44190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lugini, L.; Cecchetti, S.; Huber, V.; Luciani, F.; Macchia, G.; Spadaro, F.; Paris, L.; Abalsamo, L.; Colone, M.; Molinari, A.; et al. Immune surveillance properties of human NK cell-derived exosomes. J. Immunol. 2012, 189, 2833–2842. [Google Scholar] [CrossRef] [Green Version]
- Vickers, K.C.; Landstreet, S.R.; Levin, M.G.; Shoucri, B.M.; Toth, C.L.; Taylor, R.C.; Palmisano, B.T.; Tabet, F.; Cui, H.L.; Rye, K.A.; et al. MicroRNA-223 coordinates cholesterol homeostasis. Proc. Natl. Acad. Sci. USA 2014, 111, 14518–14523. [Google Scholar] [CrossRef]
- Ibrahim, A.G.E.; Cheng, K.; Marban, E. Exosomes as Critical Agents of Cardiac Regeneration Triggered by Cell Therapy. Stem Cell Rep. 2014, 2, 606–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, U.; George, A.; Bhutani, S.; Ghosh-Choudhary, S.; Maxwell, J.T.; Brown, M.E.; Mehta, Y.; Platt, M.O.; Liang, Y.; Sahoo, S. Experimental, systems, and computational approaches to understanding the microRNA-mediated reparative potential of cardiac progenitor cell–derived exosomes from pediatric patients. Circ. Res. 2017, 120, 701–712. [Google Scholar] [CrossRef]
- Zhang, Y.B.; Li, J.; Gao, W.; Xie, N. Exosomes as Anticancer Drug Delivery Vehicles: Prospects and Challenges. Front. Biosci. 2022, 27, 293. [Google Scholar] [CrossRef] [PubMed]
- Butreddy, A.; Kommineni, N.; Dudhipala, N. Exosomes as Naturally Occurring Vehicles for Delivery of Biopharmaceuticals: Insights from Drug Delivery to Clinical Perspectives. Nanomaterials 2021, 11, 1481. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Jang, H.; Cho, H.; Choi, J.; Hwang, K.Y.; Choi, Y.; Kim, S.H.; Yang, Y. Recent Advances in Exosome-Based Drug Delivery for Cancer Therapy. Cancers 2021, 13, 4435. [Google Scholar] [CrossRef] [PubMed]
- Gomari, H.; Forouzandeh Moghadam, M.; Soleimani, M.J.O. Targeted cancer therapy using engineered exosome as a natural drug delivery vehicle. OncoTargets Ther. 2018, 11, 5753–5762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- del Pozo-Acebo, L.; López de las Hazas, M.; Tomé-Carneiro, J.; Gil-Cabrerizo, P.; San-Cristobal, R.; Busto, R.; García-Ruiz, A.; Dávalos, A. Bovine milk-derived exosomes as a drug delivery vehicle for miRNA-based therapy. Int. J. Mol. Sci. 2021, 22, 1105. [Google Scholar] [CrossRef]
- Tian, T.; Zhang, H. Surface functionalized exosomes as targeted drug delivery vehicles for cerebral ischemia therapy. Biomaterials 2018, 150, 137–149. [Google Scholar] [CrossRef] [PubMed]
- ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/home (accessed on 29 June 2023).

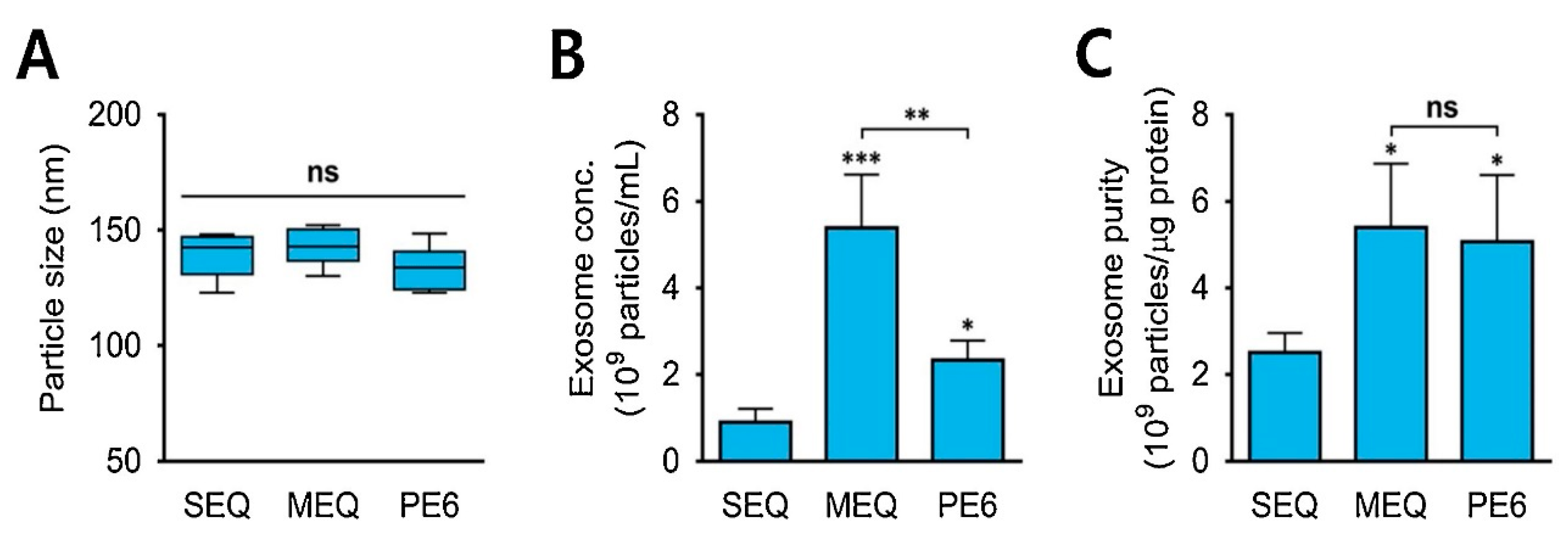



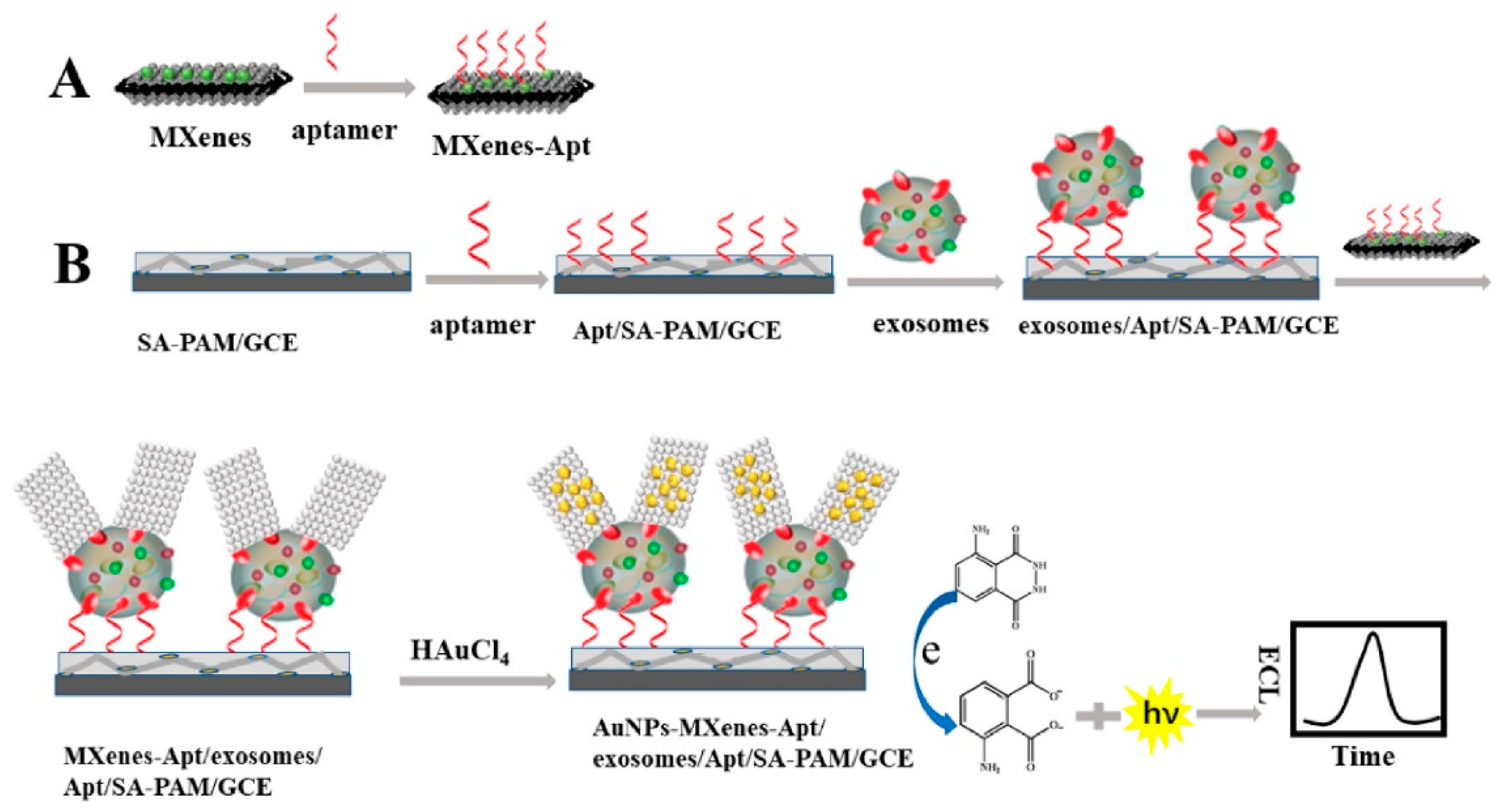
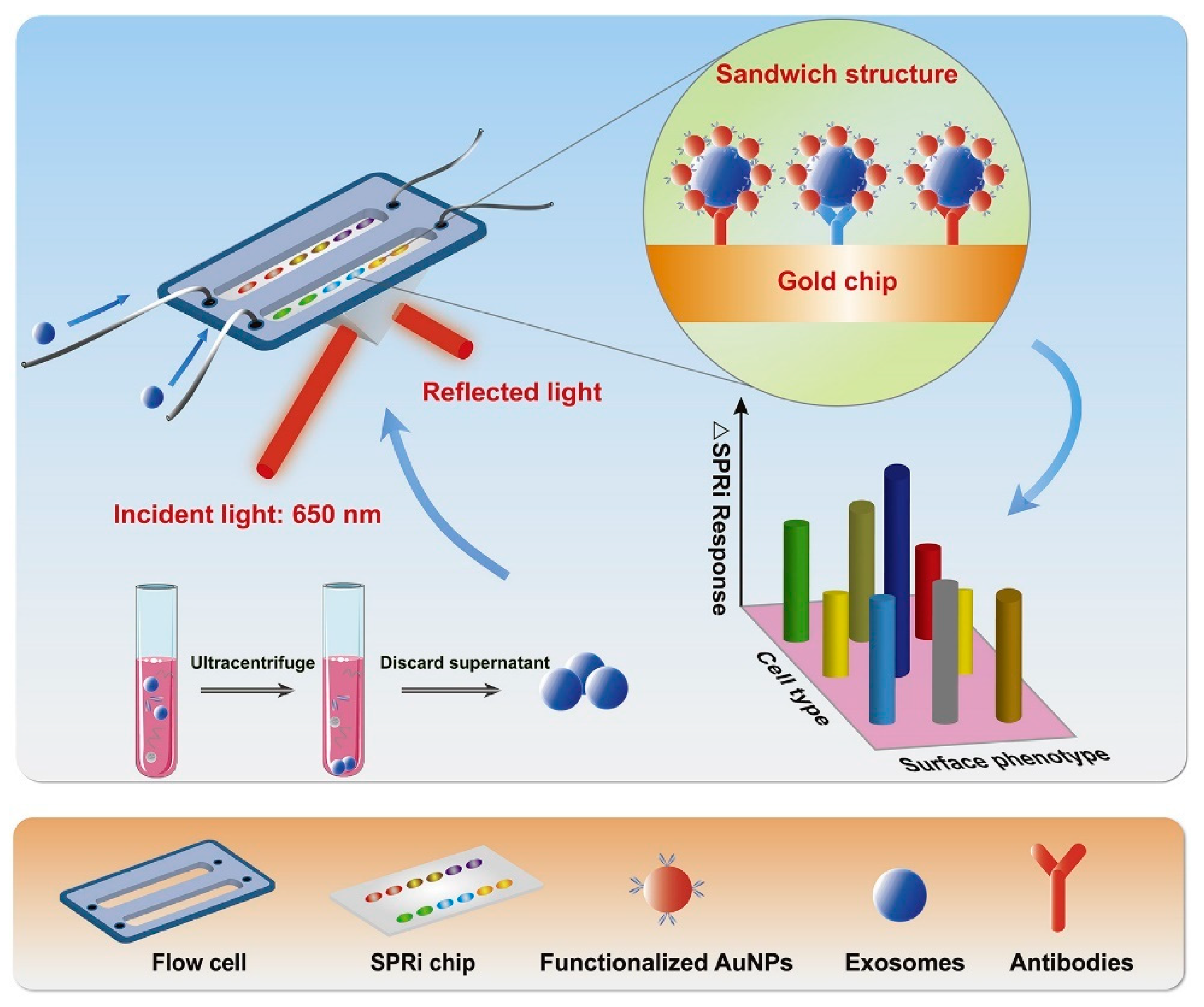
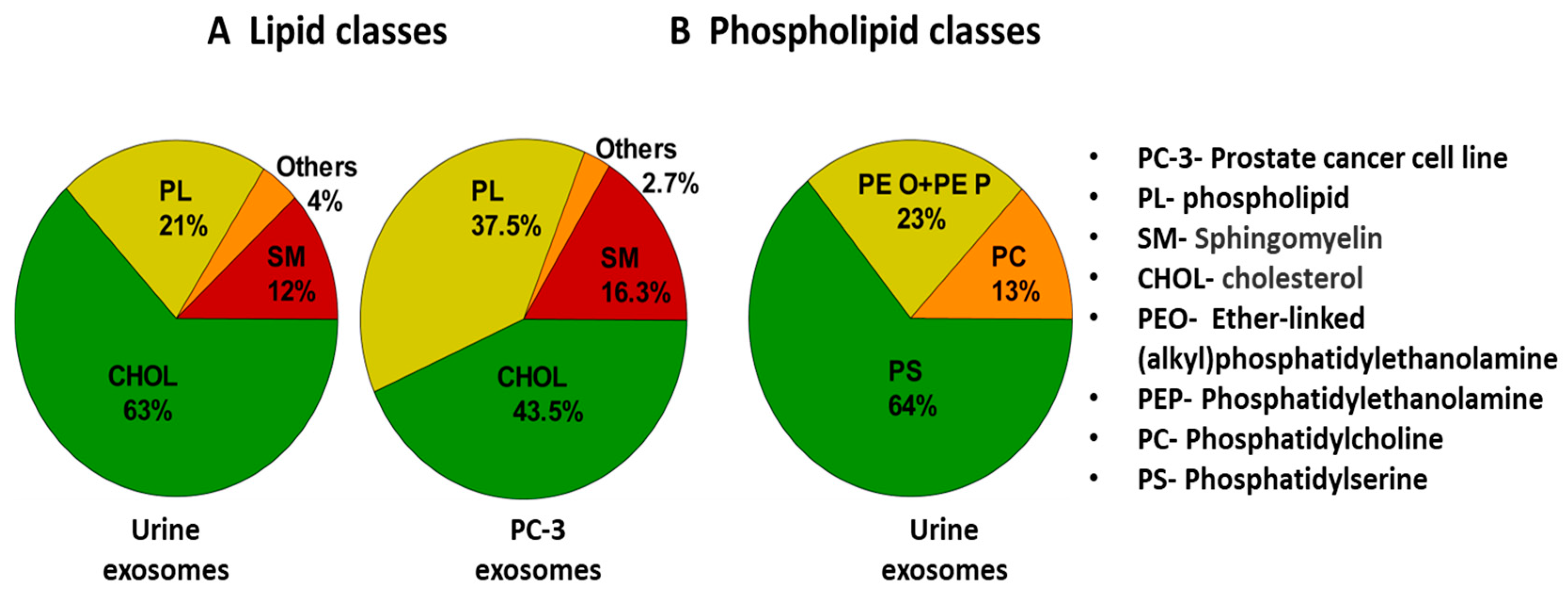
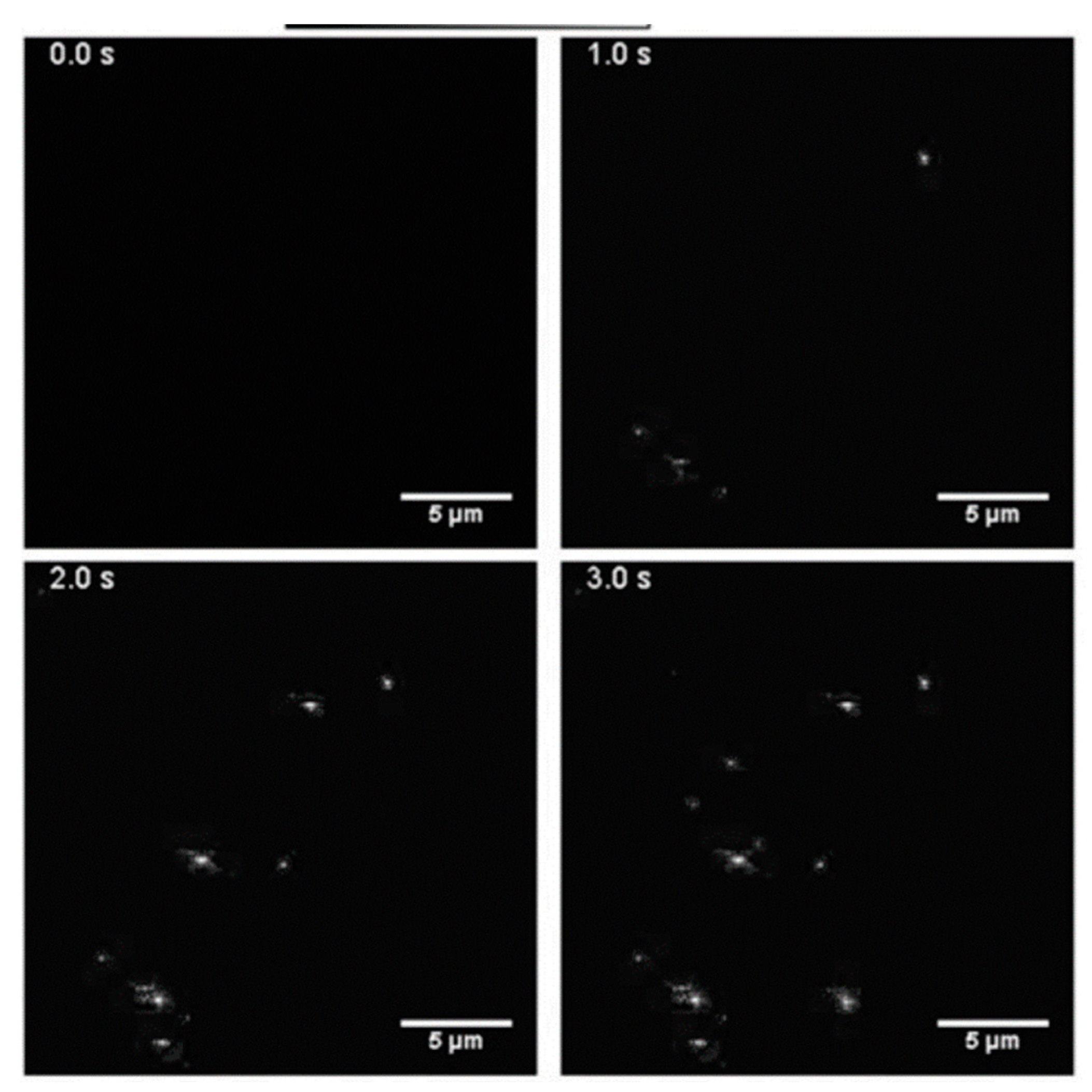

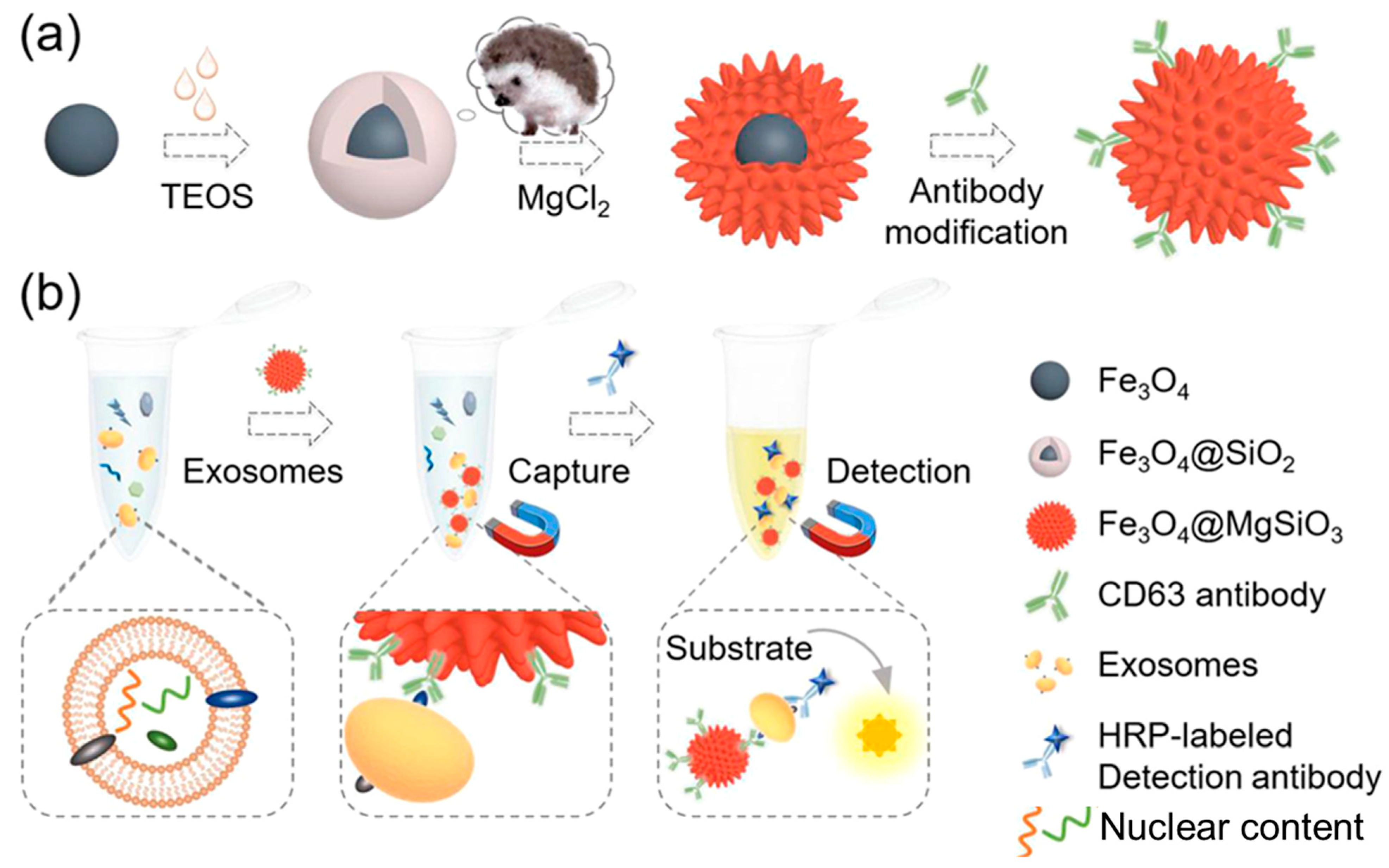
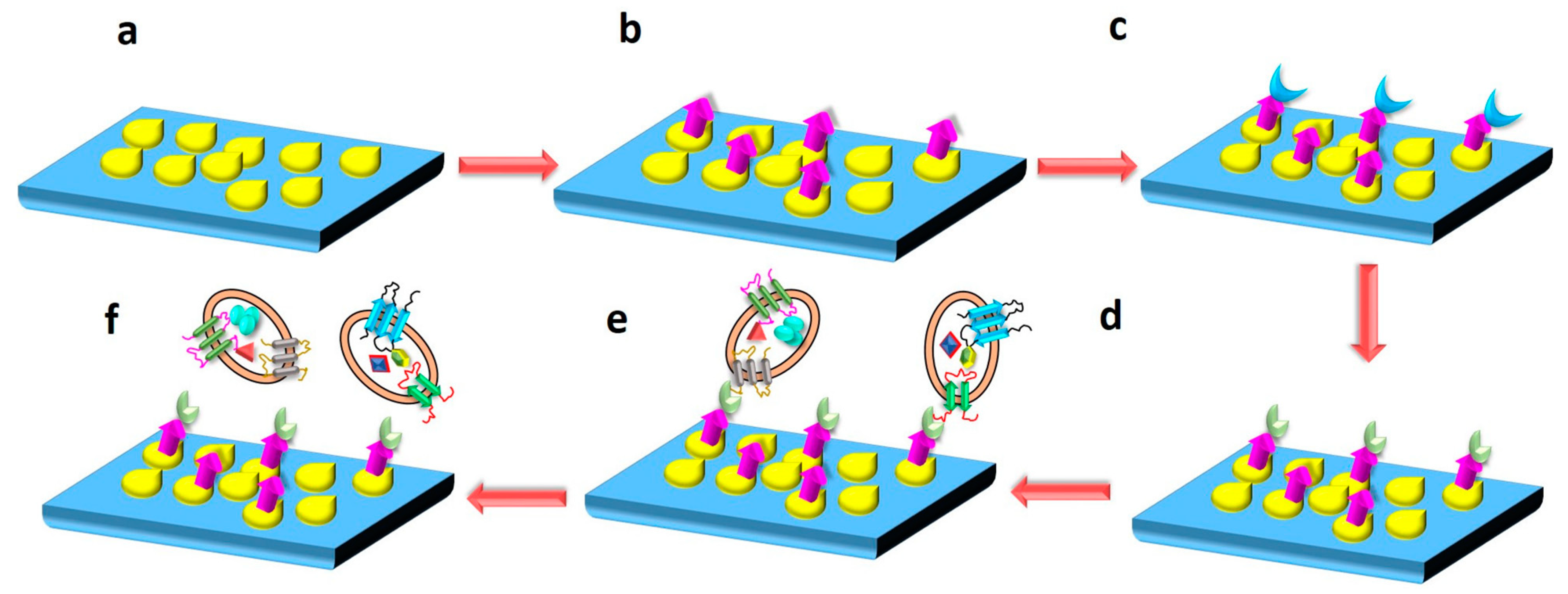
| Isolation Techniques | Advantages | Disadvantages | References |
|---|---|---|---|
| Ultracentrifugation |
|
| [33] |
| Precipitation |
|
| [77,116] |
| Immunoaffinity |
|
| [79,117] |
| Ultrafiltration |
|
| [89] |
| Size exclusion chromatography |
|
| [88] |
| Microfluidic separation |
|
| [100,110] |
| Charge-based isolation |
|
| [103] |
| Disease | Biomarker(s) | Nanoplatform Used | Antibody/Aptamer | Method of Detection | Isolated Exosome Concentration (Particles/mL) | Capture Efficiency (%) | Ref. |
|---|---|---|---|---|---|---|---|
| Parkinson’s disease | miR-128 | NA | p-FoxO3a (Ser253) | NA | NA | NA | [176] |
| Parkinson’s disease | 23 exosomal proteins | NA | Rabbit anti-flotillin, rabbit anti-Tsg101, rabbit anti-syntenin 1 | Immuno-typing | NA | NA | [181] |
| Amyotrophic lateral sclerosis | miR-23c and miR-192-5p | NA | HRP-conjugated goat anti-mouse antibody | NA | NA | NA | [188] |
| Amyotrophic lateral sclerosis | miR-15a-5p and miR-193a-5p | NA | NA | NA | NA | NA | [189] |
| Amyotrophic lateral sclerosis | miR-342-3p and miR-1254 | NA | anti-β-actin | NA | NA | NA | [187] |
| Alzheimer’s disease | Aβ1-42 and P-S96-tau | Fe3O4@Au@aptamer | Anti-CD63 antibody | NA | NA | NA | [12] |
| Alzheimer’s disease | miR-135a, miR-193b and miR-384 | NA | NA | NA | NA | NA | [15] |
| Prostate cancer | Prostate specific membrane antigen (PSMA) | Fe3O4@SiO2@TiO2 | Anti-PSMA, anti-CD9, and CD63 | Fluorescence | 3.21 × 1010 | 91.5% | [193] |
| Prostate cancer | Tetraspanin | Ag/IO/GRP | Dye-tetraspanin antibody | Magnetofluoro-immunosensing | NA | NA | [194] |
| Six different cancers | CD9, CD63, CD81 and TSG101 | NA | NA | SERS profiling using AI | 109–1010 | ~90 | [197] |
| Breast cancer | CD9 | MNPs@PEI@MUA | Biotin | SERS | NA | ~91 | [195] |
| Breast cancer | PD-L1 and miR-21 | NA | NA | NA | NA | NA | [196] |
| Colorectal cancer | CD63 | Magnetic beads coated with carbon nanomaterial | Anti-CD63 antibody | Aptamagnetic-fluorescence sensing | 1457 | NA | [198] |
| Disease | Exosome | Mode of Administration | Administration Dosage and Duration | Clinical Trial Phase | Recruitment Status | Ref. @ |
|---|---|---|---|---|---|---|
| Knee Osteoarthritis | MSC-derived exosomes | Intra-articular | 3–5 × 1011 particles/dose | Phase I | Not yet recruiting | NCT05060107 |
| Type I Diabetes Mellitus (T1DM) | Blood-derived exosomes | Intravenous | 120–160 mg/dL | Phase I | Unknown | NCT02138331 |
| Decompensated Liver Cirrhosis | Umbilical cord-derived MSC exosomes | Not specified | 40 mg in three weeks | Phase II | Recruiting | NCT05871463 |
| Skin Rejuvenation | MSC-derived exosomes | Intravenous | Not specified | Phase I/II | Recruiting | NCT05813379 |
| Colon cancer | Curcumin-conjugated plant exosomes | Oral | 3.6 g (gm) taken daily for 7 days | Phase I | Unknown | NCT01294072 |
| Pancreatic Adenocarcinoma | MSC-derived Exosomes with KRAS G12D siRNA | Not specified | 15–20 min on days 1, 4, and 10 | Phase I | Recruiting | NCT01294072 |
| Alzheimer’s Disease | Allogenic Adipose MSC- derived exosomes | Nasal | 5–20 μg for 12 weeks | Phase I/II | Unknown | NCT04388982 |
| Alzheimer’s Disease | Blood neuro-exosomal synaptic proteins | Not specified | Not specified | Not specified | Not yet recruiting | NCT05163626 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sonbhadra, S.; Mehak; Pandey, L.M. Biogenesis, Isolation, and Detection of Exosomes and Their Potential in Therapeutics and Diagnostics. Biosensors 2023, 13, 802. https://doi.org/10.3390/bios13080802
Sonbhadra S, Mehak, Pandey LM. Biogenesis, Isolation, and Detection of Exosomes and Their Potential in Therapeutics and Diagnostics. Biosensors. 2023; 13(8):802. https://doi.org/10.3390/bios13080802
Chicago/Turabian StyleSonbhadra, Smrity, Mehak, and Lalit M. Pandey. 2023. "Biogenesis, Isolation, and Detection of Exosomes and Their Potential in Therapeutics and Diagnostics" Biosensors 13, no. 8: 802. https://doi.org/10.3390/bios13080802
APA StyleSonbhadra, S., Mehak, & Pandey, L. M. (2023). Biogenesis, Isolation, and Detection of Exosomes and Their Potential in Therapeutics and Diagnostics. Biosensors, 13(8), 802. https://doi.org/10.3390/bios13080802





