Extraordinarily Stable Hairpin-Based Biosensors for Rapid Detection of DNA Ligases
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Oligonucleotides
2.3. Fluorescence Spectroscopic Examinations
2.4. Preparation of DNA Probes
2.5. DNA Ligase-Catalyzed Ligation and Dissociation of DNA Probes
3. Results and Discussion
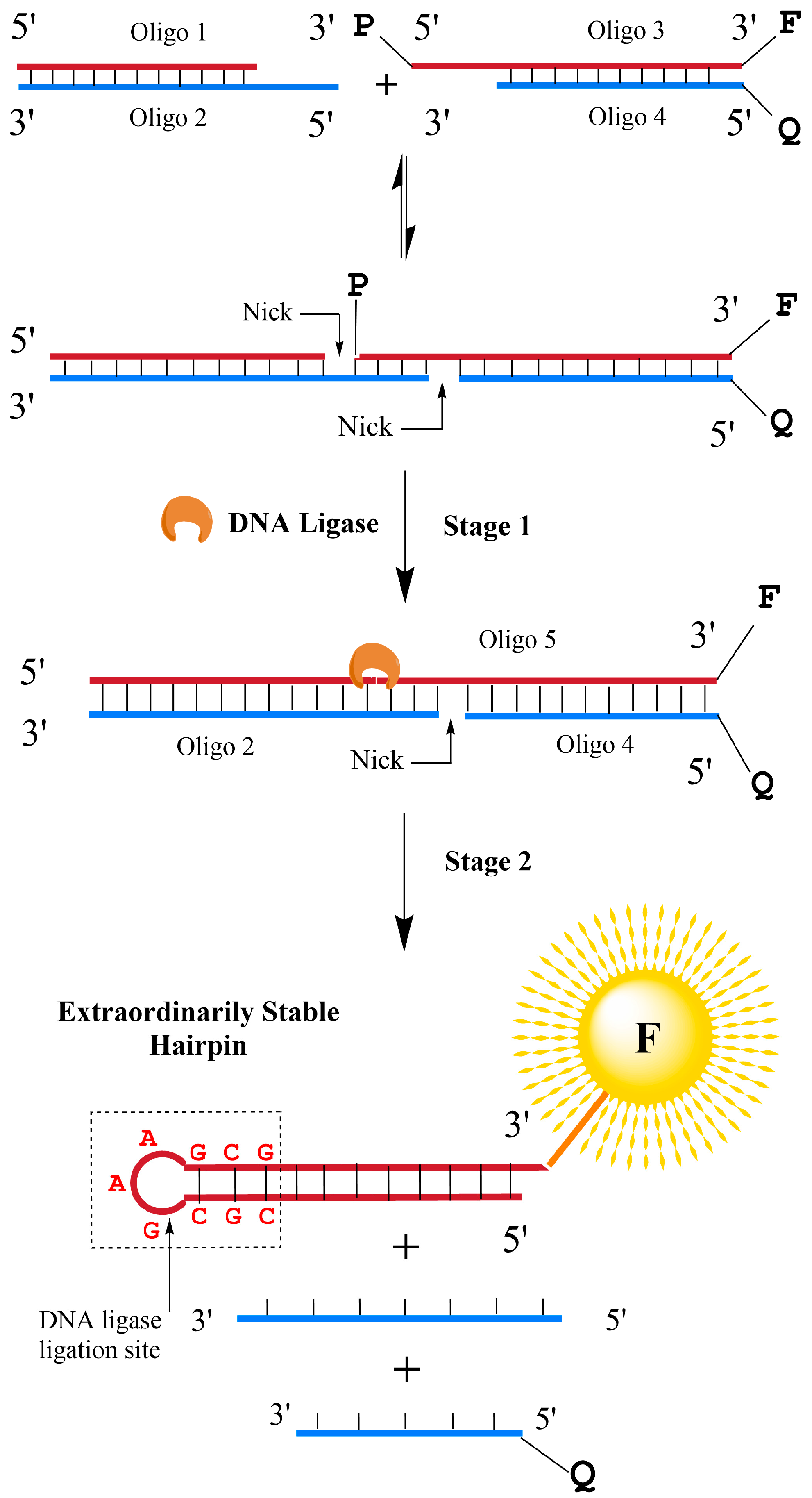
3.1. Design Strategies of the DNA Probes Targeting DNA Ligases
3.2. Selection of DNA Ligase
3.3. Detection of T4 DNA Ligase Using the DNA Probes
3.4. Optimization of Experimental Conditions for Effective Detecting of DNA Ligase
3.5. Measurement of the Limit of Detection (LoD) of DNA Probes
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lohman, G.J.S.; Tabor, S.; Nichols, N.M. DNA Ligases. Curr. Protoc. Mol. Biol. 2011, 94, 3.14.1–3.14.7. [Google Scholar] [CrossRef] [PubMed]
- Tomkinson, A.E.; Vijayakumar, S.; Pascal, J.M.; Ellenberger, T. DNA ligases: Structure, reaction mechanism, and function. ChemInform 2006, 37, 687–699. [Google Scholar] [CrossRef]
- Timson, D.J.; Singleton, M.R.; Wigley, D.B. DNA ligases in the repair and replication of DNA. Mutat. Res. 2000, 460, 301–318. [Google Scholar] [CrossRef] [PubMed]
- Doherty, A.J.; Suh, S.W. Structural and mechanistic conservation in DNA ligases. Nucleic Acids Res. 2000, 28, 4051–4058. [Google Scholar] [CrossRef] [PubMed]
- Shuman, S. DNA ligases: Progress and prospects. J. Biol. Chem. 2009, 284, 17365–17369. [Google Scholar] [CrossRef] [PubMed]
- Martin, I.V.; MacNeill, S.A. ATP-dependent DNA ligases. Genome Biol. 2002, 3, reviews3005.1. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, A.; Smith, A.; Bullard, D.; Lavesa-Curto, M.; Sayer, H.; Bonner, A.; Hemmings, A.; Bowater, R. Analysis of ligation and DNA binding by Escherichia coli DNA ligase (LigA). Biochim. Biophys. Acta (BBA) Proteins Proteom. 2005, 1749, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, A.; Sayer, H.; Bullard, D.; Smith, A.; Day, J.; Kieser, T.; Bowater, R. NAD+-dependent DNA ligases of Mycobacterium tuberculosis and Streptomyces coelicolor. Proteins Struct. Funct. Bioinf. 2003, 51, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, N.; Dube, D.; Pandey, J.; Singh, B.; Kukshal, V.; Ramachandran, R.; Tripathi, R.P. NAD+-Dependent DNA Ligase: A novel target waiting for the right inhibitor. Med. Res. Rev. 2008, 28, 545–568. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.K.; Krishna, S.; Chandra, S.; Shameem, M.; Desh-mukh, A.L.; Banerjee, D. Human DNA Ligases: A Comprehensive New Look for Cancer Therapy. Med. Res. Rev. 2014, 34, 567–595. [Google Scholar] [CrossRef] [PubMed]
- Tomkinson, A.E.; Naila, T.; Khattri Bhandari, S. Altered DNA ligase activity in human disease. Mutagenesis 2020, 35, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Maiko, T.; Yoshizumi, I.; Hirokazu, N. Biotechnological Uses of Archaeal Proteins. Archaea Int. Microbiol. J. 2015, 2015, 267570. [Google Scholar] [CrossRef]
- Zhu, W.; Wang, X.; Dai, L.; Yang, W.; Li, Y.; Liu, R. A Novel Label-Free Fluorescence Strategy Based on Dumbbell Probe for Sensitive Detection of DNA Ligase. Open Access Libr. J. 2021, 8, 1–9. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, Q.; Wang, Z.; Chen, X.; Xu, L.; Hu, J.; Pei, R. Label-free detection of T4 DNA ligase and polynucleotide kinase activity based on toehold-mediated strand displacement and split G-quadruplex probes. Sens. Actuators B Chem. 2015, 214, 50–55. [Google Scholar] [CrossRef]
- Hirao, I.; Nishimura, Y.; Tagawa, Y.; Watanabe, K.; Miura, K. Extraordinarily stable mini-hairpins: Electrophoretical and thermal properties of the various sequence variants of d(GCFAAAGC) and their effect on DNA sequencing. Nucleic Acids Res. 1992, 20, 3891–3896. [Google Scholar] [CrossRef] [PubMed]
- Yoshizawa, S.; Ueda, T.; Ishido, Y.; Miura, K.; Watanabe, I. Nuclease resistance of an extraordinarily thermostable mini-hairpin DNA fragment, d(GCGAAGC) and its application to in vitro protein synthesis. Nucleic Acids Res. 1994, 22, 2217–2221. [Google Scholar] [CrossRef] [PubMed]
- Antao, V.P.; Lai, S.Y.; Tinoco, I., Jr. A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res. 1991, 19, 5901–5905. [Google Scholar] [CrossRef] [PubMed]
- Armbruster, D.A.; Pry, T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 2008, 29 (Suppl. S1), S49–S52. [Google Scholar] [PubMed]




| Name | Structure | Stem Length | Stem GC Content | Tm (°C) |
|---|---|---|---|---|
| Hairpin 1 (extraordinarily stable hairpin used in our biosensor) |  | 3 bp | 100% | 88.5 |
| Hairpin 2 (extraordinarily stable hairpin) |  | 2 bp | 100% | 76.5 |
| Hairpin 3 (ordinary hairpin) |  | 4 bp | 75% | 44.2 |
| Name | Sequence (5′ to 3′) | Modification |
|---|---|---|
| Oligo 1 | AAACTCCACGC | |
| Oligo 2 | CTTCGCGTGGAGTTT | |
| Oligo 3 | GAAGCGTGGAGTTTA | 5′ P, 3′ Cy3 |
| Oligo 4 | TAAACTCCACG | 5′ BHQ2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Z.; Kou, R.; Ni, K.; Song, R.; Li, Y.; Li, T.; Zhang, H. Extraordinarily Stable Hairpin-Based Biosensors for Rapid Detection of DNA Ligases. Biosensors 2023, 13, 875. https://doi.org/10.3390/bios13090875
Wu Z, Kou R, Ni K, Song R, Li Y, Li T, Zhang H. Extraordinarily Stable Hairpin-Based Biosensors for Rapid Detection of DNA Ligases. Biosensors. 2023; 13(9):875. https://doi.org/10.3390/bios13090875
Chicago/Turabian StyleWu, Ziang, Roujuan Kou, Kun Ni, Rui Song, Yuxuan Li, Tianhu Li, and Hao Zhang. 2023. "Extraordinarily Stable Hairpin-Based Biosensors for Rapid Detection of DNA Ligases" Biosensors 13, no. 9: 875. https://doi.org/10.3390/bios13090875
APA StyleWu, Z., Kou, R., Ni, K., Song, R., Li, Y., Li, T., & Zhang, H. (2023). Extraordinarily Stable Hairpin-Based Biosensors for Rapid Detection of DNA Ligases. Biosensors, 13(9), 875. https://doi.org/10.3390/bios13090875





