Integrated Droplet-Based Digital Loop-Mediated Isothermal Amplification Microfluidic Chip with Droplet Generation, Incubation, and Continuous Fluorescence Detection
Abstract
1. Introduction
2. Materials and Methods
2.1. Chip Design and Fabrication
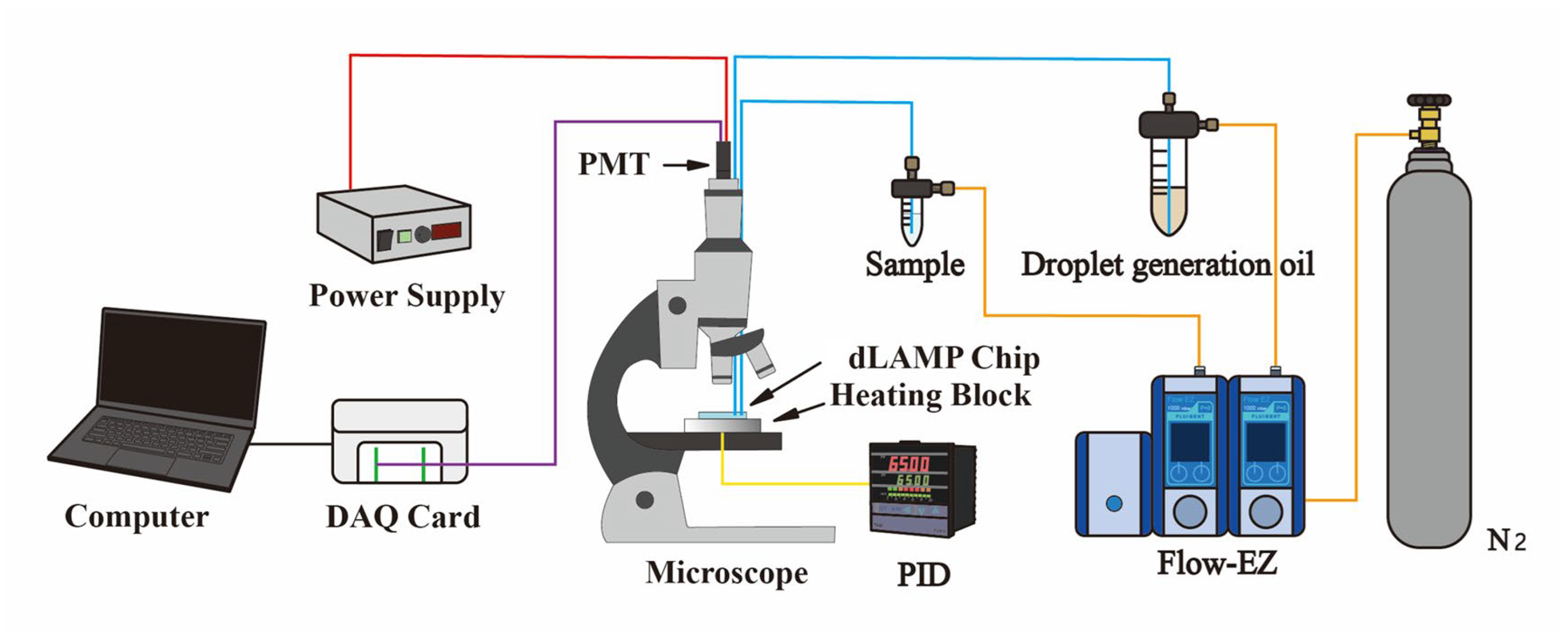
2.2. Experimental Setup and Reagent Preparation
2.3. Experimental Procedure
2.4. Data Analysis
3. Results and Discussion
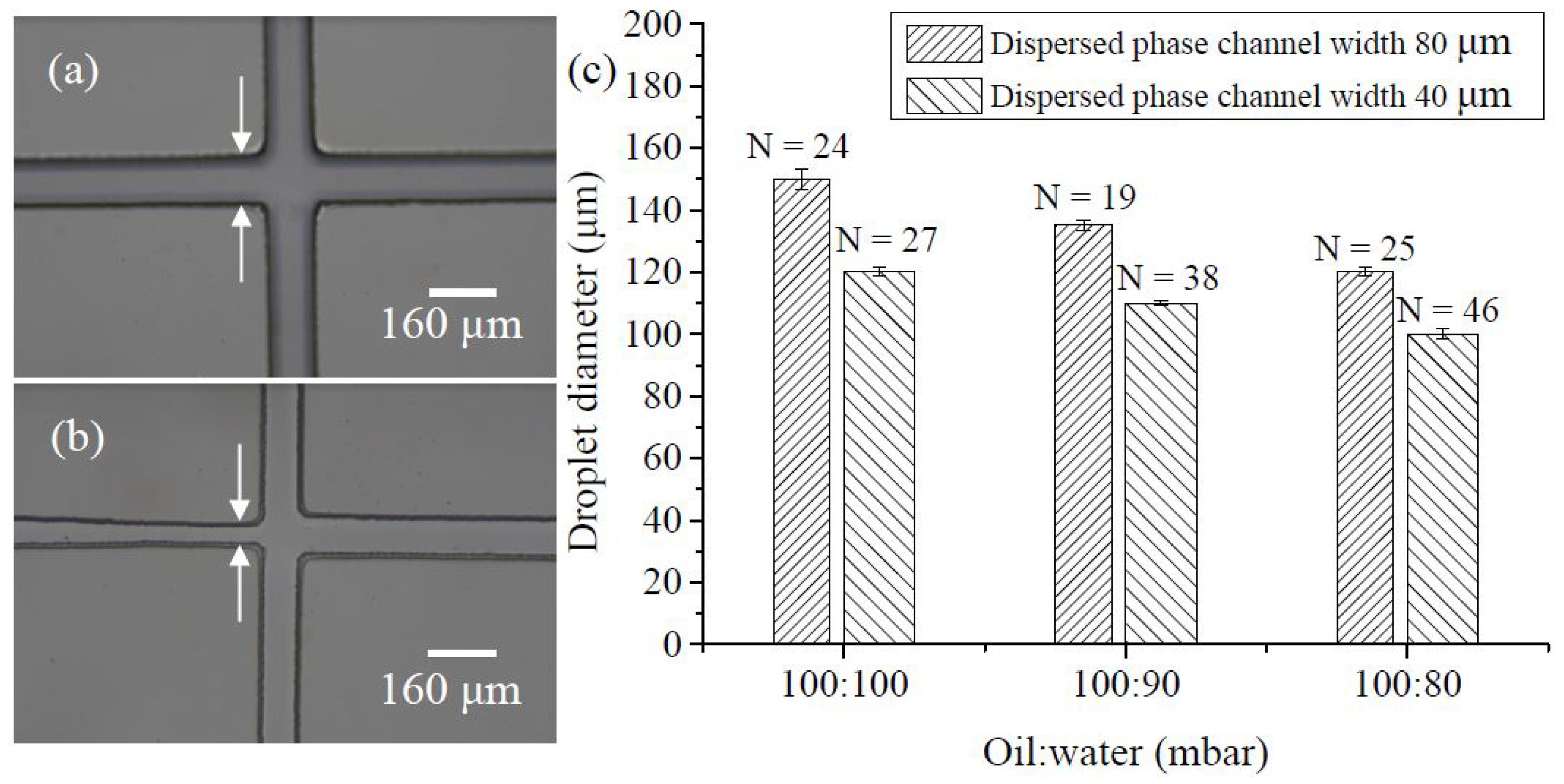
3.1. Droplet Preparation
3.2. Droplet Stability during Incubation
3.3. Droplet Fluorescent Detection and Counting
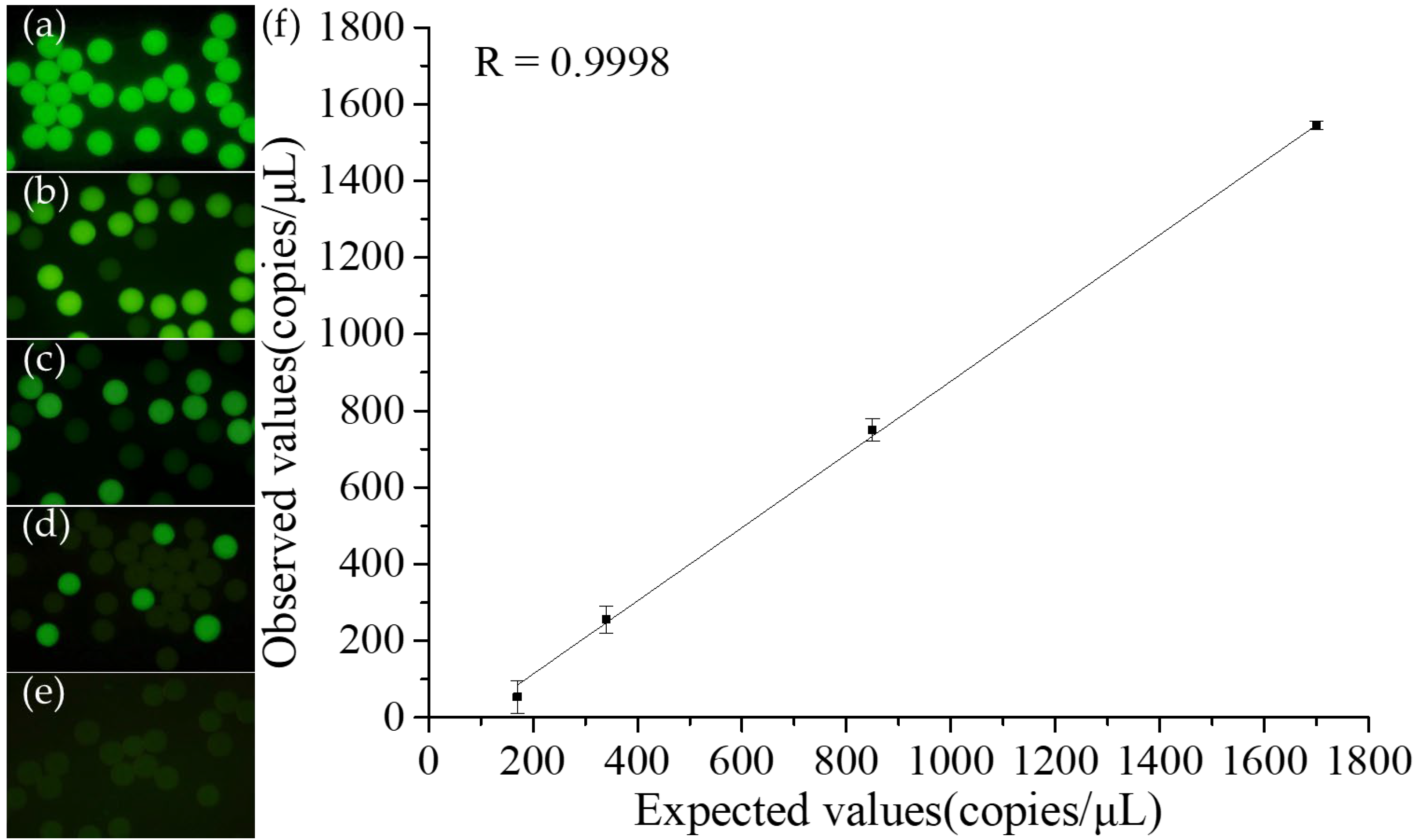
3.4. Quantitative Detection of Staphylococcus Aureus
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mullis, K.B. The Unusual Origin of the Polymerase Chain Reaction. Sci. Am. 1990, 262, 56–65. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Shen, F.; McCalla, S.E.; Kreutz, J.E.; Karymov, M.A.; Ismagilov, R.F. Mechanistic Evaluation of the Pros and Cons of Digital RT-LAMP for HIV-1 Viral Load Quantification on a Microfluidic Device and Improved Efficiency via a Two-Step Digital Protocol. Anal. Chem. 2013, 85, 1540–1546. [Google Scholar] [CrossRef]
- Schoepp, N.G.; Schlappi, T.S.; Curtis, M.S.; Butkovich, S.S.; Miller, S.; Humphries, R.M.; Ismagilov, R.F. Rapid pathogen-specific phenotypic antibiotic susceptibility testing using digital LAMP quantification in clinical samples. Sci. Transl. Med. 2017, 9, eaal3693. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Lin, X.; Urmann, K.; Li, L.; Xie, X.; Jiang, S.; Hoffmann, M.R. Smartphone-Based in-Gel Loop-Mediated Isothermal Amplification (gLAMP) System Enables Rapid Coliphage MS2 Quantification in Environmental Waters. Environ. Sci. Technol. 2018, 52, 6399–6407. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, S.S.; Ball, C.S.; Langevin, S.A.; Fang, Y.; Coffey, L.L.; Meagher, R.J. Surveillance for Western Equine Encephalitis, St. Louis Encephalitis, and West Nile Viruses Using Reverse Transcription Loop-Mediated Isothermal Amplification. PLoS ONE 2016, 11, e0147962. [Google Scholar] [CrossRef] [PubMed]
- Hayashida, K.; Garcia, A.; Moonga, L.C.; Sugi, T.; Takuya, K.; Kawase, M.; Kodama, F.; Nagasaka, A.; Ishiguro, N.; Takada, A.; et al. Field-deployable multiplex detection method of SARS-CoV-2 and influenza virus using loop-mediated isothermal amplification and DNA chromatography. PLoS ONE 2023, 18, e0285861. [Google Scholar] [CrossRef] [PubMed]
- Khorosheva, E.M.; Karymov, M.A.; Selck, D.A.; Ismagilov, R.F. Lack of correlation between reaction speed and analytical sensitivity in isothermal amplification reveals the value of digital methods for optimization: Validation using digital real-time RT-LAMP. Nucleic Acids Res. 2015, 44, e10. [Google Scholar] [CrossRef] [PubMed]
- Gansen, A.; Herrick, A.M.; Dimov, I.K.; Lee, L.P.; Chiu, D.T. Digital LAMP in a sample self-digitization (SD) chip. Lab A Chip 2012, 12, 2247–2254. [Google Scholar] [CrossRef]
- Rong, N.; Chen, K.; Shao, J.; Ouyang, Q.; Luo, C. A 3D Scalable Chamber-Array Chip for Digital LAMP. Anal. Chem. 2023, 95, 7830–7838. [Google Scholar] [CrossRef]
- Zhu, Q.; Gao, Y.; Yu, B.; Ren, H.; Qiu, L.; Han, S.; Jin, W.; Jin, Q.; Mu, Y. Self-priming compartmentalization digital LAMP for point-of-care. Lab A Chip 2012, 12, 4755–4763. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.-D.; Chang, W.-H.; Luo, K.; Wang, C.-H.; Liu, S.-Y.; Yen, W.-H.; Lee, G.-B. Digital quantification of DNA via isothermal amplification on a self-driven microfluidic chip featuring hydrophilic film-coated polydimethylsiloxane. Biosens. Bioelectron. 2018, 99, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Lyu, W.; Zhang, J.; Yu, Y.; Xu, L.; Shen, F. Slip formation of a high-density droplet array for nucleic acid quantification by digital LAMP with a random-access system. Lab A Chip 2021, 21, 3086–3093. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Lyu, W.; Yu, M.; Wang, Q.; Qu, H.; Ismagilov, R.F.; Han, X.; Lai, D.; Shen, F. Self-partitioning SlipChip for slip-induced droplet formation and human papillomavirus viral load quantification with digital LAMP. Biosens. Bioelectron. 2020, 155, 112107. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Tian, J.; Chao, Y.; Chien, Y.-S.; Luo, R.-H.; Guo, J.-Y.; Li, S.; Chou, Y.-J.; Shum, H.C.; Chen, C.-F. Hand-Powered Microfluidics for Parallel Droplet Digital Loop-Mediated Isothermal Amplification Assays. ACS Sens. 2021, 6, 2868–2874. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Liu, L.; Ye, Z.; Gong, J.; Hao, P.; Ping, J.; Ying, Y. TriD-LAMP: A pump-free microfluidic chip for duplex droplet digital loop-mediated isothermal amplification analysis. Anal. Chim. Acta 2022, 1233, 340513. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, S.-A.; Shamsaei, D.; Eitzmann, D.R.; Anderson, J.L. Digital Droplet Loop-Mediated Isothermal Amplification Featuring a Molecular Beacon Assay, 3D Printed Droplet Generation, and Smartphone Imaging for Sequence-Specific DNA Detection. Anal. Chem. 2022, 94, 11949–11956. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Lan, X.; Ren, L.; Yang, M.; Wei, B.; Wang, Y. Design of a Digital LAMP Detection Platform Based on Droplet Microfluidic Technology. Micromachines 2023, 14, 1077. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Yan, S.; Zhang, X.; Ma, P.; Du, W.; Feng, X.; Liu, B.-F. Monte Carlo Modeling-Based Digital Loop-Mediated Isothermal Amplification on a Spiral Chip for Absolute Quantification of Nucleic Acids. Anal. Chem. 2017, 89, 3716–3723. [Google Scholar] [CrossRef]
- Ma, Y.-D.; Luo, K.; Chang, W.-H.; Lee, G.-B. A microfluidic chip capable of generating and trapping emulsion droplets for digital loop-mediated isothermal amplification analysis. Lab A Chip 2018, 18, 296–303. [Google Scholar] [CrossRef]
- Pan, Y.; Ma, T.; Meng, Q.; Mao, Y.; Chu, K.; Men, Y.; Pan, T.; Li, B.; Chu, J. Droplet digital PCR enabled by microfluidic impact printing for absolute gene quantification. Talanta 2020, 211, 120680. [Google Scholar] [CrossRef] [PubMed]
- Hess, D.; Yang, T.; Stavrakis, S. Droplet-based optofluidic systems for measuring enzyme kinetics. Anal. Bioanal. Chem. 2020, 412, 3265–3283. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Li, J.; Zhang, Z.; Li, M.; Zhao, S.; Li, Z.; Peng, N. Smartphone-Based Droplet Digital LAMP Device with Rapid Nucleic Acid Isolation for Highly Sensitive Point-of-Care Detection. Anal. Chem. 2020, 92, 2258–2265. [Google Scholar] [CrossRef]
- Liu, F.; Xu, T.; Liu, W.; Zheng, X.; Xu, J.; Ma, B. Spontaneous droplet generation via surface wetting. Lab A Chip 2020, 20, 3544–3551. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Sun, Y.; Liu, C.; Duan, X.; Tang, W.; Li, Z. Precise Quantitation of MicroRNA in a Single Cell with Droplet Digital PCR Based on Ligation Reaction. Anal. Chem. 2016, 88, 11384–11389. [Google Scholar] [CrossRef] [PubMed]
- Ou, C.-Y.; Vu, T.; Grunwald, J.T.; Toledano, M.; Zimak, J.; Toosky, M.; Shen, B.; Zell, J.A.; Gratton, E.; Abram, T.J.; et al. An ultrasensitive test for profiling circulating tumor DNA using integrated comprehensive droplet digital detection. Lab A Chip 2019, 19, 993–1005. [Google Scholar] [CrossRef]
- Clark, I.C.; Fontanez, K.M.; Meltzer, R.H.; Xue, Y.; Hayford, C.; May-Zhang, A.; D’Amato, C.; Osman, A.; Zhang, J.Q.; Hettige, P.; et al. Microfluidics-free single-cell genomics with templated emulsification. Nat. Biotechnol. 2023, 41, 1557–1566. [Google Scholar] [CrossRef]
- Zilionis, R.; Nainys, J.; Veres, A.; Savova, V.; Zemmour, D.; Klein, A.M.; Mazutis, L. Single-cell barcoding and sequencing using droplet microfluidics. Nat. Protoc. 2017, 12, 44–73. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, Y.-H.; Hung, Y.-T.; Chang, W.; Chiou, C.-C. Integrated Droplet-Based Digital Loop-Mediated Isothermal Amplification Microfluidic Chip with Droplet Generation, Incubation, and Continuous Fluorescence Detection. Biosensors 2024, 14, 334. https://doi.org/10.3390/bios14070334
Lin Y-H, Hung Y-T, Chang W, Chiou C-C. Integrated Droplet-Based Digital Loop-Mediated Isothermal Amplification Microfluidic Chip with Droplet Generation, Incubation, and Continuous Fluorescence Detection. Biosensors. 2024; 14(7):334. https://doi.org/10.3390/bios14070334
Chicago/Turabian StyleLin, Yen-Heng, Yuan-Ting Hung, Wei Chang, and Chiuan-Chian Chiou. 2024. "Integrated Droplet-Based Digital Loop-Mediated Isothermal Amplification Microfluidic Chip with Droplet Generation, Incubation, and Continuous Fluorescence Detection" Biosensors 14, no. 7: 334. https://doi.org/10.3390/bios14070334
APA StyleLin, Y.-H., Hung, Y.-T., Chang, W., & Chiou, C.-C. (2024). Integrated Droplet-Based Digital Loop-Mediated Isothermal Amplification Microfluidic Chip with Droplet Generation, Incubation, and Continuous Fluorescence Detection. Biosensors, 14(7), 334. https://doi.org/10.3390/bios14070334




