Fluorogenic RNA-Based Biosensors of Small Molecules: Current Developments, Uses, and Perspectives
Abstract
1. Introduction
2. Reporting and Sensing Modules
2.1. Light-Up Aptamers Discovery and Properties
2.2. Choice and Development of Sensing Aptamers
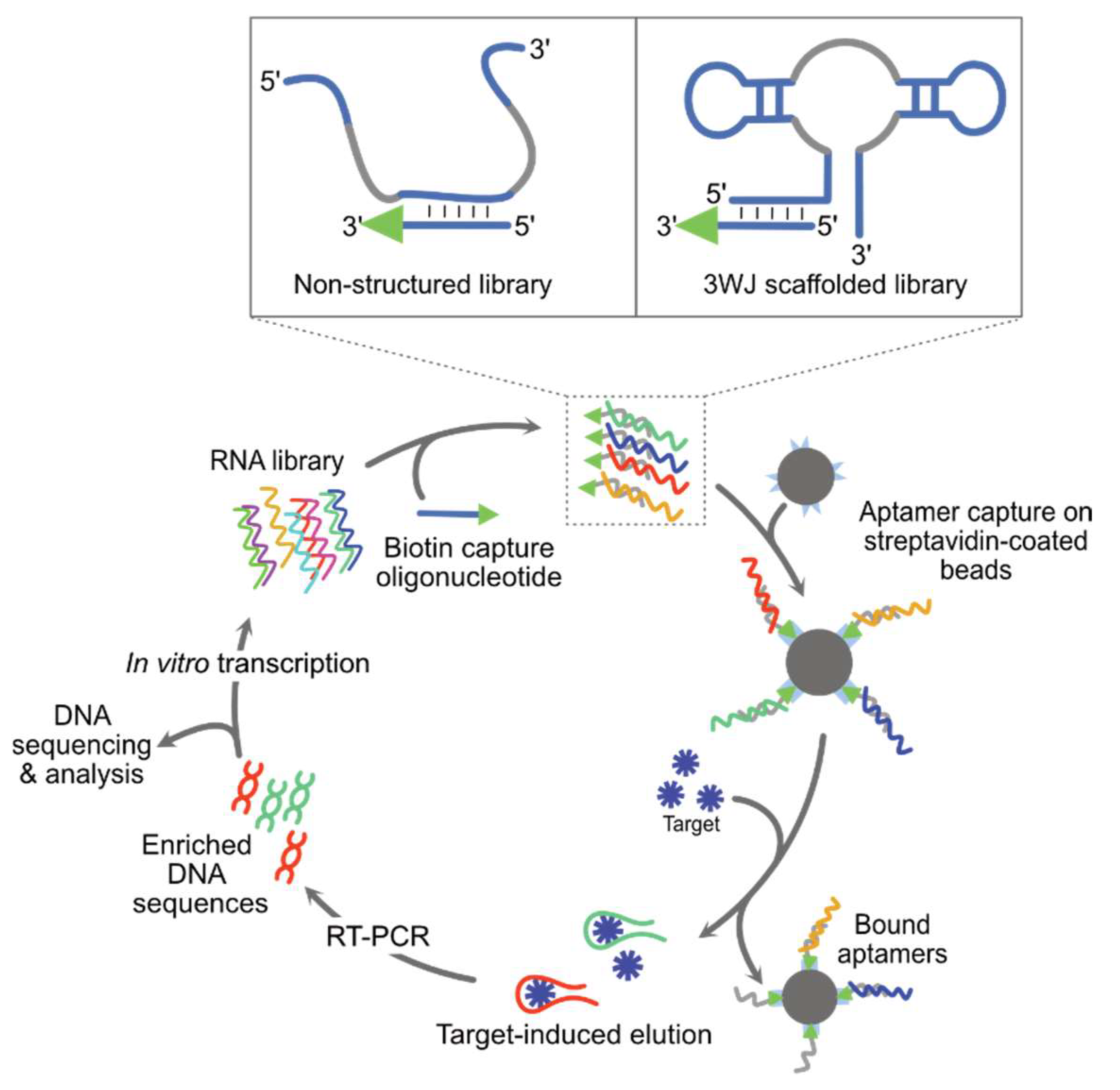
2.2.1. Repurposing Natural Structure-Switching Aptamers
2.2.2. Reprogramming the Specificity of Natural Structure-Switching Aptamers
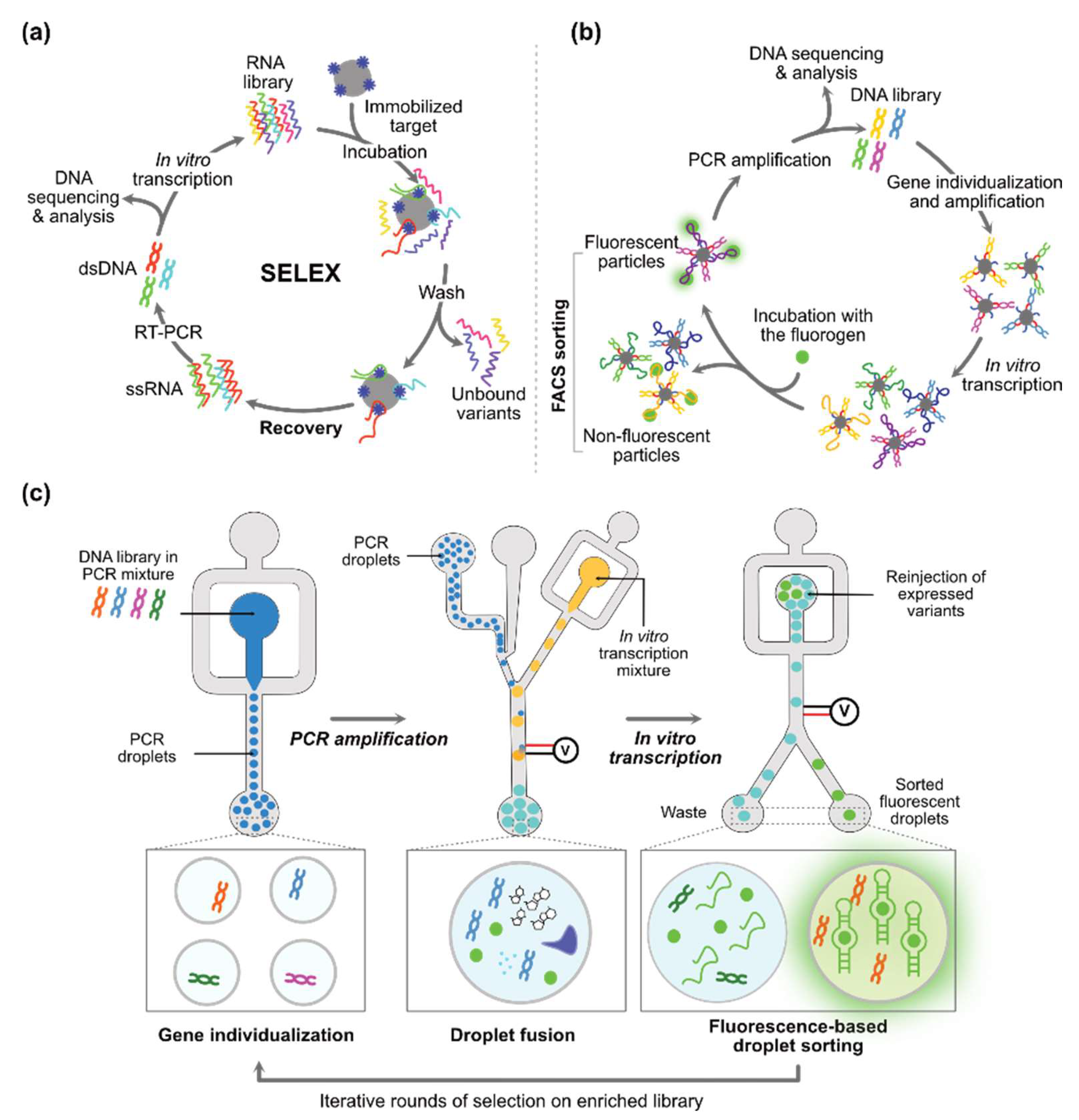
2.2.3. De Novo Discovery of Synthetic Structure-Switching Aptamers
3. Functional Connection of the Sensing with the Reporting Modules
3.1. Direct Allosteric Control
3.2. Strand Displacement-Mediated Control
3.3. Mixed Control
4. Applications of FRBs
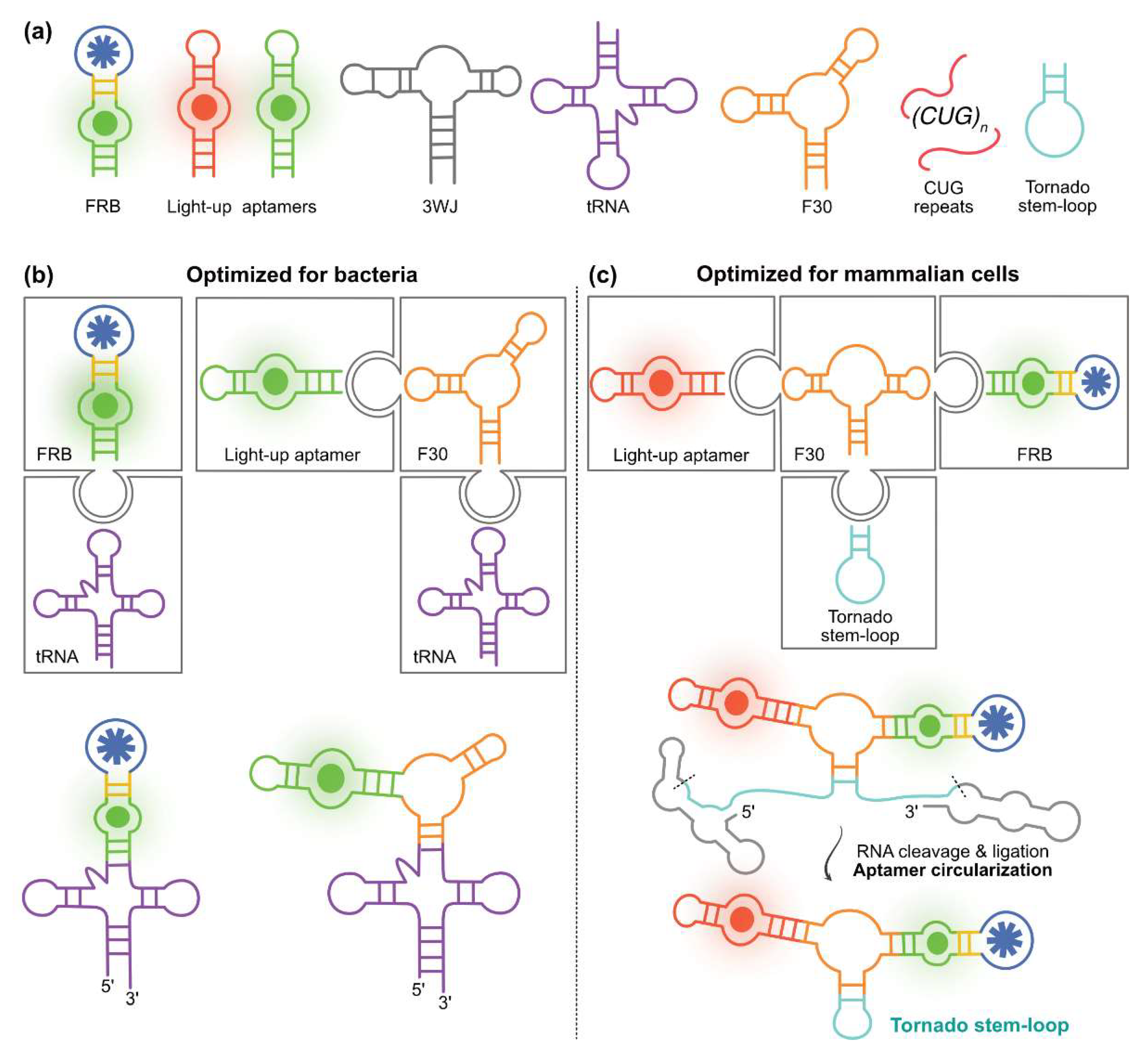
4.1. Exploiting RNA Modularity to Extend FRBs Properties
4.2. Intracellular In Vivo FRB Applications
4.3. Using FRB as Extracellular In Vitro Reporters
5. Exploring Alternative Chemistries
| Aptamer | Chemistry | Fluorogen | Evaluated as FRB of Small Molecule | Target Small Molecule | Other Target | Ref. |
|---|---|---|---|---|---|---|
| AO-binding | DNA | Auramine O | Yes | Serotonin | [157] | |
| AptII-Mini 3-4 | DNA | Hoechst | No | [174] | ||
| BBR4S3 | DNA | Berberine | No | [175] | ||
| CV-30S | DNA | Crystal Violet | No | [176] | ||
| DAP-10-42 | DNA | Dapoxyl | Yes | ATP | [156,177] | |
| DIR 2-1 | DNA | DIR | No | [178] | ||
| Guanidine | RNA/Hachimoji | DFHBI-T | Yes | Guanidine | [170] | |
| L-Mango-III | L-RNA | TO1-Biotin | No | MicroRNA-155 | [167] | |
| L-Spinach | L-RNA | DFHBI | Yes | Guanine | [168] | |
| Lettuce | DNA | DFHBI-T | No | SARS-CoV2 RNA | [158] | |
| Spinach-Hachimoji | DNA/Hachimoji | DFHBI-T | No | [169] | ||
| 2′F-Broccoli | RNA/2′ Fluoropyrimidine | DFHBI-T | Yes | K+ | [164] | |
| 2′F-Spinach | RNA/2′ Fluoropyrimidine | DFHBI-T | Yes | Pb2+ | [163] |
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 2′FY | 2′ fluorinated pyrimidine |
| 3WJ | 3-way junction |
| 5-HT | 5-hydroxytryptamine |
| 5-HTP | 5-hydroxytryptophan |
| ADP | Adenosine diphosphate |
| AMP | Adenosine monophosphate |
| AO | Auramine O |
| apta-FRET | aptamer-based Förster resonance energy transfer |
| ATP | Adenosine triphosphate |
| BC | Benzopyrylium Coumarin |
| BFR | Blue fluorescent RNA |
| BRET | Bioluminescence Resonance Energy Transfer |
| c-di-GMP | cyclic-dimeric guanosine monophosphate |
| c-AMP | cyclic adenosine monophosphate |
| Cbl | Cobalamin |
| c-diA | cyclic di-AMP |
| cGAMP | cyclic GMP-AMP |
| cGAS | cyclic GMP-AMP synthase |
| CHARGE | Catalytic Hairpin Assembly RNA circuit that is Genetically Encoded |
| CM | Communication Module |
| CV | Crystal violet |
| DAP | Dapoxyl DNA aptamer |
| DFAMF | 3,5-difluoro-4-hydroxybenzylidene imidazolinone-2-acrylate methyl |
| DFHBI | 3,5-difluoro-4-hydroxybenzylidene imidazolinone |
| DFHBI1-T | 3,5-difluoro-4-hydroxybenzylidene-1-trifluoroethyl-imidazolinone |
| DFHO | 3,5-difluoro-4-hydroxybenzylidene-imidazolinone-2-oxime |
| DIR | Dimethylindole red |
| DMHBI | 3,5-dimethoxy-4-hydroxybenzylidene imidazolone |
| DNA | Deoxyribonucleic acid |
| DNB | Dinitroaniline-binding aptamer |
| dsDNA | double stranded DNA |
| FACS | Fluorescence-activated Cell Sorting |
| FLARE | Fluorogenic Aptamer-based RNA condensates |
| FMN | Flavin mononucleotide |
| FP | Fluorescent protein |
| FRB | Fluorogenic RNA-based biosensor |
| FRET | Fluorescence Resonance Energy Transfer |
| GEMM | Genes for the Environment, for Membranes and for Motility |
| GFP | Green Fluorescent Protein |
| GRAP | Gene-linked RNA aptamer particle |
| GTP | Guanosine 5′-triphosphate |
| HBC | 4-((2-hydroxyethyl)(methyl)amino)-benzylidene)-cyanophenylacetonitrile |
| IC50 | Half-maximal inhibitory concentration |
| IVT | In vitro transcription |
| MG | Malachite green |
| DIR-2 | Dimethylindole red 2 |
| PCR | Polymerase chain reaction |
| ppGpp | Guanosine tetraphosphate |
| RAPID | RNA Aptamer In Droplets |
| RhoBAST | Rhodamine Binding Aptamer for Super-resolution Imaging Techniques |
| RNA | Ribonucleic acid |
| SAH | S-Adenosyl-L-homocysteine |
| SAHA | Suberoylanilide hydroxamic acid |
| SAM | S-adenosylmethionine |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| SELEX | Systematic Evolution of Ligands by EXponential enrichment |
| SerBLAS | Serotonin-responsive binary light-up aptameric sensor |
| SiRA | Silicon Rhodamine-Binding Aptamer |
| SR-DN | Sulforhodamine B-dinitroaniline |
| SRB | Sulforhodamine B |
| TMR | Tetramethylrhodamine |
| TMR-DN | Tetramethylrhodamine dinitroaniline |
| TO | Thiazole orange |
| TORNADO | Twister-optimized RNA for durable overexpression |
| TPP | Thiamine pyrophosphate |
| tRNA | transfert RNA |
| µIVC | Microfluidic-assisted in vitro compartmentalization |
| XNA | Xeno nucleic acid |
| xpt-pbuX | Xanthine phosphoribosyltransferase- xanthine-specific purine permease |
References
- Huang, C.-W.; Lin, C.; Nguyen, M.K.; Hussain, A.; Bui, X.-T.; Ngo, H.H. A Review of Biosensor for Environmental Monitoring: Principle, Application, and Corresponding Achievement of Sustainable Development Goals. Bioengineered 2023, 14, 58–80. [Google Scholar] [CrossRef] [PubMed]
- Meliana, C.; Liu, J.; Show, P.L.; Low, S.S. Biosensor in Smart Food Traceability System for Food Safety and Security. Bioengineered 2024, 15, 2310908. [Google Scholar] [CrossRef]
- Visconti, G.; Boccard, J.; Feinberg, M.; Rudaz, S. From Fundamentals in Calibration to Modern Methodologies: A Tutorial for Small Molecules Quantification in Liquid Chromatography–Mass Spectrometry Bioanalysis. Anal. Chim. Acta 2023, 1240, 340711. [Google Scholar] [CrossRef]
- Malik, P.; Katyal, V.; Malik, V.; Asatkar, A.; Inwati, G.; Mukherjee, T.K. Nanobiosensors: Concepts and Variations. ISRN Nanomater. 2013, 2013, 1–9. [Google Scholar] [CrossRef]
- Mao, Y.; Huang, C.; Zhou, X.; Han, R.; Deng, Y.; Zhou, S. Genetically Encoded Biosensor Engineering for Application in Directed Evolution. J. Microbiol. Biotechnol. 2023, 33, 1257–1267. [Google Scholar] [CrossRef]
- Clark, L.C.; Lyons, C. Electrode Systems for Continuous Monitoring in Cardiovascular Surgery. Ann. N. Y. Acad. Sci. 1962, 102, 29–45. [Google Scholar] [CrossRef]
- Wang, J. Electrochemical Glucose Biosensors. Chem. Rev. 2008, 108, 814–825. [Google Scholar] [CrossRef]
- Koyappayil, A.; Lee, M.-H. Ultrasensitive Materials for Electrochemical Biosensor Labels. Sensors 2020, 21, 89. [Google Scholar] [CrossRef]
- Shimomura, O.; Johnson, F.H.; Saiga, Y. Extraction, Purification and Properties of Aequorin, a Bioluminescent Protein from the Luminous Hydromedusan, Aequorea. J. Cell. Comp. Physiol. 1962, 59, 223–239. [Google Scholar] [CrossRef]
- Tamura, T.; Hamachi, I. Recent Progress in Design of Protein-Based Fluorescent Biosensors and Their Cellular Applications. ACS Chem. Biol. 2014, 9, 2708–2717. [Google Scholar] [CrossRef]
- Nasu, Y.; Shen, Y.; Kramer, L.; Campbell, R.E. Structure- and Mechanism-Guided Design of Single Fluorescent Protein-Based Biosensors. Nat. Chem. Biol. 2021, 17, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Ding, N.; Zhou, S.; Deng, Y. Transcription-Factor-Based Biosensor Engineering for Applications in Synthetic Biology. ACS Synth. Biol. 2021, 10, 911–922. [Google Scholar] [CrossRef] [PubMed]
- Funk, M.A.; Leitner, J.; Gerner, M.C.; Hammerler, J.M.; Salzer, B.; Lehner, M.; Battin, C.; Gumpelmair, S.; Stiasny, K.; Grabmeier-Pfistershammer, K.; et al. Interrogating Ligand-Receptor Interactions Using Highly Sensitive Cellular Biosensors. Nat. Commun. 2023, 14, 7804. [Google Scholar] [CrossRef] [PubMed]
- Reja, S.I.; Minoshima, M.; Hori, Y.; Kikuchi, K. Recent Advancements of Fluorescent Biosensors Using Semisynthetic Probes. Biosens. Bioelectron. 2024, 247, 115862. [Google Scholar] [CrossRef] [PubMed]
- Broch, F.; Gautier, A. Illuminating Cellular Biochemistry: Fluorogenic Chemogenetic Biosensors for Biological Imaging. ChemPlusChem 2020, 85, 1487–1497. [Google Scholar] [CrossRef] [PubMed]
- Gautier, A. Fluorescence-Activating and Absorption-Shifting Tags for Advanced Imaging and Biosensing. Acc. Chem. Res. 2022, 55, 3125–3135. [Google Scholar] [CrossRef] [PubMed]
- Broch, F.; El Hajji, L.; Pietrancosta, N.; Gautier, A. Engineering of Tunable Allosteric-like Fluorogenic Protein Sensors. ACS Sens. 2023, 8, 3933–3942. [Google Scholar] [CrossRef] [PubMed]
- Childs-Disney, J.L.; Yang, X.; Gibaut, Q.M.R.; Tong, Y.; Batey, R.T.; Disney, M.D. Targeting RNA Structures with Small Molecules. Nat. Rev. Drug Discov. 2022, 21, 736–762. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Zhou, D.; Steitz, T.A.; Polikanov, Y.S.; Gagnon, M.G. Ribosome-Targeting Antibiotics: Modes of Action, Mechanisms of Resistance, and Implications for Drug Design. Annu. Rev. Biochem. 2018, 87, 451–478. [Google Scholar] [CrossRef]
- Mikaeeli Kangarshahi, B.; Naghib, S.M.; Rabiee, N. DNA/RNA-Based Electrochemical Nanobiosensors for Early Detection of Cancers. Crit. Rev. Clin. Lab. Sci. 2024, 1–23. [Google Scholar] [CrossRef]
- Xu, R.; Ouyang, L.; Chen, H.; Zhang, G.; Zhe, J. Recent Advances in Biomolecular Detection Based on Aptamers and Nanoparticles. Biosensors 2023, 13, 474. [Google Scholar] [CrossRef]
- Cai, R.; Yin, F.; Zhang, Z.; Tian, Y.; Zhou, N. Functional Chimera Aptamer and Molecular Beacon Based Fluorescent Detection of Staphylococcus Aureus with Strand Displacement-Target Recycling Amplification. Anal. Chim. Acta 2019, 1075, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Ma, D.; Zhang, J.; Kang, X.; Wu, Y.; Zhao, Y.; Yi, L.; Xi, Z. Development of a Universal RNA Dual-Terminal Labeling Method for Sensing RNA-Ligand Interactions. CCS Chem. 2022, 5, 221–233. [Google Scholar] [CrossRef]
- Klöcker, N.; Weissenboeck, F.P.; Rentmeister, A. Covalent Labeling of Nucleic Acids. Chem. Soc. Rev. 2020, 49, 8749–8773. [Google Scholar] [CrossRef] [PubMed]
- Wicks, S.L.; Hargrove, A.E. Fluorescent Indicator Displacement Assays to Identify and Characterize Small Molecule Interactions with RNA. Methods 2019, 167, 3–14. [Google Scholar] [CrossRef]
- Braselmann, E.; Wierzba, A.J.; Polaski, J.T.; Chromiński, M.; Holmes, Z.E.; Hung, S.-T.; Batan, D.; Wheeler, J.R.; Parker, R.; Jimenez, R.; et al. A Multicolor Riboswitch-Based Platform for Imaging of RNA in Live Mammalian Cells. Nat. Chem. Biol. 2018, 14, 964–971. [Google Scholar] [CrossRef]
- Sunbul, M.; Jäschke, A. SRB-2: A Promiscuous Rainbow Aptamer for Live-Cell RNA Imaging. Nucleic Acids Res. 2018, 46, e110. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, D.; Su, N.; Bao, B.; Xie, X.; Zuo, F.; Yang, L.; Wang, H.; Jiang, L.; Lin, Q.; et al. Visualizing RNA Dynamics in Live Cells with Bright and Stable Fluorescent RNAs. Nat. Biotechnol. 2019, 37, 1287–1293. [Google Scholar] [CrossRef]
- Neubacher, S.; Hennig, S. RNA Structure and Cellular Applications of Fluorescent Light-Up Aptamers. Angew. Chem. Int. Ed. 2019, 58, 1266–1279. [Google Scholar] [CrossRef]
- Swetha, P.; Fan, Z.; Wang, F.; Jiang, J.-H. Genetically Encoded Light-up RNA Aptamers and Their Applications for Imaging and Biosensing. J. Mater. Chem. B 2020, 8, 3382–3392. [Google Scholar] [CrossRef]
- Braselmann, E.; Rathbun, C.; Richards, E.M.; Palmer, A.E. Illuminating RNA Biology: Tools for Imaging RNA in Live Mammalian Cells. Cell Chem. Biol. 2020, 27, 891–903. [Google Scholar] [CrossRef] [PubMed]
- Ryckelynck, M. Development and Applications of Fluorogen/Light-Up RNA Aptamer Pairs for RNA Detection and More. In RNA Tagging; Heinlein, M., Ed.; Methods in Molecular Biology; Springer US: New York, NY, USA, 2020; Volume 2166, pp. 73–102. ISBN 978-1-07-160711-4. [Google Scholar]
- Lu, X.; Kong, K.Y.S.; Unrau, P.J. Harmonizing the Growing Fluorogenic RNA Aptamer Toolbox for RNA Detection and Imaging. Chem. Soc. Rev. 2023, 52, 4071–4098. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhang, S. Recent Development of Fluorescent Light-Up RNA Aptamers. Crit. Rev. Anal. Chem. 2022, 52, 1644–1661. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, L.; Jäschke, A.; Sunbul, M. A Color-Shifting Near-Infrared Fluorescent Aptamer–Fluorophore Module for Live-Cell RNA Imaging. Angew. Chem. Int. Ed. 2021, 60, 21441–21448. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Svensen, N.; Song, W.; Kim, H.; Zhang, S.; Li, X.; Jaffrey, S.R. Self-Assembly of Intracellular Multivalent RNA Complexes Using Dimeric Corn and Beetroot Aptamers. J. Am. Chem. Soc. 2022, 144, 5471–5477. [Google Scholar] [CrossRef] [PubMed]
- Passalacqua, L.F.M.; Starich, M.R.; Link, K.A.; Wu, J.; Knutson, J.R.; Tjandra, N.; Jaffrey, S.R.; Ferré-D’Amaré, A.R. Co-Crystal Structures of the Fluorogenic Aptamer Beetroot Show That Close Homology May Not Predict Similar RNA Architecture. Nat. Commun. 2023, 14, 2969. [Google Scholar] [CrossRef]
- Filonov, G.S.; Moon, J.D.; Svensen, N.; Jaffrey, S.R. Broccoli: Rapid Selection of an RNA Mimic of Green Fluorescent Protein by Fluorescence-Based Selection and Directed Evolution. J. Am. Chem. Soc. 2014, 136, 16299–16308. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Kim, H.; Litke, J.L.; Wu, J.; Jaffrey, S.R. Fluorophore-Promoted RNA Folding and Photostability Enables Imaging of Single Broccoli-Tagged mRNAs in Live Mammalian Cells. Angew. Chem. Int. Ed. 2020, 59, 4511–4518. [Google Scholar] [CrossRef]
- Steinmetzger, C.; Palanisamy, N.; Gore, K.R.; Höbartner, C. A Multicolor Large Stokes Shift Fluorogen-Activating RNA Aptamer with Cationic Chromophores. Chem.–Eur. J. 2019, 25, 1931–1935. [Google Scholar] [CrossRef]
- Mieczkowski, M.; Steinmetzger, C.; Bessi, I.; Lenz, A.-K.; Schmiedel, A.; Holzapfel, M.; Lambert, C.; Pena, V.; Höbartner, C. Large Stokes Shift Fluorescence Activation in an RNA Aptamer by Intermolecular Proton Transfer to Guanine. Nat. Commun. 2021, 12, 3549. [Google Scholar] [CrossRef]
- Song, W.; Filonov, G.S.; Kim, H.; Hirsch, M.; Li, X.; Moon, J.D.; Jaffrey, S.R. Imaging RNA Polymerase III Transcription Using a Photostable RNA–Fluorophore Complex. Nat. Chem. Biol. 2017, 13, 1187–1194. [Google Scholar] [CrossRef] [PubMed]
- Warner, K.D.; Sjekloća, L.; Song, W.; Filonov, G.S.; Jaffrey, S.R.; Ferré-D’Amaré, A.R. A Homodimer Interface without Base Pairs in an RNA Mimic of Red Fluorescent Protein. Nat. Chem. Biol. 2017, 13, 1195–1201. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Constantin, T.P.; Sloane, K.L.; Waggoner, A.S.; Bruchez, M.P.; Armitage, B.A. Fluoromodules Consisting of a Promiscuous RNA Aptamer and Red or Blue Fluorogenic Cyanine Dyes: Selection, Characterization, and Bioimaging. J. Am. Chem. Soc. 2017, 139, 9001–9009. [Google Scholar] [CrossRef] [PubMed]
- Shelke, S.A.; Shao, Y.; Laski, A.; Koirala, D.; Weissman, B.P.; Fuller, J.R.; Tan, X.; Constantin, T.P.; Waggoner, A.S.; Bruchez, M.P.; et al. Structural Basis for Activation of Fluorogenic Dyes by an RNA Aptamer Lacking a G-Quadruplex Motif. Nat. Commun. 2018, 9, 4542. [Google Scholar] [CrossRef] [PubMed]
- Arora, A.; Sunbul, M.; Jäschke, A. Dual-Colour Imaging of RNAs Using Quencher- and Fluorophore-Binding Aptamers. Nucleic Acids Res. 2015, 43, e144. [Google Scholar] [CrossRef] [PubMed]
- Autour, A.; Westhof, E.; Ryckelynck, M. iSpinach: A Fluorogenic RNA Aptamer Optimized for in Vitro Applications. Nucleic Acids Res. 2016, 44, 2491–2500. [Google Scholar] [CrossRef]
- Fernandez-Millan, P.; Autour, A.; Ennifar, E.; Westhof, E.; Ryckelynck, M. Crystal Structure and Fluorescence Properties of the iSpinach Aptamer in Complex with DFHBI. RNA 2017, 23, 1788–1795. [Google Scholar] [CrossRef] [PubMed]
- Dolgosheina, E.V.; Jeng, S.C.Y.; Panchapakesan, S.S.S.; Cojocaru, R.; Chen, P.S.K.; Wilson, P.D.; Hawkins, N.; Wiggins, P.A.; Unrau, P.J. RNA Mango Aptamer-Fluorophore: A Bright, High-Affinity Complex for RNA Labeling and Tracking. ACS Chem. Biol. 2014, 9, 2412–2420. [Google Scholar] [CrossRef] [PubMed]
- Trachman, R.J.; Demeshkina, N.A.; Lau, M.W.L.; Panchapakesan, S.S.S.; Jeng, S.C.Y.; Unrau, P.J.; Ferré-D’Amaré, A.R. Structural Basis for High-Affinity Fluorophore Binding and Activation by RNA Mango. Nat. Chem. Biol. 2017, 13, 807–813. [Google Scholar] [CrossRef]
- Autour, A.; Jeng, S.C.Y.; Cawte, A.D.; Abdolahzadeh, A.; Galli, A.; Panchapakesan, S.S.S.; Rueda, D.; Ryckelynck, M.; Unrau, P.J. Fluorogenic RNA Mango Aptamers for Imaging Small Non-Coding RNAs in Mammalian Cells. Nat. Commun. 2018, 9, 656. [Google Scholar] [CrossRef]
- Trachman, R.J.; Abdolahzadeh, A.; Andreoni, A.; Cojocaru, R.; Knutson, J.R.; Ryckelynck, M.; Unrau, P.J.; Ferré-D’Amaré, A.R. Crystal Structures of the Mango-II RNA Aptamer Reveal Heterogeneous Fluorophore Binding and Guide Engineering of Variants with Improved Selectivity and Brightness. Biochemistry 2018, 57, 3544–3548. [Google Scholar] [CrossRef] [PubMed]
- Trachman, R.J.; Autour, A.; Jeng, S.C.Y.; Abdolahzadeh, A.; Andreoni, A.; Cojocaru, R.; Garipov, R.; Dolgosheina, E.V.; Knutson, J.R.; Ryckelynck, M.; et al. Structure and Functional Reselection of the Mango-III Fluorogenic RNA Aptamer. Nat. Chem. Biol. 2019, 15, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Grate, D.; Wilson, C. Laser-Mediated, Site-Specific Inactivation of RNA Transcripts. Proc. Natl. Acad. Sci. USA 1999, 96, 6131–6136. [Google Scholar] [CrossRef] [PubMed]
- Baugh, C.; Grate, D.; Wilson, C. A Crystal Structure of the Malachite Green Aptamer. J. Mol. Biol. 2000, 301, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Bouhedda, F.; Fam, K.T.; Collot, M.; Autour, A.; Marzi, S.; Klymchenko, A.; Ryckelynck, M. A Dimerization-Based Fluorogenic Dye-Aptamer Module for RNA Imaging in Live Cells. Nat. Chem. Biol. 2020, 16, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Zuo, F.; Jiang, L.; Su, N.; Zhang, Y.; Bao, B.; Wang, L.; Shi, Y.; Yang, H.; Huang, X.; Li, R.; et al. Imaging the Dynamics of Messenger RNA with a Bright and Stable Green Fluorescent RNA. Nat. Chem. Biol. 2024, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kong, K.Y.S.; Jeng, S.C.Y.; Rayyan, B.; Unrau, P.J. RNA Peach and Mango: Orthogonal Two-Color Fluorogenic Aptamers Distinguish Nearly Identical Ligands. RNA 2021, 27, 604–615. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Chen, X.; Li, C.; Song, Q.; Li, H.; Zhu, L.; Yang, Y.; Ren, A. Structure-Based Investigation of Fluorogenic Pepper Aptamer. Nat. Chem. Biol. 2021, 17, 1289–1295. [Google Scholar] [CrossRef] [PubMed]
- Sunbul, M.; Lackner, J.; Martin, A.; Englert, D.; Hacene, B.; Grün, F.; Nienhaus, K.; Nienhaus, G.U.; Jäschke, A. Super-Resolution RNA Imaging Using a Rhodamine-Binding Aptamer with Fast Exchange Kinetics. Nat. Biotechnol. 2021, 39, 686–690. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Z.; Xiao, Y.; Jiang, H.; Zuo, X.; Li, X.; Fang, X. Structural Mechanisms for Binding and Activation of a Contact-Quenched Fluorophore by RhoBAST. Nat. Commun. 2024, 15, 4206. [Google Scholar] [CrossRef]
- Wirth, R.; Gao, P.; Nienhaus, G.U.; Sunbul, M.; Jäschke, A. SiRA: A Silicon Rhodamine-Binding Aptamer for Live-Cell Super-Resolution RNA Imaging. J. Am. Chem. Soc. 2019, 141, 7562–7571. [Google Scholar] [CrossRef] [PubMed]
- Paige, J.S.; Wu, K.Y.; Jaffrey, S.R. RNA Mimics of Green Fluorescent Protein. Science 2011, 333, 642–646. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Suslov, N.B.; Li, N.-S.; Shelke, S.A.; Evans, M.E.; Koldobskaya, Y.; Rice, P.A.; Piccirilli, J.A. A G-Quadruplex–Containing RNA Activates Fluorescence in a GFP-like Fluorophore. Nat. Chem. Biol. 2014, 10, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Strack, R.L.; Disney, M.D.; Jaffrey, S.R. A Superfolding Spinach2 Reveals the Dynamic Nature of Trinucleotide Repeat–Containing RNA. Nat. Methods 2013, 10, 1219–1224. [Google Scholar] [CrossRef] [PubMed]
- Dey, S.K.; Filonov, G.S.; Olarerin-George, A.O.; Jackson, B.T.; Finley, L.W.S.; Jaffrey, S.R. Repurposing an Adenine Riboswitch into a Fluorogenic Imaging and Sensing Tag. Nat. Chem. Biol. 2022, 18, 180–190. [Google Scholar] [CrossRef] [PubMed]
- Truong, L.; Kooshapur, H.; Dey, S.K.; Li, X.; Tjandra, N.; Jaffrey, S.R.; Ferré-D’Amaré, A.R. The Fluorescent Aptamer Squash Extensively Repurposes the Adenine Riboswitch Fold. Nat. Chem. Biol. 2022, 18, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.H.; Szostak, J.W. Isolation of High-Affinity GTP Aptamers from Partially Structured RNA Libraries. Proc. Natl. Acad. Sci. USA 2002, 99, 11616–11621. [Google Scholar] [CrossRef]
- Endoh, T.; Ohyama, T.; Sugimoto, N. RNA-Capturing Microsphere Particles (R-CAMPs) for Optimization of Functional Aptamers. Small 2019, 15, 1805062. [Google Scholar] [CrossRef]
- Endoh, T.; Sugimoto, N. Signaling Aptamer Optimization through Selection Using RNA-Capturing Microsphere Particles. Anal. Chem. 2020, 92, 7955–7963. [Google Scholar] [CrossRef] [PubMed]
- Bart, J.; Tiggelaar, R.; Yang, M.; Schlautmann, S.; Zuilhof, H.; Gardeniers, H. Room-Temperature Intermediate Layer Bonding for Microfluidic Devices. Lab. Chip 2009, 9, 3481. [Google Scholar] [CrossRef] [PubMed]
- Bouhedda, F.; Cubi, R.; Baudrey, S.; Ryckelynck, M. μIVC-Seq: A Method for Ultrahigh-Throughput Development and Functional Characterization of Small RNAs. In Small Non-Coding RNAs; Rederstorff, M., Ed.; Methods in Molecular Biology; Springer US: New York, NY, USA, 2021; Volume 2300, pp. 203–237. ISBN 978-1-07-161385-6. [Google Scholar]
- Truong, J.; Hsieh, Y.-F.; Truong, L.; Jia, G.; Hammond, M.C. Designing Fluorescent Biosensors Using Circular Permutations of Riboswitches. Methods 2018, 143, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Chen, W.; Reheman, Z.; Jiang, H.; Wu, J.; Li, X. Genetically Encoded RNA-Based Sensors with Pepper Fluorogenic Aptamer. Nucleic Acids Res. 2023, 51, 8322–8336. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Shi, J.; Mudiyanselage, A.P.K.K.K.; Wu, R.; Zhao, B.; Zhou, M.; You, M. Genetically Encoded RNA-Based Sensors for Intracellular Imaging of Silver Ions. Chem. Commun. 2019, 55, 707–710. [Google Scholar] [CrossRef] [PubMed]
- Kavita, K.; Breaker, R.R. Discovering Riboswitches: The Past and the Future. Trends Biochem. Sci. 2023, 48, 119–141. [Google Scholar] [CrossRef] [PubMed]
- Greenlee, E.B.; Stav, S.; Atilho, R.M.; Brewer, K.I.; Harris, K.A.; Malkowski, S.N.; Mirihana Arachchilage, G.; Perkins, K.R.; Sherlock, M.E.; Breaker, R.R. Challenges of Ligand Identification for the Second Wave of Orphan Riboswitch Candidates. RNA Biol. 2018, 15, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, Z.; Lünse, C.E.; Corbino, K.A.; Ames, T.D.; Nelson, J.W.; Roth, A.; Perkins, K.R.; Sherlock, M.E.; Breaker, R.R. Detection of 224 Candidate Structured RNAs by Comparative Analysis of Specific Subsets of Intergenic Regions. Nucleic Acids Res. 2017, 45, 10811–10823. [Google Scholar] [CrossRef] [PubMed]
- Porter, E.B.; Marcano-Velázquez, J.G.; Batey, R.T. The Purine Riboswitch as a Model System for Exploring RNA Biology and Chemistry. Biochim. Biophys. Acta BBA-Gene Regul. Mech. 2014, 1839, 919–930. [Google Scholar] [CrossRef]
- Mandal, M.; Breaker, R.R. Adenine Riboswitches and Gene Activation by Disruption of a Transcription Terminator. Nat. Struct. Mol. Biol. 2004, 11, 29–35. [Google Scholar] [CrossRef]
- Bose, D.; Su, Y.; Marcus, A.; Raulet, D.H.; Hammond, M.C. An RNA-Based Fluorescent Biosensor for High-Throughput Analysis of the cGAS-cGAMP-STING Pathway. Cell Chem. Biol. 2016, 23, 1539–1549. [Google Scholar] [CrossRef] [PubMed]
- Porter, E.B.; Polaski, J.T.; Morck, M.M.; Batey, R.T. Recurrent RNA Motifs as Scaffolds for Genetically Encodable Small-Molecule Biosensors. Nat. Chem. Biol. 2017, 13, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.Y.; Lau, H.L.; Kwok, C.K. Capture-SELEX: Selection Strategy, Aptamer Identification, and Biosensing Application. Biosensors 2022, 12, 1142. [Google Scholar] [CrossRef]
- Boussebayle, A.; Groher, F.; Suess, B. RNA-Based Capture-SELEX for the Selection of Small Molecule-Binding Aptamers. Methods 2019, 161, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Boussebayle, A.; Torka, D.; Ollivaud, S.; Braun, J.; Bofill-Bosch, C.; Dombrowski, M.; Groher, F.; Hamacher, K.; Suess, B. Next-Level Riboswitch Development—Implementation of Capture-SELEX Facilitates Identification of a New Synthetic Riboswitch. Nucleic Acids Res. 2019, 47, 4883–4895. [Google Scholar] [CrossRef]
- Kraus, L.; Duchardt-Ferner, E.; Bräuchle, E.; Fürbacher, S.; Kelvin, D.; Marx, H.; Boussebayle, A.; Maurer, L.-M.; Bofill-Bosch, C.; Wöhnert, J.; et al. Development of a Novel Tobramycin Dependent Riboswitch. Nucleic Acids Res. 2023, 51, 11375–11385. [Google Scholar] [CrossRef] [PubMed]
- Kramat, J.; Kraus, L.; Gunawan, V.J.; Smyej, E.; Froehlich, P.; Weber, T.E.; Spiehl, D.; Koeppl, H.; Blaeser, A.; Suess, B. Sensing Levofloxacin with an RNA Aptamer as a Bioreceptor. Biosensors 2024, 14, 56. [Google Scholar] [CrossRef]
- Legen, T.; Mayer, G. Modular Approach for Rapid Identification of RNA-Based Sensors. ACS Sens. 2024, 9, 753–758. [Google Scholar] [CrossRef]
- Mohsen, M.G.; Midy, M.K.; Balaji, A.; Breaker, R.R. Exploiting Natural Riboswitches for Aptamer Engineering and Validation. Nucleic Acids Res. 2023, 51, 966–981. [Google Scholar] [CrossRef]
- Citartan, M. The Dynamicity of Light-up Aptamers in One-Pot in Vitro Diagnostic Assays. Analyst 2021, 147, 10–21. [Google Scholar] [CrossRef]
- Li, Y.; Arce, A.; Lucci, T.; Rasmussen, R.A.; Lucks, J.B. Dynamic RNA Synthetic Biology: New Principles, Practices and Potential. RNA Biol. 2023, 20, 817–829. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.K.; Alam, K.K.; Verosloff, M.S.; Capdevila, D.A.; Desmau, M.; Clauer, P.R.; Lee, J.W.; Nguyen, P.Q.; Pastén, P.A.; Matiasek, S.J.; et al. Cell-Free Biosensors for Rapid Detection of Water Contaminants. Nat. Biotechnol. 2020, 38, 1451–1459. [Google Scholar] [CrossRef] [PubMed]
- Thavarajah, W.; Silverman, A.D.; Verosloff, M.S.; Kelley-Loughnane, N.; Jewett, M.C.; Lucks, J.B. Point-of-Use Detection of Environmental Fluoride via a Cell-Free Riboswitch-Based Biosensor. ACS Synth. Biol. 2020, 9, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, R.S.; Batey, R.T. SPRINT: A Cas13a-Based Platform for Detection of Small Molecules. Nucleic Acids Res. 2020, 48, e101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhou, W.; Lin, X.; Khan, M.R.; Deng, S.; Zhou, M.; He, G.; Wu, C.; Deng, R.; He, Q. Light-up RNA Aptamer Signaling-CRISPR-Cas13a-Based Mix-and-Read Assays for Profiling Viable Pathogenic Bacteria. Biosens. Bioelectron. 2021, 176, 112906. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xue, T.; Wang, M.; Ledesma-Amaro, R.; Lu, Y.; Hu, X.; Zhang, T.; Yang, M.; Li, Y.; Xiang, J.; et al. CRISPR-Cas13a Cascade-Based Viral RNA Assay for Detecting SARS-CoV-2 and Its Mutations in Clinical Samples. Sens. Actuators B Chem. 2022, 362, 131765. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Song, Y.; Jang, H.; Moon, J.; Kang, H.; Huh, Y.-M.; Son, H.Y.; Rho, H.W.; Park, M.; Lim, E.-K.; et al. Elution-Free DNA Detection Using CRISPR/Cas9-Mediated Light-up Aptamer Transcription: Toward All-in-One DNA Purification and Detection Tube. Biosens. Bioelectron. 2023, 225, 115085. [Google Scholar] [CrossRef] [PubMed]
- Autour, A.; Bouhedda, F.; Cubi, R.; Ryckelynck, M. Optimization of Fluorogenic RNA-Based Biosensors Using Droplet-Based Microfluidic Ultrahigh-Throughput Screening. Methods 2019, 161, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Husser, C.; Vuilleumier, S.; Ryckelynck, M. FluorMango, an RNA-Based Fluorogenic Biosensor for the Direct and Specific Detection of Fluoride. Small 2023, 19, 2205232. [Google Scholar] [CrossRef]
- You, M.; Litke, J.L.; Jaffrey, S.R. Imaging Metabolite Dynamics in Living Cells Using a Spinach-Based Riboswitch. Proc. Natl. Acad. Sci. 2015, 112, E256–E265. [Google Scholar] [CrossRef]
- Wang, T.; Simmel, F.C. Switchable Fluorescent Light-Up Aptamers Based on Riboswitch Architectures. Angew. Chem. Int. Ed. 2023, 62, e202302858. [Google Scholar] [CrossRef] [PubMed]
- Geraci, I.; Autour, A.; Pietruschka, G.; Shiian, A.; Borisova, M.; Mayer, C.; Ryckelynck, M.; Mayer, G. Fluorogenic RNA-Based Biosensor to Sense the Glycolytic Flux in Mammalian Cells. ACS Chem. Biol. 2022, 17, 1164–1173. [Google Scholar] [CrossRef] [PubMed]
- Sett, A.; Zara, L.; Dausse, E.; Toulmé, J.-J. A Malachite Green Light-up Aptasensor for the Detection of Theophylline. Talanta 2021, 232, 122417. [Google Scholar] [CrossRef]
- You, M.; Litke, J.L.; Wu, R.; Jaffrey, S.R. Detection of Low-Abundance Metabolites in Live Cells Using an RNA Integrator. Cell Chem. Biol. 2019, 26, 471–481.e3. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Breaker, R.R. Rational Design of Allosteric Ribozymes. Chem. Biol. 1997, 4, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Karunanayake Mudiyanselage, A.P.K.K.; Yu, Q.; Leon-Duque, M.A.; Zhao, B.; Wu, R.; You, M. Genetically Encoded Catalytic Hairpin Assembly for Sensitive RNA Imaging in Live Cells. J. Am. Chem. Soc. 2018, 140, 8739–8745. [Google Scholar] [CrossRef]
- Wu, R.; Karunanayake Mudiyanselage, A.P.K.K.; Shafiei, F.; Zhao, B.; Bagheri, Y.; Yu, Q.; McAuliffe, K.; Ren, K.; You, M. Genetically Encoded Ratiometric RNA-Based Sensors for Quantitative Imaging of Small Molecules in Living Cells. Angew. Chem. Int. Ed. 2019, 58, 18271–18275. [Google Scholar] [CrossRef]
- Paige, J.S.; Nguyen-Duc, T.; Song, W.; Jaffrey, S.R. Fluorescence Imaging of Cellular Metabolites with RNA. Science 2012, 335, 1194. [Google Scholar] [CrossRef]
- Furutani, C.; Shinomiya, K.; Aoyama, Y.; Yamada, K.; Sando, S. Modular Blue Fluorescent RNA Sensors for Label-Free Detection of Target Molecules. Mol. Biosyst. 2010, 6, 1569. [Google Scholar] [CrossRef]
- Stojanovic, M.N.; Kolpashchikov, D.M. Modular Aptameric Sensors. J. Am. Chem. Soc. 2004, 126, 9266–9270. [Google Scholar] [CrossRef]
- Kellenberger, C.A.; Wilson, S.C.; Hickey, S.F.; Gonzalez, T.L.; Su, Y.; Hallberg, Z.F.; Brewer, T.F.; Iavarone, A.T.; Carlson, H.K.; Hsieh, Y.-F.; et al. GEMM-I Riboswitches from Geobacter Sense the Bacterial Second Messenger Cyclic AMP-GMP. Proc. Natl. Acad. Sci. USA 2015, 112, 5383–5388. [Google Scholar] [CrossRef] [PubMed]
- Kellenberger, C.A.; Wilson, S.C.; Sales-Lee, J.; Hammond, M.C. RNA-Based Fluorescent Biosensors for Live Cell Imaging of Second Messengers Cyclic Di-GMP and Cyclic AMP-GMP. J. Am. Chem. Soc. 2013, 135, 4906–4909. [Google Scholar] [CrossRef] [PubMed]
- Mukkayyan, N.; Poon, R.; Sander, P.N.; Lai, L.-Y.; Zubair-Nizami, Z.; Hammond, M.C.; Chatterjee, S.S. In Vivo Detection of Cyclic-Di-AMP in Staphylococcus Aureus. ACS Omega 2022, 7, 32749–32753. [Google Scholar] [CrossRef] [PubMed]
- Kellenberger, C.A.; Chen, C.; Whiteley, A.T.; Portnoy, D.A.; Hammond, M.C. RNA-Based Fluorescent Biosensors for Live Cell Imaging of Second Messenger Cyclic Di-AMP. J. Am. Chem. Soc. 2015, 137, 6432–6435. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, S.; Luo, Y.; Zhou, J.; Dayie, T.K.; Sintim, H.O. Nanomolar Fluorescent Detection of C-Di-GMP Using a Modular Aptamer Strategy. Chem. Commun. 2012, 48, 9059–9061. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.C.; Wilson, S.C.; Hammond, M.C. Next-Generation RNA-Based Fluorescent Biosensors Enable Anaerobic Detection of Cyclic Di-GMP. Nucleic Acids Res. 2016, 44, e139. [Google Scholar] [CrossRef]
- Ketterer, S.; Gladis, L.; Kozica, A.; Meier, M. Engineering and Characterization of Fluorogenic Glycine Riboswitches. Nucleic Acids Res. 2016, 44, 5983–5992. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.D.; Wu, J.; Dey, S.K.; Litke, J.L.; Li, X.; Kim, H.; Jaffrey, S.R. Naturally Occurring Three-Way Junctions Can Be Repurposed as Genetically Encoded RNA-Based Sensors. Cell Chem. Biol. 2021, 28, 1569–1580.e4. [Google Scholar] [CrossRef] [PubMed]
- Allchin, E.R.; Rosch, J.C.; Stoneman, A.D.; Kim, H.; Lippmann, E.S. Generalized Strategy for Engineering Mammalian Cell-Compatible RNA-Based Biosensors from Random Sequence Libraries. ACS Sens. 2023, 8, 2079–2086. [Google Scholar] [CrossRef]
- Xu, J.; Cotruvo, J.A., Jr. The czcD (NiCo) Riboswitch Responds to Iron(II). Biochemistry 2020, 59, 1508–1516. [Google Scholar] [CrossRef]
- Abatemarco, J.; Sarhan, M.F.; Wagner, J.M.; Lin, J.-L.; Liu, L.; Hassouneh, W.; Yuan, S.-F.; Alper, H.S.; Abate, A.R. RNA-Aptamers-in-Droplets (RAPID) High-Throughput Screening for Secretory Phenotypes. Nat. Commun. 2017, 8, 332. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Wu, R.; Zhao, B.; Zeinert, R.; Chien, P.; You, M. Live-Cell Imaging of Guanosine Tetra- and Pentaphosphate (p)ppGpp with RNA-Based Fluorescent Sensors. Angew. Chem. Int. Ed. Engl. 2021, 60, 24070–24074. [Google Scholar] [CrossRef] [PubMed]
- Shafiei, F.; McAuliffe, K.; Bagheri, Y.; Sun, Z.; Yu, Q.; Wu, R.; You, M. Paper-Based Fluorogenic RNA Aptamer Sensors for Label-Free Detection of Small Molecules. Anal. Methods 2020, 12, 2674–2681. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Hickey, S.F.; Keyser, S.G.L.; Hammond, M.C. In Vitro and In Vivo Enzyme Activity Screening via RNA-Based Fluorescent Biosensors for S-Adenosyl-l-Homocysteine (SAH). J. Am. Chem. Soc. 2016, 138, 7040–7047. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Jaffrey, S.R. A Fluorogenic RNA-Based Sensor Activated by Metabolite-Induced RNA Dimerization. Cell Chem. Biol. 2019, 26, 1725–1731. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Mo, L.; Litke, J.L.; Dey, S.K.; Suter, S.R.; Jaffrey, S.R. Imaging Intracellular S-Adenosyl Methionine Dynamics in Live Mammalian Cells with a Genetically Encoded Red Fluorescent RNA-Based Sensor. J. Am. Chem. Soc. 2020, 142, 14117–14124. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Li, H.; Xie, X.; Wang, H.; Jiang, Y.; Li, T.; Zhang, B.; Jiang, X.; Cao, Y.; Zhang, R.; et al. Imaging Intracellular Metabolite and Protein Changes in Live Mammalian Cells with Bright Fluorescent RNA-Based Genetically Encoded Sensors. Biosens. Bioelectron. 2023, 235, 115411. [Google Scholar] [CrossRef] [PubMed]
- Endoh, T.; Tan, J.-H.; Chen, S.-B.; Sugimoto, N. Cladogenetic Orthogonal Light-Up Aptamers for Simultaneous Detection of Multiple Small Molecules in Cells. Anal. Chem. 2022, 95, 976–985. [Google Scholar] [CrossRef] [PubMed]
- Jepsen, M.D.E.; Sparvath, S.M.; Nielsen, T.B.; Langvad, A.H.; Grossi, G.; Gothelf, K.V.; Andersen, E.S. Development of a Genetically Encodable FRET System Using Fluorescent RNA Aptamers. Nat. Commun. 2018, 9, 18. [Google Scholar] [CrossRef]
- Yuan, S.-F.; Alper, H.S. Improving Spinach2-and Broccoli-Based Biosensors for Single and Double Analytes. Biotechnol. Notes 2020, 1, 2–8. [Google Scholar] [CrossRef]
- Ponchon, L.; Dardel, F. Recombinant RNA Technology: The tRNA Scaffold. Nat. Methods 2007, 4, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Filonov, G.S.; Kam, C.W.; Song, W.; Jaffrey, S.R. In-Gel Imaging of RNA Processing Using Broccoli Reveals Optimal Aptamer Expression Strategies. Chem. Biol. 2015, 22, 649–660. [Google Scholar] [CrossRef] [PubMed]
- Litke, J.L.; Jaffrey, S.R. Highly Efficient Expression of Circular RNA Aptamers in Cells Using Autocatalytic Transcripts. Nat. Biotechnol. 2019, 37, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Luo, Y.; Wang, X.; Li, S.; Luo, M.; Yin, M.; Zuo, Y.; Li, G.; Yao, J.; Yang, H.; et al. A Protein-Independent Fluorescent RNA Aptamer Reporter System for Plant Genetic Engineering. Nat. Commun. 2020, 11, 3847. [Google Scholar] [CrossRef] [PubMed]
- Poudyal, R.R.; Sieg, J.P.; Portz, B.; Keating, C.D.; Bevilacqua, P.C. RNA Sequence and Structure Control Assembly and Function of RNA Condensates. RNA N. Y. N. 2021, 27, 1589–1601. [Google Scholar] [CrossRef] [PubMed]
- Ji, R.; Wang, L.; Shang, Y.; Du, S.; Xiao, Y.; Dong, W.; Cui, L.; Gao, R.; Ren, K. RNA Condensate as a Versatile Platform for Improving Fluorogenic RNA Aptamer Properties and Cell Imaging. J. Am. Chem. Soc. 2024, 146, 4402–4411. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Karunanayake Mudiyanselage, A.P.K.K.; Ren, K.; Sun, Z.; Tian, Q.; Zhao, B.; Bagheri, Y.; Lutati, D.; Keshri, P.; You, M. Ratiometric Fluorogenic RNA-Based Sensors for Imaging Live-Cell Dynamics of Small Molecules. ACS Appl. Bio Mater. 2020, 3, 2633–2642. [Google Scholar] [CrossRef] [PubMed]
- Mumbleau, M.M.; Chevance, F.; Hughes, K.; Hammond, M.C. Investigating the Effect of RNA Scaffolds on the Multicolor Fluorogenic Aptamer Pepper in Different Bacterial Species. ACS Synth. Biol. 2024, 13, 1093–1099. [Google Scholar] [CrossRef] [PubMed]
- Poppleton, E.; Urbanek, N.; Chakraborty, T.; Griffo, A.; Monari, L.; Göpfrich, K. RNA Origami: Design, Simulation and Application. RNA Biol. 2023, 20, 510–524. [Google Scholar] [CrossRef]
- Mi, L.; Yu, Q.; Karunanayake Mudiyanselage, A.P.K.K.; Wu, R.; Sun, Z.; Zheng, R.; Ren, K.; You, M. Genetically Encoded RNA-Based Bioluminescence Resonance Energy Transfer (BRET) Sensors. ACS Sens. 2023, 8, 308–316. [Google Scholar] [CrossRef]
- Su, Y.; Hammond, M.C. RNA-Based Fluorescent Biosensors for Live Cell Imaging of Small Molecules and RNAs. Curr. Opin. Biotechnol. 2020, 63, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Ren, A.; Wang, X.C.; Kellenberger, C.A.; Rajashankar, K.R.; Jones, R.A.; Hammond, M.C.; Patel, D.J. Structural Basis for Molecular Discrimination by a 3′,3′-cGAMP Sensing Riboswitch. Cell Rep. 2015, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wright, T.A.; Jiang, L.; Park, J.J.; Anderson, W.A.; Chen, G.; Hallberg, Z.F.; Nan, B.; Hammond, M.C. Second Messengers and Divergent HD-GYP Phosphodiesterases Regulate 3’,3’-cGAMP Signaling. Mol. Microbiol. 2020, 113, 222–236. [Google Scholar] [CrossRef]
- Greenberg, M.V.C.; Bourc’his, D. The Diverse Roles of DNA Methylation in Mammalian Development and Disease. Nat. Rev. Mol. Cell Biol. 2019, 20, 590–607. [Google Scholar] [CrossRef]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, Writing and Erasing mRNA Methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Schapira, M.; Arrowsmith, C.H.; Barsyte-Lovejoy, D. Protein Arginine Methylation: From Enigmatic Functions to Therapeutic Targeting. Nat. Rev. Drug Discov. 2021, 20, 509–530. [Google Scholar] [CrossRef]
- Zheng, L.; Song, Q.; Xu, X.; Shen, X.; Li, C.; Li, H.; Chen, H.; Ren, A. Structure-Based Insights into Recognition and Regulation of SAM-Sensing Riboswitches. Sci. China Life Sci. 2023, 66, 31–50. [Google Scholar] [CrossRef]
- Guo, J.U.; Bartel, D.P. RNA G-Quadruplexes Are Globally Unfolded in Eukaryotic Cells and Depleted in Bacteria. Science 2016, 353, aaf5371. [Google Scholar] [CrossRef]
- Hao, X.; Huang, Y.; Qiu, M.; Yin, C.; Ren, H.; Gan, H.; Li, H.; Zhou, Y.; Xia, J.; Li, W.; et al. Immunoassay of S-Adenosylmethionine and S-Adenosylhomocysteine: The Methylation Index as a Biomarker for Disease and Health Status. BMC Res. Notes 2016, 9, 498. [Google Scholar] [CrossRef]
- Khodr, R.; Husser, C.; Ryckelynck, M. Direct Fluoride Monitoring Using a Fluorogenic RNA-Based Biosensor. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2024; Volume 696, pp. 85–107. ISBN 978-0-443-23643-3. [Google Scholar]
- Boyd, M.A.; Thavarajah, W.; Lucks, J.B.; Kamat, N.P. Robust and Tunable Performance of a Cell-Free Biosensor Encapsulated in Lipid Vesicles. Sci. Adv. 2023, 9, eadd6605. [Google Scholar] [CrossRef]
- Chang, T.; Li, G.; Chang, D.; Amini, R.; Zhu, X.; Zhao, T.; Gu, J.; Li, Z.; Li, Y. An RNA-Cleaving DNAzyme That Requires an Organic Solvent to Function. Angew. Chem. Int. Ed. 2023, 62, e202310941. [Google Scholar] [CrossRef]
- Nakano, S.-I.; Sugimoto, N. The Structural Stability and Catalytic Activity of DNA and RNA Oligonucleotides in the Presence of Organic Solvents. Biophys. Rev. 2016, 8, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Shimada, I.; Kimura, R.; Hyuga, M. Light-up Fluorophore–DNA Aptamer Pair for Label-Free Turn-on Aptamer Sensors. Chem. Commun. 2016, 52, 4041–4044. [Google Scholar] [CrossRef]
- Song, G.; Zou, Z.; Feng, A.; Li, C.; Liang, M.; Wang, F.; Liu, X. Light-Up Aptameric Sensor of Serotonin for Point-of-Care Use. Anal. Chem. 2023, 95, 9076–9082. [Google Scholar] [CrossRef]
- VarnBuhler, B.S.; Moon, J.; Dey, S.K.; Wu, J.; Jaffrey, S.R. Detection of SARS-CoV-2 RNA Using a DNA Aptamer Mimic of Green Fluorescent Protein. ACS Chem. Biol. 2022, 17, 840–853. [Google Scholar] [CrossRef]
- Egli, M.; Manoharan, M. Chemistry, Structure and Function of Approved Oligonucleotide Therapeutics. Nucleic Acids Res. 2023, 51, 2529–2573. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.; Park, S.V.; Oh, S.S. Ionic Liquid-Caged Nucleic Acids Enable Active Folding-Based Molecular Recognition with Hydrolysis Resistance. Nucleic Acids Res. 2023, 52, gkad1093. [Google Scholar] [CrossRef]
- Hollenstein, M. Enzymatic Synthesis of Base-Modified Nucleic Acids; Springer Nature: Singapore, 2023. [Google Scholar]
- Chan, K.Y.; Kinghorn, A.B.; Hollenstein, M.; Tanner, J.A. Chemical Modifications for a Next Generation of Nucleic Acid Aptamers. Chembiochem Eur. J. Chem. Biol. 2022, 23, e202200006. [Google Scholar] [CrossRef]
- Savage, J.C.; Shinde, P.; Bächinger, H.P.; Davare, M.A.; Shinde, U. A Ribose Modification of Spinach Aptamer Accelerates Lead(ii) Cation Association in Vitro. Chem. Commun. 2019, 55, 5882–5885. [Google Scholar] [CrossRef]
- Savage, J.C.; Shinde, P.; Yao, Y.; Davare, M.A.; Shinde, U. A Broccoli Aptamer Chimera Yields a Fluorescent K + Sensor Spanning Physiological Concentrations. Chem. Commun. 2021, 57, 1344–1347. [Google Scholar] [CrossRef]
- DasGupta, S.; Shelke, S.A.; Li, N.; Piccirilli, J.A. Spinach RNA Aptamer Detects Lead(ii) with High Selectivity. Chem. Commun. 2015, 51, 9034–9037. [Google Scholar] [CrossRef]
- Klußmann, S.; Nolte, A.; Bald, R.; Erdmann, V.A.; Fürste, J.P. Mirror-Image RNA That Binds D-Adenosine. Nat. Biotechnol. 1996, 14, 1112–1115. [Google Scholar] [CrossRef]
- Zhong, W.; Sczepanski, J.T. A Mirror Image Fluorogenic Aptamer Sensor for Live-Cell Imaging of MicroRNAs. ACS Sens. 2019, 4, 566–570. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, T.F. Mirror-Image T7 Transcription of Chirally Inverted Ribosomal and Functional RNAs. Science 2022, 378, 405–412. [Google Scholar] [CrossRef]
- Hoshika, S.; Leal, N.A.; Kim, M.-J.; Kim, M.-S.; Karalkar, N.B.; Kim, H.-J.; Bates, A.M.; Watkins, N.E.; SantaLucia, H.A.; Meyer, A.J.; et al. Hachimoji DNA and RNA: A Genetic System with Eight Building Blocks. Science 2019, 363, 884–887. [Google Scholar] [CrossRef]
- Manna, S.; Kimoto, M.; Truong, J.; Bommisetti, P.; Peitz, A.; Hirao, I.; Hammond, M.C. Systematic Mutation and Unnatural Base Pair Incorporation Improves Riboswitch-Based Biosensor Response Time. ACS Sens. 2023, 8, 4468–4472. [Google Scholar] [CrossRef]
- Svensen, N.; Jaffrey, S.R. Fluorescent RNA Aptamers as a Tool to Study RNA-Modifying Enzymes. Cell Chem. Biol. 2016, 23, 415–425. [Google Scholar] [CrossRef]
- Ji, D.; Feng, H.; Liew, S.W.; Kwok, C.K. Modified Nucleic Acid Aptamers: Development, Characterization, and Biological Applications. Trends Biotechnol. 2023, 41, 1360–1384. [Google Scholar] [CrossRef]
- Dunn, M.R.; Chaput, J.C. An In Vitro Selection Protocol for Threose Nucleic Acid (TNA) Using DNA Display. Curr. Protoc. Nucleic Acid. Chem. 2014, 57, 9.8.1–9.8.19. [Google Scholar] [CrossRef]
- Sando, S.; Narita, A.; Aoyama, Y. Light-Up Hoechst–DNA Aptamer Pair: Generation of an Aptamer-Selective Fluorophore from a Conventional DNA-Staining Dye. Available online: https://chemistry-europe.onlinelibrary.wiley.com/doi/10.1002/cbic.200700325 (accessed on 4 June 2024).
- Wang, J.; Zhang, Y.; Wang, H.; Chen, Y.; Xu, L.; Gao, T.; Pei, R. Selection and Analysis of DNA Aptamers to Berberine to Develop a Label-Free Light-up Fluorescent Probe. New J. Chem. 2016, 40, 9768–9773. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, J.; Zhang, Y.; Xu, L.; Gao, T.; Wang, B.; Pei, R. Selection and Characterization of a DNA Aptamer to Crystal Violet. Photochem. Photobiol. Sci. 2018, 17, 800–806. [Google Scholar] [CrossRef] [PubMed]
- Connelly, R.P.; Madalozzo, P.F.; Mordeson, J.E.; Pratt, A.D.; Gerasimova, Y.V. Promiscuous Dye Binding by a Light-up Aptamer: Application for Label-Free Multi-Wavelength Biosensing. Chem. Commun. 2021, 57, 3672–3675. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, J.; Wang, Q.; Chen, X.; Liu, M.; Chen, H.; Pei, R. Selection and Characterization of Dimethylindole Red DNA Aptamers for the Development of Light-up Fluorescent Probes. Talanta 2017, 168, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Husser, C.; Baudrey, S.; Ryckelynck, M. High-Throughput Development and Optimization of RNA-Based Fluorogenic Biosensors of Small Molecules Using Droplet-Based Microfluidics. Methods Mol. Biol. Clifton NJ 2023, 2570, 243–269. [Google Scholar] [CrossRef]
- Mudgal, S.; Manikandan, K.; Mukherjee, A.; Krishnan, A.; Sinha, K.M. Cyclic Di-AMP: Small Molecule with Big Roles in Bacteria. Microb. Pathog. 2021, 161, 105264. [Google Scholar] [CrossRef] [PubMed]
- Imler, J.-L.; Cai, H.; Meignin, C.; Martins, N. Evolutionary Immunology to Explore Original Antiviral Strategies. Philos. Trans. R. Soc. B Biol. Sci. 2024, 379, 20230068. [Google Scholar] [CrossRef] [PubMed]
- Rangel, A.E.; Hariri, A.A.; Eisenstein, M.; Soh, H.T. Engineering Aptamer Switches for Multifunctional Stimulus-Responsive Nanosystems. Adv. Mater. 2020, 32, 2003704. [Google Scholar] [CrossRef]
- Kraemer, S.; Vaught, J.D.; Bock, C.; Gold, L.; Katilius, E.; Keeney, T.R.; Kim, N.; Saccomano, N.A.; Wilcox, S.K.; Zichi, D.; et al. From SOMAmer-Based Biomarker Discovery to Diagnostic and Clinical Applications: A SOMAmer-Based, Streamlined Multiplex Proteomic Assay. PLoS ONE 2011, 6, e26332. [Google Scholar] [CrossRef]

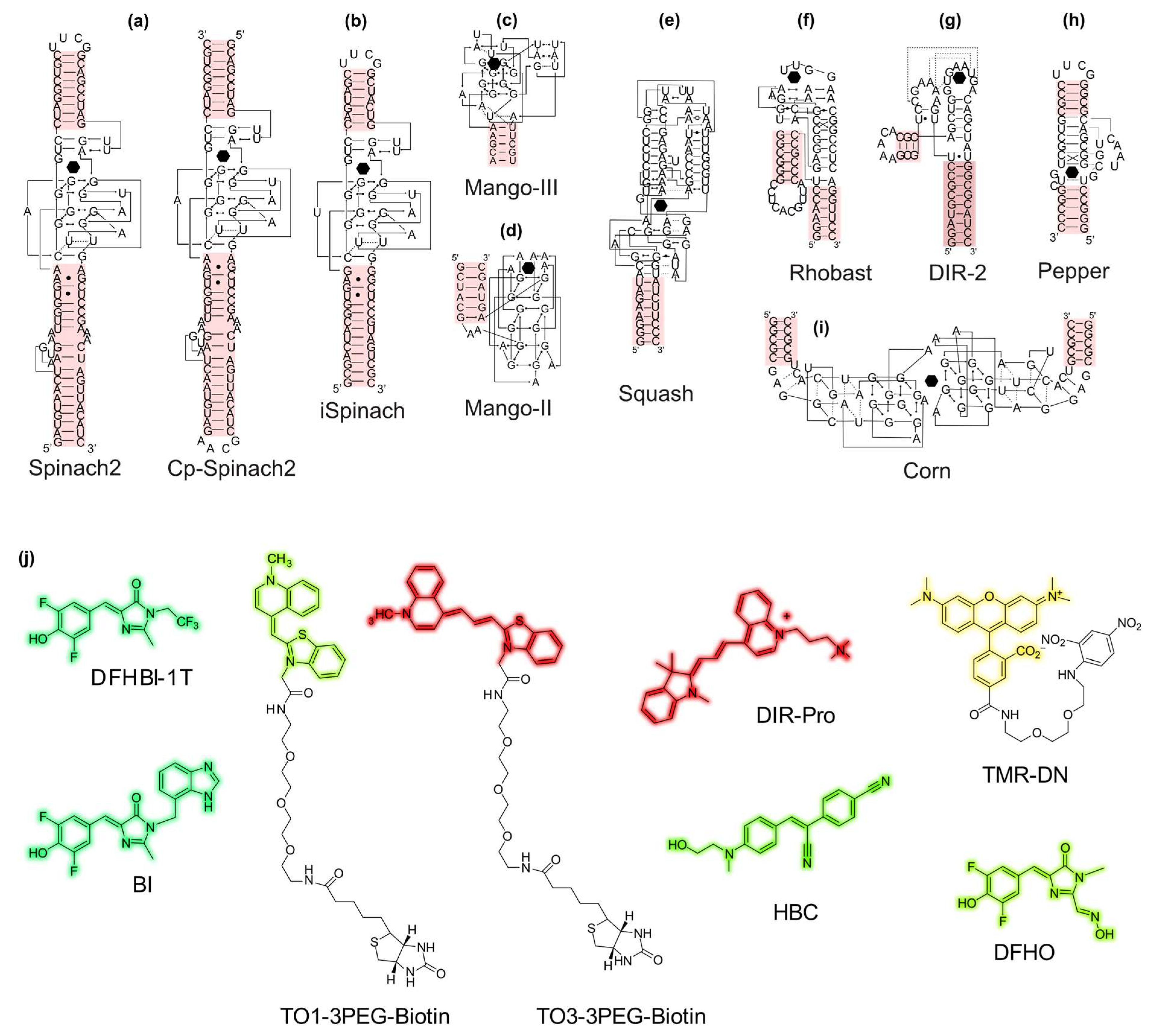


| Aptamers | Fluorogen | Complex | Ref. | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Name | G4 | #P a | Name | Ex/Em | KD | Turn-On | Brightness c | PDB | D/S d |
| BeCA | no | 2 | BC-6 | 478/686 | 230 nM | 51 | n.d. | n.d. | [35] |
| Beetroot | yes | 1/2 b | DFAME | 514/619 | 460 nM | 40 | 3825 | 8EYU | [36,37] |
| Broccoli | yes | 2 | DFHBI-1T | 470/505 | 305 nM | 800 | 27,800 | n.d. | [38] |
| BI | 470/505 | 51 nM | 1100 | 22,500 | n.d. | [39] | |||
| Chili | yes | 2 | DMHBI+ | 413/542 | 63 nM | n.d. | 8400 | 7OAW | [40,41] |
| DMHBO+ | 456/592 | 12 nM | n.d. | 2100 | 7OAX | ||||
| Corn | yes | 1/2 b | DFHO | 505/545 | 70 nM | 417 | 7250 | 5BJO | [42,43] |
| DIR-2 | no | 4 | DIR-Pro | 600/658 | 252 nM | 20 | 54,000 | 6DB8 | [44,45] |
| DNB | n.d. | 2 | TMR-DN | 555/582 | 350 nM | 73 | 42,430 | n.d. | [46] |
| SR-DN | 572/591 | 800 nM | 56 | 49,240 | n.d. | ||||
| iSpinach | yes | 2 | DFHBI | 442/503 | 920 nM | n.d. | n.d. | 5OB3 | [47,48] |
| Mango | yes | 1 | TO1-B | 510/535 | 4 nM | 1300 | 11,000 | 5V3F | [49,50] |
| Mango-II | yes | 1 | TO1-B | 510/535 | 0.7 nM | 1650 | 17,000 | 6C63 | [51,52] |
| Mango-III | yes | 1 | TO1-B | 510/535 | 5.6 nM | 4400 | 43,000 | 6E8S | [51,53] |
| MGA | no | 2 | MG | 350/630 | 117 nM | 2360 | 28,000 | 1F1T | [54,55] |
| o-Coral | no | 5 | Gemini-561 | 580/596 | 73 nM | 13 | 82,000 | n.d. | [56] |
| Okra | n.d. | 2 | ACE | 468/505 | 1.2 nM | 664 | 22,320 | n.d. | [57] |
| Peach | yes | 1 | TO3-B | 637/658 | 19.6 nM | 50 | 17,800 | n.d. | [58] |
| Pepper | no | 2 | HBC530 | 485/530 | 3.5 nM | 3595 | 43,098 | 7EOK | [28,59] |
| HBC620 | 577/620 | 6.1 nM | 12,600 | ||||||
| RhoBAST | no | 4 | TMR-DN | 564/590 | 15.1 nM | 26 | 55,000 | 8JY0 | [60,61] |
| Riboglow | no | 4 | Cbl-5xPEG-Atto590 | 594/622 | 34 nM | 5 | 37,200 | n.d. | [26] |
| Cbl-Cy5 | 646/662 | 3 nM | 2.7 | 65,750 | |||||
| SiRA | no | 3 | SiR-PEG3-NH2 | 649/662 | 430 nM | 7 | 84,280 | n.d. | [62] |
| Spinach | yes | 2 | DFHBI | 452/496 | 562 nM | 2000 | 17,500 | 4KZD | [63,64] |
| Spinach2 | yes | 2 | DFHBI | 454/498 | 430 nM | 770 | 29,100 | n.d. | [65] |
| Squash | no | 3 | DFHO | 505/545 | 53 nM | 550 | 14,760 | 7KVU | [66,67] |
| Target | Light-Up 1 (Fluorogen) | Light-Up 2 (Fluorogen) a | Sensing Aptamer Type | Functional Connection Type | Connection Design | Applications | Ref. |
|---|---|---|---|---|---|---|---|
| Adenine | Spinach (DFHBI) | Purine riboswitch | Strand displacement | Trial-Error | Proof-of-concept | [102] | |
| DNB (SR-DN) | Broccoli (DFHBI-1T) | Purine riboswitch | Allosteric | Trial-Error | Live-cell ratiometric imaging | [109] | |
| Spinach (DFHBI) | Synthetic | Allosteric | Trial-Error | Proof-of-concept | [110] | ||
| Adenosine 5-diphosphate (ADP) | Spinach (DFHBI) | Synthetic | Allosteric | Trial-Error | Live-cell imaging | [110] | |
| BFR (Hoechst 1C) | Synthetic | Strand displacement | Trial-Error | Proof-of-concept | [111] | ||
| Adenosine triphosphate (ATP) | MGA (MG) | Synthetic | Allosteric | Trial-Error | Proof-of-concept | [112] | |
| Ag2+ | Pepper (HBC530) | CC mismatch | Allosteric | Rational design | Live-cell entry imaging | [76] | |
| Broccoli (DFHBI-1T) | CC mismatch | Allosteric | Rational design | Live-cell entry imaging | [77] | ||
| Cyclic AMP (cAMP) | Broccoli (DFHBI-1T) | Synthetic | Ribozyme | Trial-Error | Proof-of-concept | [106] | |
| Cyclic AMP-GMP (3′-5′ cGAMP) | Spinach (DFHBI) | GEMM-Ib | Allosteric | Trial-Error | Live-cell imaging and riboswitch discovery | [113] | |
| Cyclic AMP-GMP (c-AMP-GMP) | Spinach (DFHBI) | G20A GEMM-I | Allosteric | Trial-Error | Live-cell imaging and in vitro enzymatic assay | [114] | |
| Cyclic di-AMP (cdiA) | Spinach (DFHBI-1T) | yuaA | Allosteric | Trial-Error | Live-cell imaging | [115] | |
| Spinach2 (DFHBI) | yuaA | Allosteric | Trial-Error | Live-cell imaging and cytometry profiling | [116] | ||
| Cyclic di-GMP (c-di-GMP) | Spinach (DFHBI) | Vc2 | Allosteric | Trial-Error | Proof-of-concept | [117] | |
| Spinach (DFHBI) | GEMM-I | Allosteric | Trial-Error | Live-cell imaging | [114] | ||
| DNB (SR-DN) | Broccoli (DFHBI-1T) | GEMM-I | Allosteric | Trial-Error | Live-cell ratiometric imaging | [109] | |
| Spinach2 (DFHBI/DFHBI-1T) | GEMM-I | Allosteric | Trial-Error | Live-cell imaging under aerobic and anaerobic conditions | [118] | ||
| Broccoli (DFHBI-1T) | GEMM-I | Ribozyme | Trial-Error | Proof-of-concept | [106] | ||
| FMN | MGA (MG) | Synthetic | Allosteric | Trial-Error | Proof-of-concept | [112] | |
| Glycine | Spinach (DFHBI-1T) | Tandem glycine aptamer (B. subtillis) | Strand displacement | Screening | Thermodynamic characterization of a riboswitch | [119] | |
| Guanine | Spinach (DFHBI) | Synthetic | Allosteric | Trial-Error | Proof-of-concept | [110] | |
| Spinach (DFHBI) | Purine riboswitch | Strand Displacement | Trial-Error | Live-cell imaging | [102] | ||
| Broccoli (DFHBI-1T) | Purine riboswitch | Allosteric | Trial-Error | Proof-of-concept | [120] | ||
| Pepper (HBC) | Purine riboswitch | Allosteric | Trial-Error | Live-cell entry imaging | [76] | ||
| Guanosine-5′-triphosphate (GTP) | Spinach (DFHBI) | Synthetic | Allosteric | Trial-Error | Proof-of-concept | [110] | |
| L-Dopa | Broccoli (DFHBI) | Synthetic | Allosteric | Screening | Proof-of-concept | [121] | |
| L-Dopa/Dopamine | Broccoli (DFHBI-1T) | Synthetic | Allosteric | Trial-Error | Proof-of-concept post SELEX screening of aptamer | [84] | |
| Nickel (Ni2+), Cobalt (Co2+), Iron (Fe2+) | Spinach2 (DFHBI-1T) | NiCo, czcD | Allosteric | Trial-Error | Determine riboswitch ligand specificity | [122] | |
| Phenylalanine | Spinach2 (DFHBI or DFHBI-1T) | Synthetic | Allosteric | Trial-Error | Screening of secreting microbes | [123] | |
| ppGpp | Broccoli (DFHBI-1T) | (p)ppGpp riboswitch | Allosteric | Trial-Error | Live-cell imaging | [124] | |
| Broccoli (DFHBI-1T) | (p)ppGpp riboswitch | Allosteric | Trial-Error | Paper-based biosensing | [125] | ||
| S-adenosyl-homocysteine (SAH) | Spinach2 (DFHBI) | SAH riboswitch | Allosteric | Trial-Error | Live-cell imaging/In vitro methyltransferase assay | [126] | |
| S-adenosylmethionine (SAM) | Spinach (DFHBI) | SAM-I riboswitch | Strand displacement | Trial-Error | Live-cell imaging | [102] | |
| Spinach (DFHBI) | Synthetic | Allosteric | Trial-Error | Live-cell imaging | [110] | ||
| Corn (DFHO) | SAM-III circular permuted | Allosteric | Trial-Error | Live-cell imaging and drug effect monitoring | [127] | ||
| Red Broccoli (OBI) | SAM riboswitch | Allosteric | Trial-Error | Live-cell imaging | [128] | ||
| Pepper (HBC530 or HBC620) | SAM-III riboswitch | Allosteric | Trial-Error | Live-cell imaging and drug effect monitoring | [129] | ||
| Spinach (DFHBI-1T) | SAM-III riboswitch | Allosteric | Screening | Proof-of-concept | [72] | ||
| Cladogenetic B2 (Isa-5a) Cladogenetic G2 (DFHBI-1T) | SAM-III riboswitch | Allosteric | Screening from [72] | Live-cell imaging | [130] | ||
| Squash (DFHO) | Broccoli (BI) | SAM-III riboswitch | Allosteric | Selection | Accurate detection and drug effect monitoring | [66] | |
| Pepper (HBC530) | RhoBAST (TMR-DN) | SAM-III riboswitch | Allosteric | Trial-Error | Accurate detection and drug effect monitoring | [76] | |
| Corn (DFHO) | SAM-III riboswitch | Allosteric | Trial-Error | Live-cell imaging and drug effect monitoring | [120] | ||
| Spinach (DFHBI) | SAM-I riboswitch | Strand Displacement | Trial-Error | Live-cell imaging | [102] | ||
| Spinach (DFHBI-1T) | Mango (YO-3) | SAM-III riboswitch | FRET | Rational design | Proof-of-concept | [131] | |
| Spinach (DFHBI-1T) | SAM-III riboswitch | Allosteric | Screening | Accurate detection and drug effect monitoring | [72] | ||
| Tetracycline | DNB (SR-DN) | Broccoli (DFHBI-1T) | Synthetic | Allosteric | Trial-Error | Live-cell ratiometric imaging | [109] |
| Pepper (HBC530) | Synthetic | Allosteric | Trial-Error | Live-cell entry imaging | [76] | ||
| Brocolli (DFHBI-1T) | Synthetic | Allosteric | Trial-Error | Paper-based biosensing | [125] | ||
| Theophylline | MGA (MG) | Synthetic | Allosteric | Trial-Error | Proof-of-concept | [112] | |
| iSpinach (DFHBI-1T) | Synthetic | Allosteric | Screening | Proof-of-concept | [100] | ||
| Cladogenetic B2 (Isa-5a) Cladogenetic G2 (DFHBI-1T) | Synthetic | Allosteric | Screening from [72] | Live-cell imaging | [130] | ||
| Broccoli (DFHBI-1T) | Synthetic | Ribozyme | Trial-Error | Live-cell imaging and cytometry profiling | [106] | ||
| MGA (MG) | Synthetic | Kissing | Rational design | Proof-of-concept | [105] | ||
| TPP | Spinach (DFHBI) | TPP riboswitch | Strand Displacement | Trial-Error | Live-cell imaging and drug screening | [102] | |
| Broccoli (DFHBI-1T) | TPP riboswitch | Ribozyme | Trial-Error | Proof-of-concept | [106] | ||
| Broccoli (DFHBI-1T) Red Broccoli (OBI) | Synthetic | Strand displacement | Selection | Proof-of-concept | [90] | ||
| Tryptophane | Spinach2 (DFHBI or DFHBI-1T) | Synthetic | Allosteric | Trial-Error | Screening of secreting microbes | [123] | |
| Tyrosine | Broccoli or Spinach2 (DFHBI-1T) | Synthetic | Allosteric | Trial-Error | Proof-of-concept | [132] | |
| Spinach2 (DFHBI or DFHBI-1T) | Synthetic | Allosteric | Trial-Error | Screening of secreting microbes | [123] | ||
| 2′,3′-cGAMP | Spinach2 (DFHBI-1T) | G103A GEMM-II | Allosteric | Trial-Error | Live-cell imaging and characterization of cGAS-cGAMP-STING pathway | [83] | |
| 5-HTP | Broccolli (DFHBI-1T) | Synthetic | Strand displacement | Trial-Error | Proof-of-concept | [84] | |
| 5-HTP/Serotonine | Broccoli (DFHBI-1T) | Synthetic | Allosteric | Trial-Error | Live-cell imaging | [120] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kehrli, J.; Husser, C.; Ryckelynck, M. Fluorogenic RNA-Based Biosensors of Small Molecules: Current Developments, Uses, and Perspectives. Biosensors 2024, 14, 376. https://doi.org/10.3390/bios14080376
Kehrli J, Husser C, Ryckelynck M. Fluorogenic RNA-Based Biosensors of Small Molecules: Current Developments, Uses, and Perspectives. Biosensors. 2024; 14(8):376. https://doi.org/10.3390/bios14080376
Chicago/Turabian StyleKehrli, Janine, Claire Husser, and Michael Ryckelynck. 2024. "Fluorogenic RNA-Based Biosensors of Small Molecules: Current Developments, Uses, and Perspectives" Biosensors 14, no. 8: 376. https://doi.org/10.3390/bios14080376
APA StyleKehrli, J., Husser, C., & Ryckelynck, M. (2024). Fluorogenic RNA-Based Biosensors of Small Molecules: Current Developments, Uses, and Perspectives. Biosensors, 14(8), 376. https://doi.org/10.3390/bios14080376





