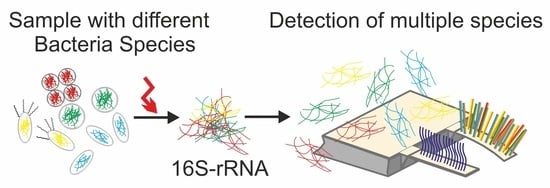
Ultra-Sensitive Biosensors for Medical Applications Based on Nanomechanics: From Detection of Synthetic Biomolecules to Analysis of Sepsis in Pediatric Patients
Abstract
:1. Introduction
1.1. Hybridization of Synthetic Oligonucleotides
1.2. Antigen Detection with Antibodies
1.3. Protein Conformation Changes
1.4. Cancer Diagnostics
1.5. Sepsis Diagnostics
2. Materials and Methods
2.1. RNA Preparation from Blood
2.2. Biosensor Preparation
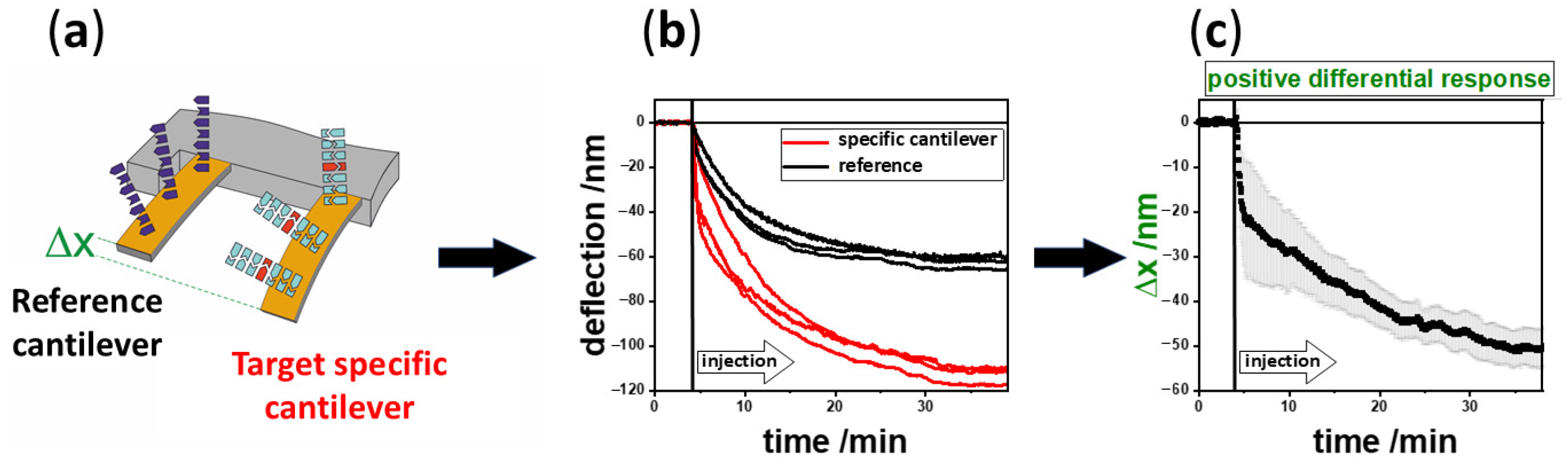
2.3. Experimental Procedure
3. Results
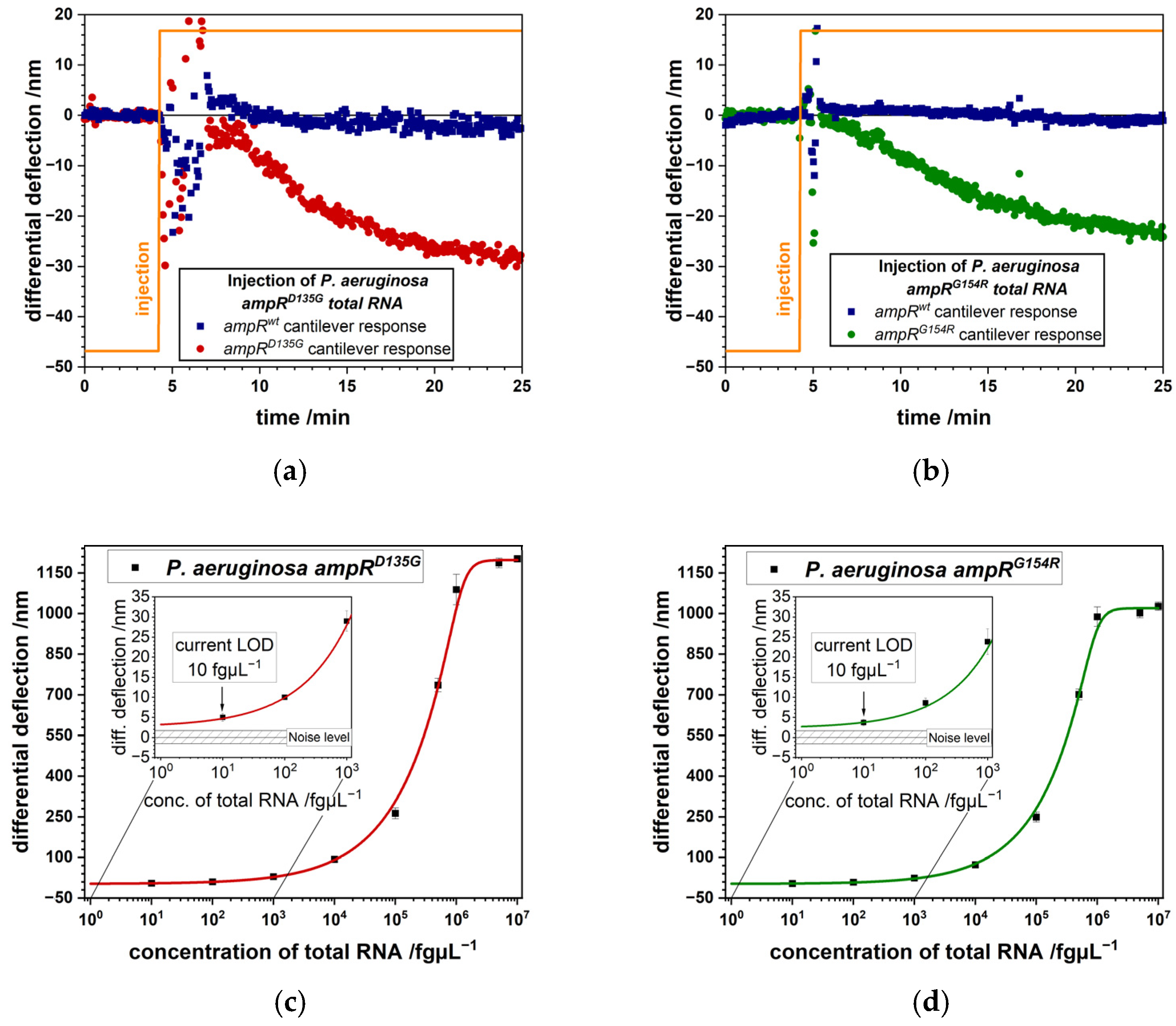
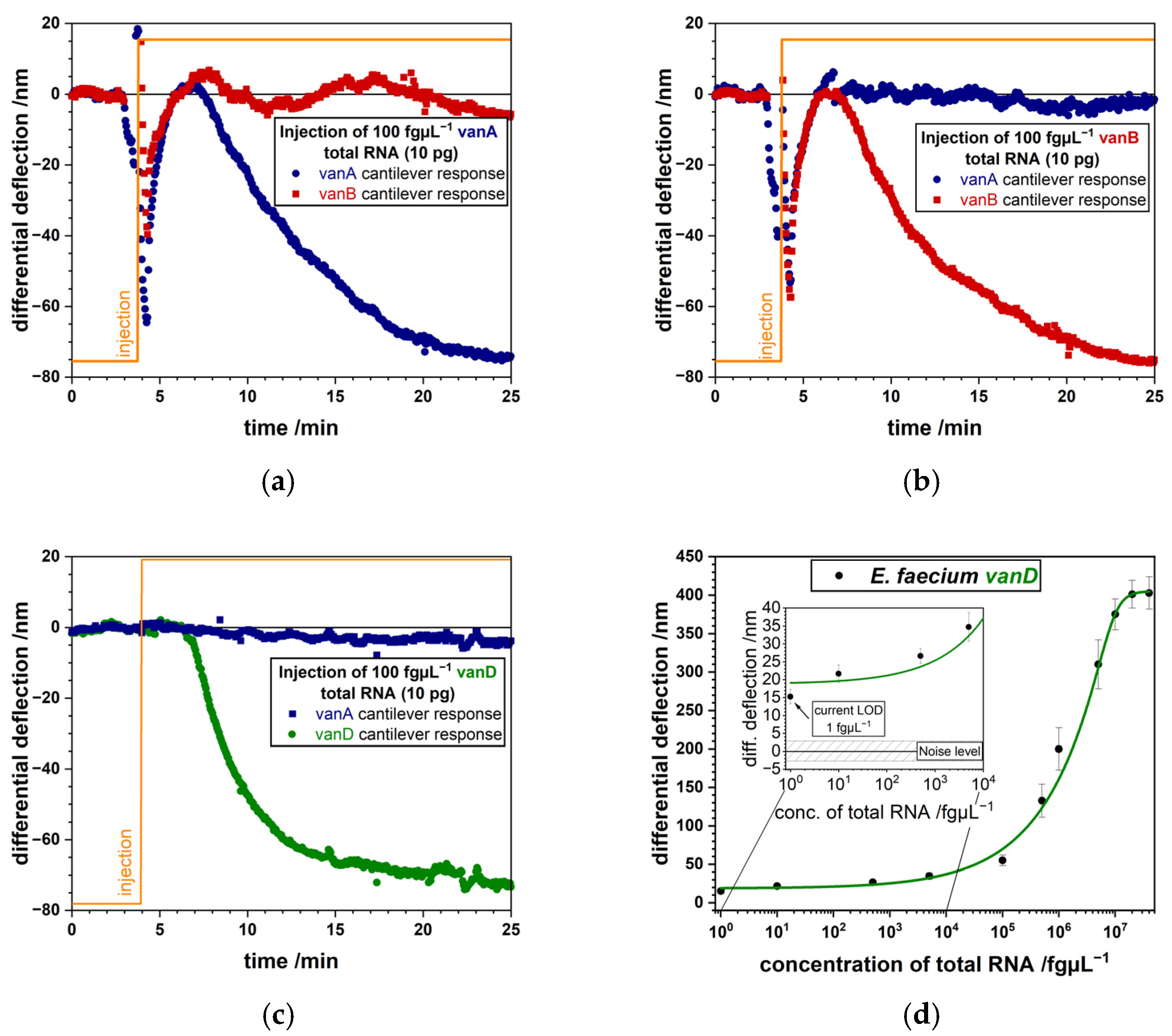
3.1. Detection of Bacterial Pathogens
3.2. Comparison of Different Age Groups
3.3. Extended Study with 233 Total RNA Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bietsch, A.; Zhang, J.Y.; Hegner, M.; Lang, H.P.; Gerber, C. Rapid functionalization of cantilever array sensors by inkjet printing. Nanotechnology 2004, 15, 873–880. [Google Scholar] [CrossRef]
- Lang, H.P.; Hegner, M.; Gerber, C. Cantilever array sensors. Mater Today 2005, 8, 30–36. [Google Scholar] [CrossRef]
- Steel, A.B.; Levicky, R.L.; Herne, T.M.; Tarlov, M.J. Immobilization of Nucleic Acids at Solid Surfaces: Effect of Oligonucleotide Length on Layer Assembly. Biophys. J. 2000, 79, 975–981. [Google Scholar] [CrossRef]
- Shih, Y.; Chen, C.; Wu, K. First-Principles Surface Stress Calculations and Multiscale Deformation Analysis of a Self-Assembled Monolayer Adsorbed on a Micro-Cantilever. Sensors 2014, 14, 7435–7450. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.A.; Sun, Z.H.; Cheng, Q.; Wang, X.X. Molecular mechanics modelling and simulation of the adsorption-induced surface stress in micro-nano-cantilever sensors. J. Phys. Conf. Ser. 2007, 61, 1266–1270. [Google Scholar] [CrossRef]
- Bennett, I.; Pyne, A.L.B.; McKendry, R.A. Cantilever Sensors for Rapid Optical Antimicrobial Sensitivity Testing. ACS Sens. 2020, 5, 3133–3139. [Google Scholar] [CrossRef]
- Zhao, Y.; Gosai, A.; Kang, K.; Shrotriya, P. Multiscale Modeling Reveals the Cause of Surface Stress Change on Microcantilevers Due to Alkanethiol SAM Adsorption. J. Chem. Inf. Model. 2020, 60, 2998–3008. [Google Scholar] [CrossRef]
- Fritz, J.; Baller, M.K.; Lang, H.P.; Rothuizen, H.; Vettiger, P.; Meyer, E.; Güntherodt, H.J.; Gerber, C.; Gimzewski, J.K. Translating Biomolecular Recognition into Nanomechanics. Science 2000, 288, 316–318. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Lang, H.P.; Yoshikawa, G.; Gerber, C. Optimization of DNA Hybridization Efficiency by pH-Driven Nanomechanical Bending. Langmuir 2012, 28, 6494–6501. [Google Scholar] [CrossRef]
- Huber, F.; Lang, H.P.; Heller, S.; Bielicki, J.A.; Gerber, C.; Meyer, E.; Egli, A. Rapid Bacteria Detection from Patients’ Blood Bypassing Classical Bacterial Culturing. Biosensors 2022, 12, 994. [Google Scholar] [CrossRef]
- Jelesarov, I.; Bosshard, H.R. Isothermal titration calorimetry and differential scanning calorimetry as complementary tools to investigate the energetics of biomolecular recognition. J. Mol. Recognit. 1999, 12, 3–18. [Google Scholar]
- Sibani Lisa Biswal, S.L.; Raorane, D.; Chaiken, A.; Birecki, H.; Majumdar, A. Nanomechanical Detection of DNA Melting on Microcantilever Surfaces. Anal. Chem. 2006, 78, 7104–7109. [Google Scholar] [CrossRef]
- McKendry, R.; Zhang, J.Y.; Arntz, Y.; Strunz, T.; Hegner, M.; Lang, H.P.; Baller, M.K.; Certa, U.; Meyer, E.; Güntherodt, H.J.; et al. Multiple label-free biodetection and quantitative DNA-binding assays on a nanomechanical cantilever array. Proc. Natl. Acad. Sci. USA 2002, 99, 9783–9788. [Google Scholar] [CrossRef] [PubMed]
- Huber, F.; Hegner, M.; Gerber, C.; Güntherodt, H.J.; Lang, H.P. Label free analysis of transcription factors using microcantilever arrays. Biosens. Bioelectron. 2006, 8, 1599–1605. [Google Scholar] [CrossRef]
- Bonnet, G.; Krichevsky, O.; Libchaber, A. Kinetics of conformational fluctuations in DNA hairpin-loops. Proc. Natl. Acad. Sci. USA 1998, 95, 8602–8606. [Google Scholar] [PubMed]
- Arntz, Y.; Seelig, J.D.; Lang, H.P.; Zhang, J.; Hunziker, P.; Ramseyer, J.P.; Meyer, E.; Hegner, M.; Gerber, C. Label-free protein assay based on a nanomechanical cantilever array. Nanotechnology 2003, 14, 86–90. [Google Scholar] [CrossRef]
- Grogan, C.; Raiteri, R.; O’Connor, G.M.; Glynn, T.J.; Cunningham, V.; Kane, M.; Charlton, M.; Leech, D. Characterisation of an antibody coated microcantilever as a potential immuno-based biosensor. Biosens. Bioelectron. 2002, 17, 201–207. [Google Scholar]
- Katarzyna, N.; Kapczynska, K.; Rybka, J.; Lipinski, T.; Grabiec, P.; Skowicki, M.; Gotszalk, T. Microcantilever array biosensors for detection and recognition of Gram-negative bacterial endotoxins. Sens. Actuators B Chem. 2014, 198, 114–124. [Google Scholar] [CrossRef]
- Backmann, N.; Zahnd, C.; Huber, F.; Bietsch, A.; Plückthun, A.; Lang, H.P.; Güntherodt, H.J.; Hegner, M.; Gerber, C. A label-free immunosensor array using single-chain antibody fragments. Proc. Natl. Acad. Sci. USA 2005, 102, 14587–14592. [Google Scholar] [CrossRef]
- Braun, T.; Backmann, N.; Vögtli, M.; Bietsch, A.; Engel, A.; Lang, H.P.; Gerber, C.; Hegner, M. Conformational Change of Bacteriorhodopsin Quantitatively Monitored by Microcantilever Sensors. Biophys. J. 2006, 90, 2970–2977. [Google Scholar] [CrossRef]
- Shelyakin, P.V.; Kovarskii, A.L.; Kasparov, V.V.; Fel’dman, T.B.; Ostrovskii, M.A. A Study of the Photoinduced Conformational Mobility of Spin-Labeled Regenerated Rhodopsin by ESR Spectroscopy. Russ. J. Phys. Chem. B 2012, 6, 694–698. [Google Scholar] [CrossRef]
- Etayash, H.; Jiang, K.; Azmi, S.; Thundat, T.; Kaur, K. Real-time Detection of Breast Cancer Cells Using Peptidefunctionalized Microcantilever Arrays. Sci. Rep. 2015, 5, 13967. [Google Scholar] [CrossRef]
- Li, C.; Ma, X.; Guan, Y.; Tang, J.; Zhang, B. Microcantilever Array Biosensor for Simultaneous Detection of Carcinoembryonic Antgens and α-Fetoprotein Based on Real-Time Monitoring of the Profile of Cantilever. ACS Sens. 2019, 4, 3034–3041. [Google Scholar] [CrossRef]
- Wang, J.; Wang, L.; Zhu, Y.; Zhang, J.; Liao, J.; Wang, S.; Yang, J.; Yang, F. A high accuracy cantilever array sensor for early liver cancer diagnosis. Biomed. Microdevices 2016, 18, 110. [Google Scholar] [CrossRef] [PubMed]
- Basua, A.K.; Basub, A.; Bhattacharyaa, S. Micro/Nano fabricated cantilever-based biosensor platform: A review and recent progress. Enzyme Microb. Technol. 2020, 139, 109558. [Google Scholar] [CrossRef] [PubMed]
- Ribas, A.; Flaherty, K.T. BRAF targeted therapy changes the treatment paradigm in melanoma. Nat. Rev. Clin. Oncol. 2011, 8, 426–433. [Google Scholar] [CrossRef]
- Huber, F.; Lang, H.P.; Backmann, N.; Rimoldi, D.; Gerber, C. Direct detection of a BRAF mutation in total RNA from melanoma cells using cantilever arrays. Nat. Nanotechnol. 2013, 8, 125–129. [Google Scholar]
- Huber, F.; Lang, H.P.; Glatz, K.; Rimoldi, D.; Meyer, E.; Gerber, C. Fast Diagnostics of BRAF Mutations in Biopsies from Malignant Melanoma. Nano Lett. 2016, 16, 5373–5377. [Google Scholar] [CrossRef]
- Rudd, K.E.; Johnson, S.C.; Agesa, K.M.; Shackelford, K.A.; Tsoi, D.; Kievlan, D.R.; Colombara, D.V.; Ikuta, K.S.; Kissoon, N.; Finfer, S.; et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet 2020, 395, 200–211. [Google Scholar] [CrossRef]
- Gorina, Y.; Kramarow, E.A. QuickStats: Sepsis-Related Death Rates Among Persons Aged ≥65 Years, by Age Group and Sex—National Vital Statistics System, United States. 2021. Available online: https://www.cdc.gov/mmwr/volumes/72/wr/mm7238a5.htm?s_cid=mm7238a5_w (accessed on 1 December 2024).
- Chun, K.; Syndergaard, C.; Damas, C.; Trubey, R.; Mukindaraj, A.; Qian, S.; Jin, X.; Breslow, S.; Niemz, A. Sepsis Pathogen Identification. J. Lab. Autom. 2015, 20, 539–561. [Google Scholar] [CrossRef]
- Cheng, M.P.; Stenstrom, R.; Paquette, K.; Stabler, S.N.; Akhter, M.; Davidson, A.C.; Gavric, M.; Lawandi, A.; Jinah, R.; Saeed, Z.; et al. Blood Culture Results Before and After Antimicrobial Administration in Patients with Severe Manifestations of Sepsis A Diagnostic Study. Ann. Intern. Med. 2019, 171, 547–555. [Google Scholar] [CrossRef]
- Behera, B.; Vishnu, G.K.A.; Chatterjee, S.; Sitaramgupta, V.S.N.V.; Sreekumar, N.; Nagabhushan, A.; Rajendran, N.; Prathik, B.H.; Pandya, H.J. Emerging technologies for antibiotic susceptibility testing. Biosens. Bioelectron. 2019, 142, 111552. [Google Scholar] [CrossRef]
- Ferran, P.V.; Villa, R.; Alvarez, M. Nanomechanical Sensors as a Tool for Bacteria Detection and Antibiotic Susceptibility Testing. Front. Mech. 2020, 6, 44. [Google Scholar] [CrossRef]
- Sturm, A.; Józwiak, G.; Verge, M.P.; Munch, L.; Cathomen, G.; Vocat, A.; Luraschi-Eggemann, A.; Orlando, C.; Fromm, K.; Delarze, E.; et al. Accurate and rapid antibiotic susceptibility testing using a machine learning-assisted nanomotion technology platform. Nat. Commun. 2024, 15, 2037. [Google Scholar] [CrossRef]
- Zhou, J.; Huang, J.B.; Huang, H.Q.; Zhao, C.; Zou, M.Q.; Liu, D.J.; Weng, X.Y.; Liu, L.W.; Qu, J.L.; Liu, L.; et al. Fiber-integrated cantilever-based nanomechanical biosensors as a tool for rapid antibiotic susceptibility testing. Biomed. Opt. Express 2023, 14, 1862–1873. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Xu, B.Z.; Zhu, Y.F.; Zhao, J.Y. Microcantilever sensors for biochemical detection. J. Semicond. 2023, 44, 023105. [Google Scholar] [CrossRef]
- Hansen, K.M.; Ji, H.F.; Wu, G.; Datar, R.; Cote, R.; Majumdar, A.; Thundat, T. Cantilever-Based Optical Deflection Assay for Discrimination of DNA Single-Nucleotide Mismatches. Anal. Chem. 2001, 73, 1567–1571. [Google Scholar] [CrossRef]
- Huber, F.; Lang, H.P.; Lang, D.; Wüthrich, D.; Hinić, V.; Gerber, C.; Egli, A.; Meyer, E. Rapid and Ultrasensitive Detection of Mutations and Genes Relevant to Antimicrobial Resistance in Bacteria. Glob. Chall. 2021, 5, 2000066. [Google Scholar] [CrossRef] [PubMed]
- Wafula Ndieyira, J.W.; Watari, M.; Barrera, A.D.; Zhou, D.; Vögtli, M.; Batchelor, M.; Cooper, M.A.; Strunz, T.; Horton, M.A.; Abell, C.; et al. Nanomechanical detection of antibiotic–mucopeptide binding in a model for superbug drug resistance. Nat. Nanotechnol. 2008, 3, 691–696. [Google Scholar] [CrossRef]
- Nakano, R.; Nakano, A.; Yano, H.; Okamoto, R. Role of AmpR in the High Expression of the Plasmid-Encoded AmpC β-Lactamase CFE-1. mSphere 2017, 2, e00192-17. [Google Scholar] [CrossRef]
- Stachowiak, J.C.; Yue, M.; Castelino, K.; Chakraborty, A.; Majumdar, A. Chemomechanics of Surface Stresses Induced by DNA Hybridization. Langmuir 2006, 22, 263–268. [Google Scholar]
- Kirn, T.J.; Weinstein, M.P. Update on blood cultures: How to obtain, process, report, and interpret. Clin. Microbiol. Infect. 2013, 19, 513–520. [Google Scholar] [PubMed]
- Liu, W.L.; Li, L.; Khan, M.A.; Zhu, F.Z. Popular molecular markers in bacteria. Mol. Gen. Microbiol. Virol. 2012, 27, 103–107. [Google Scholar]
- Zhang, J.; Lang, H.P.; Huber, F.; Bietsch, A.; Grange, W.; Certa, U.; McKendry, R.; Güntgerodt, H.J.; Hegner, M.; Gerber, C. Rapid and label-free nanomechanical detection of biomarker transcripts in human RNA. Nat. Nanotechnol. 2006, 1, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Beaulieu, L.Y.; Godin, M.; Laroche, O.; Tabard-Cossa, V.; Grütter, P. A complete analysis of the laser beam deflection systems used in cantilever-based systems. Ultramicroscopy 2007, 107, 422–430. [Google Scholar] [CrossRef]
- Huen, K.; Lizarraga, D.; Kogut, K.; Eskenazi, B.; Holland, N. Age-Related Differences in miRNA Expression in Mexican-American Newborns and Children. Int. J. Environ. Res. Public Health 2019, 4, 524. [Google Scholar] [CrossRef]
- Trevethan, R. Sensitivity, Specificity, and Predictive Values: Foundations, Pliabilities, and Pitfalls in research and Practice. Front. Public Health 2017, 5, 307. [Google Scholar] [CrossRef]
- Calderaro, A.; Chezzi, C. MALDI-TOF MS: A Reliable Tool in the Real Life of the Clinical Microbiology. Microorganisms 2024, 12, 322. [Google Scholar] [CrossRef]
- Viboud, G.; Asaro, H.; Huang, M.B. Use of matrix-assisted laser desorption ionization time of flight (MALDI-TOF) to detect antibiotic resistance in bacteria. Am. J. Clin. Pathol. 2024, 161, 317–328. [Google Scholar] [CrossRef]
- López-Cortés, X.A.; Manríquez-Troncoso, J.M.; Kandalaft-Letelier, J.; Cuadros-Orellana, S. Machine learning and matrix-assisted laser desorption/ionization time-of-flight mass spectra for antimicrobial resistance prediction: A systematic review of recent advancements and future development. J. Chromatogr. A 2024, 1734, 465262. [Google Scholar] [CrossRef]
- Gant, M.S.; Chamot-Rooke, J. Present and future perspectives on mass spectrometry for clinical microbiology. Microbes Infect. 2024, 26, 105296. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhou, M.; Li, X.; Fan, Y.; Li, J.; Lu, K.; Wen, H.; Ren, J. Recent advances of fluorescent sensors for bacteria detection-A review. Talanta 2023, 254, 124133. [Google Scholar] [CrossRef]
- Liu, S.; Lu, F.; Chen, S.; Ning, Y. Graphene oxide-based fluorescent biosensors for pathogenic bacteria detection: A review. Anal. Chim. Acta 2025, 1337, 343428. [Google Scholar] [CrossRef]
- Kang, J.; Yeom, G.; Jang, H.; Park, C.J.; Kim, M.G. Highly sensitive and universaldetection strategy based on a colorimetric assay using target-specific hetero-geneous sandwich DNA aptamer. Anal. Chim. Acta 2020, 1123, 73–80. [Google Scholar]
- Liu, Y.; Zhou, H.; Hu, Z.; Yu, G.; Yang, D.; Zhao, J. Label and label-free based surface-enhanced Raman scattering for pathogen bacteria detection: A review. Biosens. Bioelectron. 2017, 94, 131–140. [Google Scholar] [CrossRef]
- Lopez-Tellez, J.; Ramirez-Montez, S.; Ferreira, T.A.; Santos, E.M.; Rodriguez, J.A. Application of Voltammetric Sensors for Pathogen Bacteria Detection: A Review. Chemosensors 2022, 10, 424. [Google Scholar] [CrossRef]
- Templier, V.; Roux, A.; Roupioz, Y.; Livache, T. Ligands for label-free detection of whole bacteria on biosensors: A review. Trends Analyt. Chem. 2016, 79, 71–79. [Google Scholar] [CrossRef]
- Paczesny, J.; Richter, L.; Hołyst, R. Recent Progress in the Detection of Bacteria Using Bacteriophages: A Review. Viruses 2020, 12, 845. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.-Y.; Rizaev, J.A.; Pallathadka, H.; Mansouri, S.; Bokov, D.O.; Sharma, S.; Rathore, G.; Rajput, P.; Mustafa, Y.F.; Abosaoda, M.K. A review of new emergoiing biosensors based on bacteria-imprinted polymers towards pathogenic bacteria: Promising new tools for selective detection. Microchem. J. 2024, 207, 111918. [Google Scholar] [CrossRef]
- Khaleque, A.; Hossain, S.I.; Ali, R.; Aly, M.A.S.; Khan, Z.H. Bioreceptor modified electrochemical biosensors for the detection of life threating pathogenic bacteria: A review. RSC Adv. 2024, 14, 28487. [Google Scholar] [CrossRef]
- Feng, R.-M.; Liu, Y.; Liu, Z.-Q.; Wang, L.; Chen, N.; Zhao, Y.; Yi, H.-W. Advances in nucleic acid aptamer-based detection of respiratory virus and bacteria: A mini review. Virol. J. 2024, 21, 237. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Li, Y.; Chen, P.; Zhong, S.; Yang, Y. Nucleic Acid Aptamer-Based Sensors for Bacteria Detection: A Review. BioEssays 2025, 47, e202400111. [Google Scholar] [CrossRef]
- Zhao, Y.; Qian, Y.; Qi Huang, Q.; Hu, X.; Gu, W.; Xing, H. Colourimetric and SERS dual-mode aptasensor using Au@Ag and magnetic nanoparticles for the detection of Campylobacter jejuni. Talanta 2024, 270, 125585. [Google Scholar] [CrossRef] [PubMed]



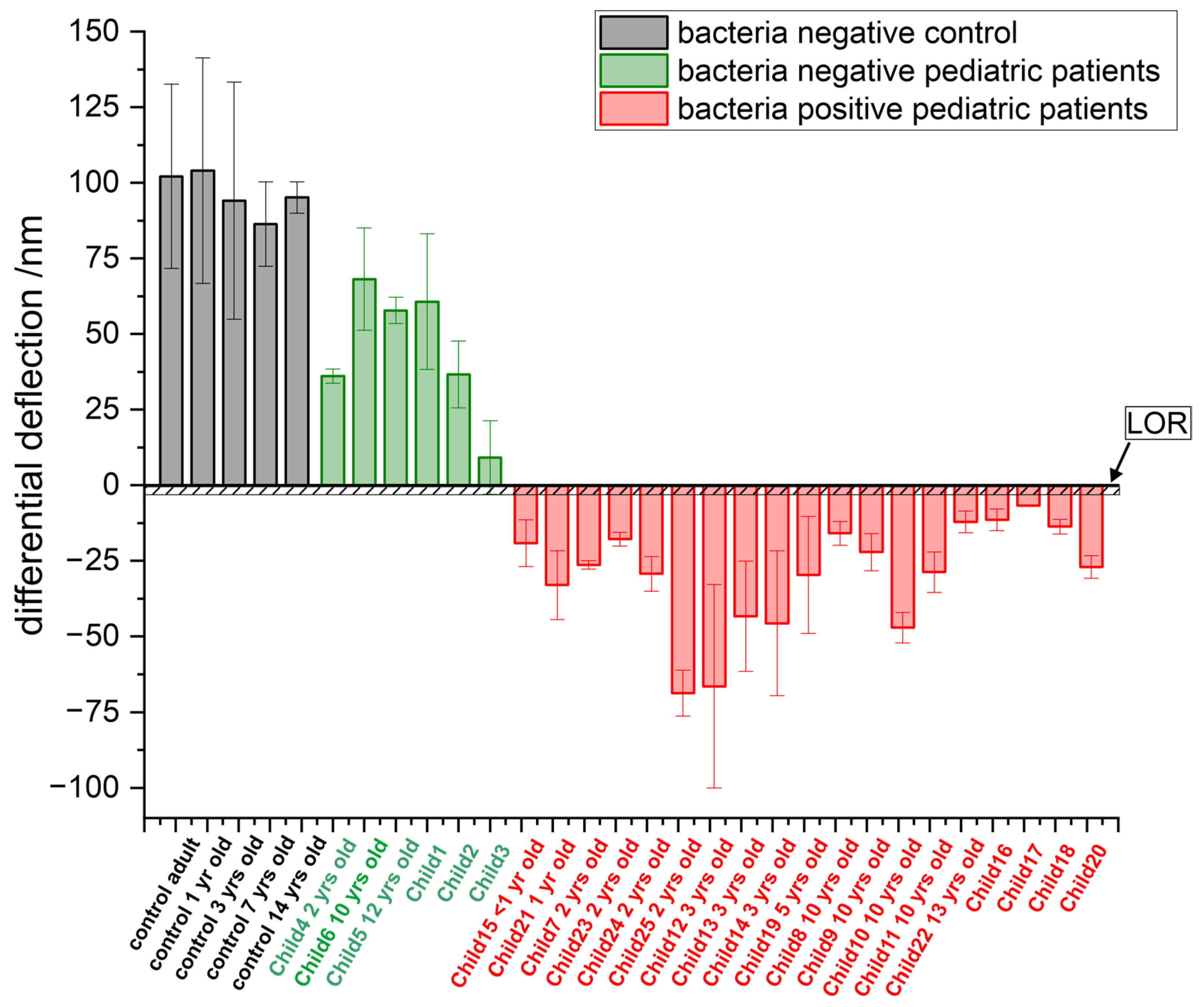
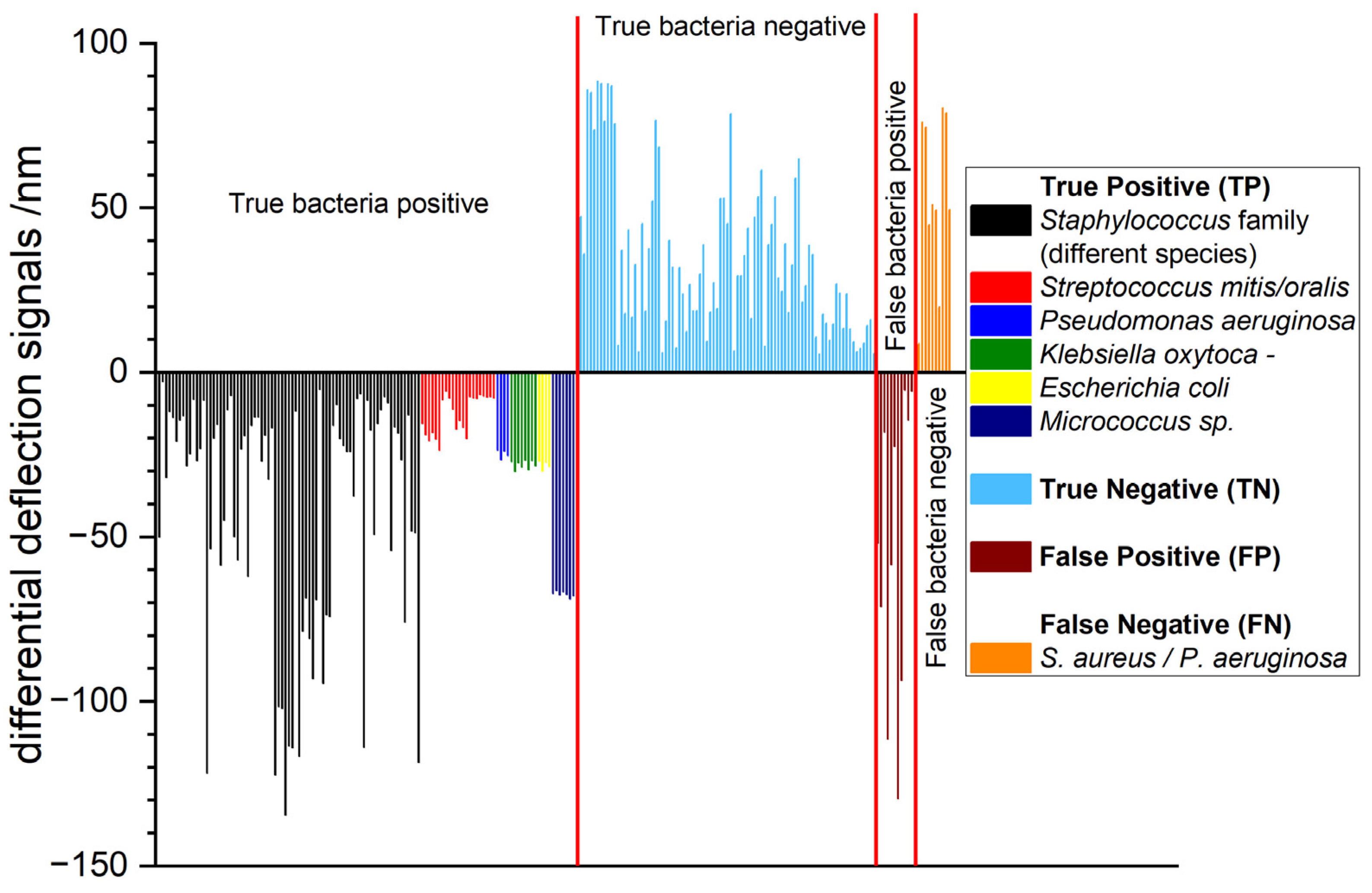
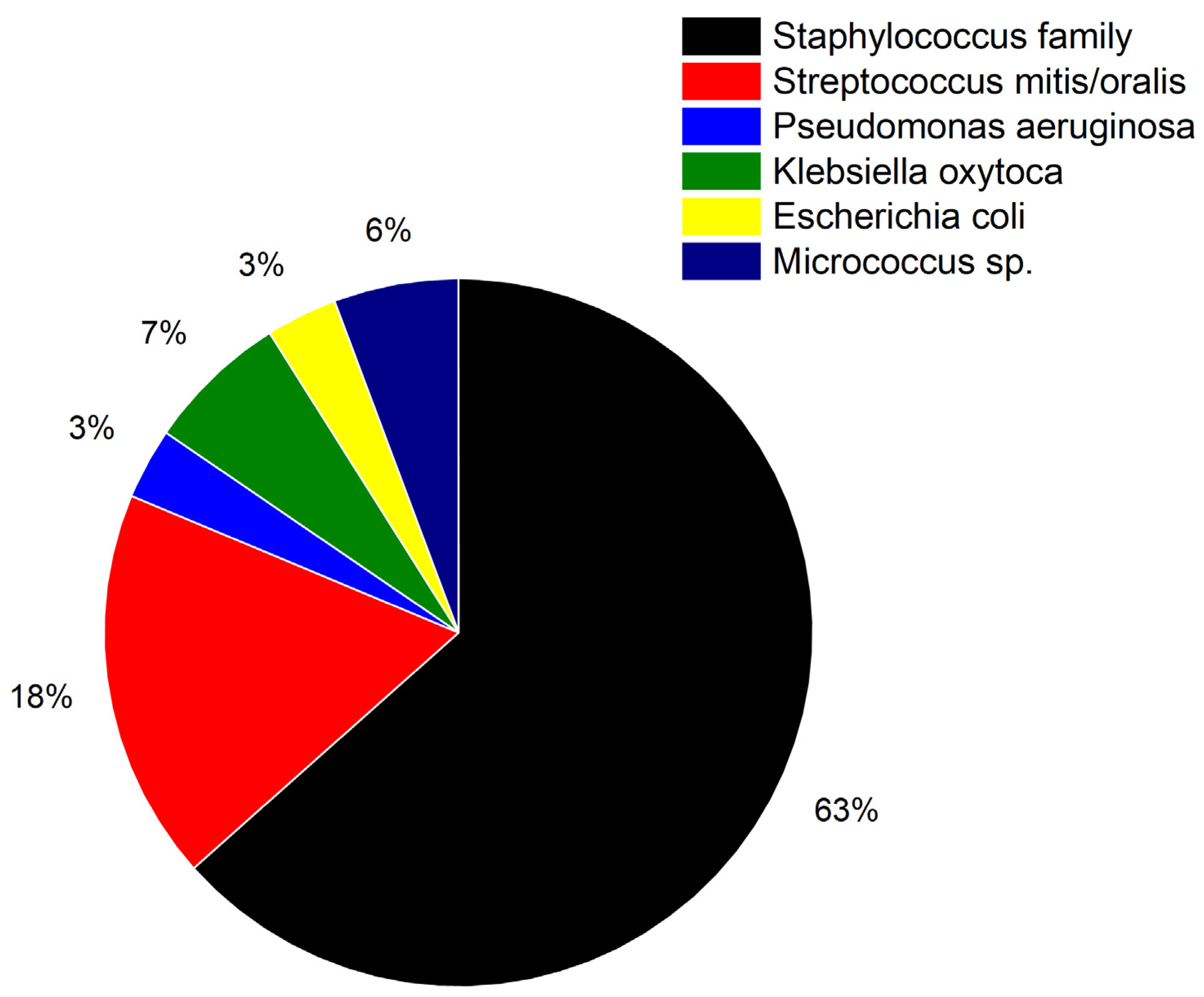
| Probe | Sequence | Use |
|---|---|---|
| 16S-rRNA | 5′GGACTACCAGGGTATCTAAT3‘ | Bacteria detection using 16S-rRNA [44]. |
| polyAC | 5′ACACACACACACACACACAC3’ | Reference sequence for non-specific binding [45]. |
| Assay | SE | SP | PPV | NPV | Diagnostic Time |
|---|---|---|---|---|---|
| Nanomechanical Sensors | 93% | 88% | 92% | 89% | 1 h |
| ELISA | 82% | 73% | 65% | 88% | 24 h |
| RT-PCR | 77% | 88% | 69% | 91% | 6 h |
| Culture | 60% | 100% | 100% | 62% | Up to 3 days |
| Assay | Limit of Detection (LOD) |
|---|---|
| Nanomechanical Sensors [10] | 5 CFU/mL |
| Immunomagnetic quantum dots | 1.0 × 103 CFU/mL |
| ELISA | 2.1 × 104 CFU/mL |
| qPCR | 125 CFU/mL |
| QCM sensor | 150 CFU/mL |
| Colorimetric aptasensor | 100 CFU/mL |
| Electrochemical impedimetric immunosensor | 1.0 × 103 CFU/mL |
| Colorimetric and SERS immunochromatographic assay | 50 CFU/mL |
| Dual-mode aptasensor | 6 CFU/mL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huber, F.; Lang, H.P.; Marten, A.; Bielicki, J.A.; Meyer, E.; Gerber, C. Ultra-Sensitive Biosensors for Medical Applications Based on Nanomechanics: From Detection of Synthetic Biomolecules to Analysis of Sepsis in Pediatric Patients. Biosensors 2025, 15, 217. https://doi.org/10.3390/bios15040217
Huber F, Lang HP, Marten A, Bielicki JA, Meyer E, Gerber C. Ultra-Sensitive Biosensors for Medical Applications Based on Nanomechanics: From Detection of Synthetic Biomolecules to Analysis of Sepsis in Pediatric Patients. Biosensors. 2025; 15(4):217. https://doi.org/10.3390/bios15040217
Chicago/Turabian StyleHuber, François, Hans Peter Lang, Andrea Marten, Julia Anna Bielicki, Ernst Meyer, and Christoph Gerber. 2025. "Ultra-Sensitive Biosensors for Medical Applications Based on Nanomechanics: From Detection of Synthetic Biomolecules to Analysis of Sepsis in Pediatric Patients" Biosensors 15, no. 4: 217. https://doi.org/10.3390/bios15040217
APA StyleHuber, F., Lang, H. P., Marten, A., Bielicki, J. A., Meyer, E., & Gerber, C. (2025). Ultra-Sensitive Biosensors for Medical Applications Based on Nanomechanics: From Detection of Synthetic Biomolecules to Analysis of Sepsis in Pediatric Patients. Biosensors, 15(4), 217. https://doi.org/10.3390/bios15040217