Advancements in Circulating Tumor Cell Detection for Early Cancer Diagnosis: An Integration of Machine Learning Algorithms with Microfluidic Technologies
Abstract
:1. Introduction
2. Microfluidic Technology and Its Significance in CTCs
2.1. Fundamentals of Microfluidic Technology
2.2. Applications of Microfluidics in CTCs Detection
3. ML and Its Application in CTCs
3.1. Fundamentals of ML
3.2. Applications of ML in CTCs Detection
4. Integration of ML and Microfluidics for Early Cancer Detection Through CTCs
4.1. Fundamentals of Tumor Analysis
4.2. Integrating ML and Microfluidics for Early Cancer Detection Through CTCs
4.3. Recent Advancements in Early Cancer Detection Technologies
5. Challenges and Limitations
5.1. Practical Considerations and Challenges
5.2. Ethical and Privacy Considerations
6. Discussion
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Li, C.; He, W.; Wang, N.; Xi, Z.; Deng, R.; Liu, X.; Kang, R.; Xie, L.; Liu, X. Application of microfluidics in detection of circulating tumor cells. Front. Bioeng. Biotechnol. 2022, 10, 907232. [Google Scholar]
- Wild, C.P.; Weiderpass, E.; Stewart, B.W. World Cancer Report; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Cai, J.; Chen, B.; He, M.; Yuan, G.; Hu, B. An Integrated Inertial-Magnetophoresis Microfluidic Chip Online-Coupled with ICP-MS for Rapid Separation and Precise Detection of Circulating Tumor Cells. Anal. Chem. 2024, 96, 14222–14229. [Google Scholar] [PubMed]
- Lyu, N.; Hassanzadeh-Barforoushi, A.; Rey Gomez, L.M.; Zhang, W.; Wang, Y. SERS biosensors for liquid biopsy towards cancer diagnosis by detection of various circulating biomarkers: Current progress and perspectives. Nano Converg. 2024, 11, 22. [Google Scholar]
- Li, L.; Jiang, H.; Zeng, B.; Wang, X.; Bao, Y.; Chen, C.; Ma, L.; Yuan, J. Liquid biopsy in lung cancer. Clin. Chim. Acta 2024, 554, 117757. [Google Scholar]
- Chen, J.; Liu, C.-Y.; Wang, X.; Sweet, E.; Liu, N.; Gong, X.; Lin, L. 3D printed microfluidic devices for circulating tumor cells (CTCs) isolation. Biosens. Bioelectron. 2020, 150, 111900. [Google Scholar]
- Lao, Z.; Ren, X.; Zhuang, D.; Xie, L.; Zhang, Y.; Li, W.; Chen, Y.; Li, P.; Tong, L.; Chu, P.K. A phenotype-independent “label-capture-release” process for isolating viable circulating tumor cells in real-time drug susceptibility testing. Innovation 2025, 100805. [Google Scholar] [CrossRef]
- Shi, J.; Zhang, H.; Cui, Y.; Xing, J.; Wang, W.; Chen, J.; Wang, S.; Yang, Z. Extracellular vesicles for breast cancer diagnosis and therapy. Extracell. Vesicle 2024, 3, 100039. [Google Scholar]
- Ashworth, T. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aust. Med. J. 1869, 14, 146. [Google Scholar]
- Williams, J.; Sanchez, S.; Hernandez, M. Research on the biophysical properties of circulating tumor cells. Camb. Sci. Adv. 2024, 2024, 13–18. [Google Scholar]
- Shi, R.; Yue, Y.; Liu, Z.; Chai, H.; Miao, P. Recent advances in integrated biophysical and biochemical microfluidic methods for circulating tumor cells isolation and analysis. Fundam. Res. 2024. [Google Scholar] [CrossRef]
- Li, W.; Guo, Z.; Zhou, Z.; Zhou, Z.; He, H.; Sun, J.; Zhou, X.; Chin, Y.R.; Zhang, L.; Yang, M. Distinguishing high-metastasis-potential circulating tumor cells through fluidic shear stress in a bloodstream-like microfluidic circulatory system. Oncogene 2024, 43, 2295–2306. [Google Scholar] [CrossRef] [PubMed]
- Tukova, A.; Zhang, W.; Rodger, A.; Wang, Y. Analytical SERS for Liquid Biopsy Biomarkers Detection. In Surface and Tip-Enhanced Raman Scattering Spectroscopy: Bridging Theory and Applications; Springer: Singapore, 2024; pp. 567–607. [Google Scholar]
- Abusamra, S.M.; Barber, R.; Sharafeldin, M.; Edwards, C.M.; Davis, J.J. The integrated on-chip isolation and detection of circulating tumour cells. Sens. Diagn. 2024, 3, 562–584. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Shen, L.; Luo, M.; Zhang, K.; Li, J.; Yang, Q.; Zhu, F.; Zhou, D.; Zheng, S.; Chen, Y. Circulating tumor cells: Biology and clinical significance. Signal Transduct. Target. Ther. 2021, 6, 404. [Google Scholar] [CrossRef] [PubMed]
- Bagheri, R.; Ghorbian, M.; Ghorbian, S. Tumor circulating biomarkers in colorectal cancer. Cancer Treat. Res. Commun. 2024, 38, 100787. [Google Scholar] [CrossRef]
- Bu, J.; Lee, T.H.; Poellmann, M.J.; Rawding, P.A.; Jeong, W.J.; Hong, R.S.; Hyun, S.H.; Eun, H.S.; Hong, S. Tri-modal liquid biopsy: Combinational analysis of circulating tumor cells, exosomes, and cell-free DNA using machine learning algorithm. Clin. Transl. Med. 2021, 11, e499. [Google Scholar] [CrossRef]
- Zhang, C.; Xu, L.; Miao, X.; Zhang, D.; Xie, Y.; Hu, Y.; Zhang, Z.; Wang, X.; Wu, X.; Liu, Z. Machine learning assisted dual-modal SERS detection for circulating tumor cells. Biosens. Bioelectron. 2025, 268, 116897. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, N.; Cui, X.; Liu, Z.; Yuan, X.; Chen, S.; Xu, H.; Yi, M.; Ti, Y.; Zheng, F. Detection of circulating tumor cells using a microfluidic chip for diagnostics and therapeutic prediction in mediastinal neuroblastoma. Eur. J. Pediatr. 2024, 184, 93. [Google Scholar] [CrossRef]
- Kilercik, M.; Özgür, E.; Şahin, Ş.; Şen Doğan, B.; Mutlu, E.; Cihan, C.; Kolay, M.; Erkal, N.; Zorlu, Ö.; Doğanca, T.S. Detection of circulating tumor cells in non-metastatic prostate cancer through integration of a microfluidic CTC enrichment system and multiparametric flow cytometry. PLoS ONE 2024, 19, e0312296. [Google Scholar] [CrossRef]
- Jiang, Y.; Yang, M.; Wang, S.; Li, X.; Sun, Y. Emerging role of deep learning-based artificial intelligence in tumor pathology. Cancer Commun. 2020, 40, 154–166. [Google Scholar] [CrossRef]
- Douglass, P.M.; O’Connor, T.; Javidi, B. Automated sickle cell disease identification in human red blood cells using a lensless single random phase encoding biosensor and convolutional neural networks. Opt. Express 2022, 30, 35965–35977. [Google Scholar] [CrossRef]
- Pasupa, K.; Vatathanavaro, S.; Tungjitnob, S. Convolutional neural networks based focal loss for class imbalance problem: A case study of canine red blood cells morphology classification. J. Ambient Intell. Humaniz. Comput. 2023, 14, 15259–15275. [Google Scholar]
- Wang, S.; Zhou, Y.; Qin, X.; Nair, S.; Huang, X.; Liu, Y. Label-free detection of rare circulating tumor cells by image analysis and machine learning. Sci. Rep. 2020, 10, 12226. [Google Scholar]
- Guo, Z.; Lin, X.; Hui, Y.; Wang, J.; Zhang, Q.; Kong, F. Circulating tumor cell identification based on deep learning. Front. Oncol. 2022, 12, 843879. [Google Scholar]
- Liu, Y.; Zhao, W.; Hodgson, J.; Egan, M.; Cooper Pope, C.N.; Hicks, G.; Nikolinakos, P.G.; Mao, L. CTC-race: Single-cell motility assay of circulating tumor cells from metastatic lung cancer patients. ACS Nano 2024, 18, 8683–8693. [Google Scholar]
- Kulkarni, M.B.; Goel, S. Microfluidic devices for synthesizing nanomaterials—A review. Nano Express 2020, 1, 32004. [Google Scholar]
- Huang, Y.; Liu, C.; Feng, Q.; Sun, J. Microfluidic synthesis of nanomaterials for biomedical applications. Nanoscale Horiz. 2023, 8, 1610–1627. [Google Scholar]
- Elvira, K.S.; Gielen, F.; Tsai, S.S.; Nightingale, A.M. Materials and methods for droplet microfluidic device fabrication. Lab Chip 2022, 22, 859–875. [Google Scholar]
- Hua, X.; Liu, X.; Zhu, Q.; Liu, Y.; Zhou, S.; Huang, P.; Li, Q.; Liu, S. Three-dimensional microfluidic chip for efficient capture of secretory autophagosomes and sensitive detection of their surface proteins. Anal. Chem. 2022, 94, 8489–8496. [Google Scholar]
- Battat, S.; Weitz, D.A.; Whitesides, G.M. Nonlinear phenomena in microfluidics. Chem. Rev. 2022, 122, 6921–6937. [Google Scholar]
- Wu, L.; Guo, Z.; Liu, W. Surface behaviors of droplet manipulation in microfluidics devices. Adv. Colloid Interface Sci. 2022, 308, 102770. [Google Scholar]
- Kronfeld, K.-P.; Köhler, J.M.; Ellinger, T. Microfluidic synthesis and properties of thermoresponsive hydrogel core–shell particles. J. Compos. Sci. 2024, 8, 162. [Google Scholar] [CrossRef]
- Rahmanizadeh, M. Design and Fabrication of Thermo-Responsive Hydrogel Particles with Tunable Swelling and Mechanical Behavior via Microfluidics Method. 2022. Available online: https://www.politesi.polimi.it/handle/10589/219778 (accessed on 26 March 2025).
- Azizipour, N.; Avazpour, R.; Sawan, M.; Rosenzweig, D.H.; Ajji, A. Uniformity of spheroids-on-a-chip by surface treatment of PDMS microfluidic platforms. Sens. Diagn. 2022, 1, 750–764. [Google Scholar] [CrossRef]
- Ramasamy, M.; Ho, B.; Phan, C.-M.; Qin, N.; Ren, C.L.; Jones, L. Inexpensive and rapid fabrication of PDMS microfluidic devices for biological testing applications using low cost commercially available 3D printers. J. Micromech. Microeng. 2023, 33, 105016. [Google Scholar] [CrossRef]
- McIntyre, D.; Lashkaripour, A.; Fordyce, P.; Densmore, D. Machine learning for microfluidic design and control. Lab Chip 2022, 22, 2925–2937. [Google Scholar] [CrossRef]
- Lashkaripour, A.; McIntyre, D.P.; Calhoun, S.G.; Krauth, K.; Densmore, D.M.; Fordyce, P.M. Design automation of microfluidic single and double emulsion droplets with machine learning. Nat. Commun. 2024, 15, 83. [Google Scholar] [CrossRef]
- Ahmed, M.G.; Abate, M.F.; Song, Y.; Zhu, Z.; Yan, F.; Xu, Y.; Wang, X.; Li, Q.; Yang, C. Isolation, detection, and antigen-based profiling of circulating tumor cells using a size-dictated immunocapture chip. Angew. Chem. Int. Ed. 2017, 56, 10681–10685. [Google Scholar] [CrossRef]
- Bialek, J.; Muthe, A.; Yankulov, S.; Kawan, F.; Gakis, G.; Theil, G. Optimizing CTC isolation techniques for molecular characterization of circulating tumor cells in clear cell renal cell carcinoma: A comparative study of EpCAM-based and density-based methods. Cancer Res. 2024, 84, 3689. [Google Scholar] [CrossRef]
- Chowdhury, T.; Cressiot, B.; Parisi, C.; Smolyakov, G.; Thiébot, B.; Trichet, L.; Fernandes, F.M.; Pelta, J.; Manivet, P. Circulating tumor cells in cancer diagnostics and prognostics by single-molecule and single-cell characterization. ACS Sens. 2023, 8, 406–426. [Google Scholar] [CrossRef]
- Gerratana, L.; Davis, A.A.; Foffano, L.; Reduzzi, C.; Rossi, T.; Medford, A.; Clifton, K.; Shah, A.N.; Bucheit, L.; Velimirovic, M. Integrating machine learning-predicted circulating tumor cells (CTCs) and circulating tumor DNA (ctDNA) in metastatic breast cancer: A proof of principle study on endocrine resistance profiling. Cancer Lett. 2025, 609, 217325. [Google Scholar] [CrossRef]
- Harb, W.; Fan, A.; Tran, T.; Danila, D.C.; Keys, D.; Schwartz, M.; Ionescu-Zanetti, C. Mutational analysis of circulating tumor cells using a novel microfluidic collection device and qPCR assay. Transl. Oncol. 2013, 6, 528–538. [Google Scholar] [CrossRef]
- Peeters, D.; De Laere, B.; Van den Eynden, G.; Van Laere, S.; Rothé, F.; Ignatiadis, M.; Sieuwerts, A.M.; Lambrechts, D.; Rutten, A.; Van Dam, P. Semiautomated isolation and molecular characterisation of single or highly purified tumour cells from CellSearch enriched blood samples using dielectrophoretic cell sorting. Br. J. Cancer 2013, 108, 1358–1367. [Google Scholar] [PubMed]
- Nagrath, S.; Sequist, L.V.; Maheswaran, S.; Bell, D.W.; Irimia, D.; Ulkus, L.; Smith, M.R.; Kwak, E.L.; Digumarthy, S.; Muzikansky, A. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007, 450, 1235–1239. [Google Scholar] [PubMed]
- Kang, H.; Xiong, Y.; Ma, L.; Yang, T.; Xu, X. Recent advances in micro-/nanostructure array integrated microfluidic devices for efficient separation of circulating tumor cells. RSC Adv. 2022, 12, 34892–34903. [Google Scholar]
- Zhao, K.; Zhao, P.; Dong, J.; Wei, Y.; Chen, B.; Wang, Y.; Pan, X.; Wang, J. Implementation of an integrated dielectrophoretic and magnetophoretic microfluidic chip for CTC isolation. Biosensors 2022, 12, 757. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, Y.; Gao, W.; Qian, G.; Chen, X.; Liu, Y.; Yu, S. A microfluidic device for enhanced capture and high activity release of heterogeneous CTCs from whole blood. Talanta 2024, 266, 125007. [Google Scholar]
- Wang, Y.; Wang, S.; Li, L.; Zou, Y.; Liu, B.; Fang, X. Microfluidics-based molecular profiling of tumor-derived exosomes for liquid biopsy. View 2023, 4, 20220048. [Google Scholar]
- Macaraniag, C.; Luan, Q.; Zhou, J.; Papautsky, I. Microfluidic techniques for isolation, formation, and characterization of circulating tumor cells and clusters. APL Bioeng. 2022, 6, 31501. [Google Scholar]
- Addanki, S.; Meas, S.; Sarli, V.N.; Singh, B.; Lucci, A. Applications of circulating tumor cells and circulating tumor DNA in precision oncology for breast cancers. Int. J. Mol. Sci. 2022, 23, 7843. [Google Scholar] [CrossRef]
- Lee, J.H.; Park, S.H.; Kang, J.; Lee, J.; Kim, S.; Kim, J.; Sohn, Y.W.; Lee, J.K. Identification of circulating tumor cells based on machine learning. Cancer Res. 2023, 83, 6692. [Google Scholar]
- Calvo-Almeida, S.; Serrano-Llabrés, I.; Cal-González, V.M.; Piairo, P.; Pires, L.R.; Diéguez, L.; González-Castro, L. Multichannel fluorescence microscopy images CTC detection: A deep learning approach. In Proceedings of the International Conference Of Computational Methods In Sciences And Engineering ICCMSE, Virtual Conference, 26–29 October 2022; AIP Conference Proceedings. AIP Publishing: College Park, MD, USA, 2024. [Google Scholar]
- Biasiolli, L.; Ansaloni, P.; Gentili, N.; Giardiello, D.; Montanari, F.; Miserendino, R.; Signorini, G.; Medoro, G. Automated identification and enumeration of CELLSEARCH Circulating Tumor Cells (CTC) with a deep learning algorithm. Cancer Res. 2024, 84, 7492. [Google Scholar]
- Guo, X.; Lin, F.; Yi, C.; Song, J.; Sun, D.; Lin, L.; Zhong, Z.; Wu, Z.; Wang, X.; Zhang, Y. Deep transfer learning enables lesion tracing of circulating tumor cells. Nat. Commun. 2022, 13, 7687. [Google Scholar] [CrossRef] [PubMed]
- Husseini-Wüsthoff, H.; Riethdorf, S.; Schneeweiss, A.; Trumpp, A.; Pantel, K.; Wikman, H.; Nielsen, M.; Werner, R. Cluster-based human-in-the-loop strategy for improving machine learning-based circulating tumor cell detection in liquid biopsy. arXiv 2024, arXiv:2411.16332. [Google Scholar]
- An, L.; Hu, H.; Song, M.; Cheng, L.; Ba, S.; Liu, Z.; Yu, Z.; Zhang, Z.; Liu, Y.; Zhou, C.-C. Unsupervised Classification for Circulating Tumor Cells. SSRN 5090990. Available online: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=5090990 (accessed on 5 February 2025).
- Al-Eidi, S.; Darwish, O.; Husari, G.; Chen, Y.; Elkhodr, M. Convolutional neural network structure to detect and localize ctc using image processing. In Proceedings of the 2022 IEEE International IOT, Electronics and Mechatronics Conference (IEMTRONICS), Toronto, OT, Canada, 1–4 June 2022; pp. 1–7. [Google Scholar]
- Park, J.; Ha, S.; Kim, J.; Song, J.-W.; Hyun, K.-A.; Kamiya, T.; Jung, H.-I. Classification of circulating tumor cell clusters by morphological characteristics using convolutional neural network-support vector machine. Sens. Actuators B Chem. 2024, 401, 134896. [Google Scholar] [CrossRef]
- Albaradei, S.; Alganmi, N.; Albaradie, A.; Alharbi, E.; Motwalli, O.; Thafar, M.A.; Gojobori, T.; Essack, M.; Gao, X. A deep learning model predicts the presence of diverse cancer types using circulating tumor cells. Sci. Rep. 2023, 13, 21114. [Google Scholar] [CrossRef]
- Nanou, A.; Stoecklein, N.H.; Doerr, D.; Driemel, C.; Terstappen, L.W.; Coumans, F.A. Training an automated circulating tumor cell classifier when the true classification is uncertain. Proc. Natl. Acad. Sci. USA 2024, 3, 48. [Google Scholar] [CrossRef]
- Pastuszak, K.; Sieczczyński, M.; Dzięgielewska, M.; Wolniak, R.; Drewnowska, A.; Korpal, M.; Zembrzuska, L.; Supernat, A.; Żaczek, A.J. Detection of circulating tumor cells by means of machine learning using Smart-Seq2 sequencing. Sci. Rep. 2024, 14, 11057. [Google Scholar] [CrossRef] [PubMed]
- Gaysar, S.; Mustafa, Z.; Zein, A. Deep Learning Algorithms for Studying the Impact of Tumor Suppressor Gene Mutations on Breast Cancer. J. Clin. Eng. 2025, 50, 35–38. [Google Scholar] [CrossRef]
- Wu, X.; Wang, H.-Y.; Shi, P.; Sun, R.; Wang, X.; Luo, Z.; Zeng, F.; Lebowitz, M.S.; Lin, W.-Y.; Lu, J.-J. Long short-term memory model–a deep learning approach for medical data with irregularity in cancer predication with tumor markers. Comput. Biol. Med. 2022, 144, 105362. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, F.; Yao, J.; Mao, C.; Zhu, M.; Qian, M.; Hu, J.; Zhong, H.; Zhou, J.; Shi, X. Single-cell metabolic fingerprints discover a cluster of circulating tumor cells with distinct metastatic potential. Nat. Commun. 2023, 14, 2485. [Google Scholar] [CrossRef]
- Ogut, M.G.; Ma, P.; Gupta, R.; Hoerner, C.R.; Fan, A.C.; El-Kaffas, A.N.; Durmus, N.G. Automated Image Analysis for Characterization of Circulating Tumor Cells and Clusters Sorted by Magnetic Levitation. Adv. Biol. 2023, 7, 2300109. [Google Scholar] [CrossRef]
- Srikanth, S.; Dubey, S.K.; Javed, A.; Goel, S. Droplet based microfluidics integrated with machine learning. Sens. Actuators A Phys. 2021, 332, 113096. [Google Scholar]
- Gangadhar, A.; Sari-Sarraf, H.; Vanapalli, S.A. Deep learning assisted holography microscopy for in-flow enumeration of tumor cells in blood. RSC Adv. 2023, 13, 4222–4235. [Google Scholar] [PubMed]
- Moallem, G.; Pore, A.A.; Gangadhar, A.; Sari-Sarraf, H.; Vanapalli, S.A. Detection of live breast cancer cells in bright-field microscopy images containing white blood cells by image analysis and deep learning. J. Biomed. Opt. 2022, 27, 76003. [Google Scholar]
- Ba, W.; Wang, S.; Shang, M.; Zhang, Z.; Wu, H.; Yu, C.; Xing, R.; Wang, W.; Wang, L.; Liu, C. Assessment of deep learning assistance for the pathological diagnosis of gastric cancer. Mod. Pathol. 2022, 35, 1262–1268. [Google Scholar] [CrossRef]
- Surappa, S.; Multani, P.; Parlatan, U.; Sinawang, P.D.; Kaifi, J.; Akin, D.; Demirci, U. Integrated “lab-on-a-chip” microfluidic systems for isolation, enrichment, and analysis of cancer biomarkers. Lab Chip 2023, 23, 2942–2958. [Google Scholar] [CrossRef] [PubMed]
- Da Col, G.; Del Ben, F.; Bulfoni, M.; Turetta, M.; Gerratana, L.; Bertozzi, S.; Beltrami, A.P.; Cesselli, D. Image analysis of circulating tumor cells and leukocytes predicts survival and metastatic pattern in breast cancer patients. Front. Oncol. 2022, 12, 725318. [Google Scholar]
- Liu, C.; Yang, H.; Feng, Y.; Liu, C.; Rui, F.; Cao, Y.; Hu, X.; Xu, J.; Fan, J.; Zhu, Q. A K-nearest neighbor model to predict early recurrence of hepatocellular carcinoma after resection. J. Clin. Transl. Hepatol. 2022, 10, 600. [Google Scholar]
- Zhou, J.; Zhu, X.; Wu, S.; Guo, J.; Zhang, K.; Xu, C.; Chen, H.; Jin, Y.; Sun, Y.; Zheng, S. Epithelial-mesenchymal transition status of circulating tumor cells in breast cancer and its clinical relevance. Cancer Biol. Med. 2020, 17, 169–180. [Google Scholar]
- Wei, H.; Natori, T.; Tanaka, T.; Aoki, S.; Yamada, T.; Aikawa, N. Detection of Circulating Tumor Cells in Blood Using Random Forest. In Proceedings of the 2024 International Conference on Electronics, Information, and Communication (ICEIC), Taipei City, Taiwan, 28–31 January 2024; pp. 1–4. [Google Scholar]
- Svensson, C.-M.; Hübler, R.; Figge, M.T. Automated classification of circulating tumor cells and the impact of interobsever variability on classifier training and performance. J. Immunol. Res. 2015, 2015, 573165. [Google Scholar]
- Mao, Y.; Yin, Z.; Schober, J.M. Iteratively training classifiers for circulating tumor cell detection. In Proceedings of the 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), Brooklyn, NY, USA, 16–19 April 2015; pp. 190–194. [Google Scholar]
- Mocan, I.; Itu, R.; Ciurte, A.; Danescu, R.; Buiga, R. Automatic Detection of Tumor Cells in Microscopic Images of Unstained Blood using Convolutional Neural Networks. In Proceedings of the 2018 IEEE 14th International Conference on Intelligent Computer Communication and Processing (ICCP), Cluj-Napoca, Romania, 6–8 September 2018; pp. 319–324. [Google Scholar]
- Ciurte, A.; Selicean, C.; Soritau, O.; Buiga, R. Automatic detection of circulating tumor cells in darkfield microscopic images of unstained blood using boosting techniques. PLoS ONE 2018, 13, e0208385. [Google Scholar] [CrossRef]
- Nissim, N.; Dudaie, M.; Barnea, I.; Shaked, N.T. Real-time stain-free classification of cancer cells and blood cells using interferometric phase microscopy and machine learning. Cytom. Part A 2021, 99, 511–523. [Google Scholar]
- Zeune, L.L.; Boink, Y.E.; van Dalum, G.; Nanou, A.; de Wit, S.; Andree, K.C.; Swennenhuis, J.F.; van Gils, S.A.; Terstappen, L.W.; Brune, C. Deep learning of circulating tumour cells. Nat. Mach. Intell. 2020, 2, 124–133. [Google Scholar]
- Li, X.; Chen, M.; Xu, J.; Wu, D.; Ye, M.; Wang, C.; Liu, W. Interpretatively automated identification of circulating tumor cells from human peripheral blood with high performance. Front. Bioeng. Biotechnol. 2023, 11, 1013107. [Google Scholar]
- Boya, M.; Ozkaya-Ahmadov, T.; Swain, B.E.; Chu, C.-H.; Asmare, N.; Civelekoglu, O.; Liu, R.; Lee, D.; Tobia, S.; Biliya, S. High throughput, label-free isolation of circulating tumor cell clusters in meshed microwells. Nat. Commun. 2022, 13, 3385. [Google Scholar] [PubMed]
- Yalamanchili, S.; Yenuga, P.; Burla, N.; Jonnadula, H.; Bolem, S.C. MRI Brain Tumor Analysis on Improved VGG-16 and Efficient NetB7 Models. J. Image Graph. 2024, 12, 103–116. [Google Scholar]
- Reis, H.C.; Turk, V. Advanced brain tumor analysis: A novel strategy for segmentation and classification using modern computational methods. Neural Comput. Appl. 2025, 37, 4697–4731. [Google Scholar]
- Caro-Vegas, C.; Ramirez, C.; Landis, J.; Adimora, A.A.; Strickler, H.; French, A.L.; Ofotokun, I.; Fischl, M.; Seaberg, E.C.; Wang, C.-c.J. Molecular profiling of breast and lung cancer in women with HIV reveals high tumor mutational burden. Aids 2022, 36, 567–571. [Google Scholar]
- Philip, P.A.; Azar, I.; Xiu, J.; Hall, M.J.; Hendifar, A.E.; Lou, E.; Hwang, J.J.; Gong, J.; Feldman, R.; Ellis, M. Molecular characterization of KRAS wild-type tumors in patients with pancreatic adenocarcinoma. Clin. Cancer Res. 2022, 28, 2704–2714. [Google Scholar]
- Shahzad, M.; Ali, F.; Shirazi, S.H.; Rasheed, A.; Ahmad, A.; Shah, B.; Kwak, D. Blood cell image segmentation and classification: A systematic review. PeerJ Comput. Sci. 2024, 10, e1813. [Google Scholar] [CrossRef]
- Akbarnataj, K.; Maleki, S.; Rezaeian, M.; Haki, M.; Shamloo, A. Novel size-based design of spiral microfluidic devices with elliptic configurations and trapezoidal cross-section for ultra-fast isolation of circulating tumor cells. Talanta 2023, 254, 124125. [Google Scholar]
- Wang, J.; Meng, X.; Yu, M.; Li, X.; Chen, Z.; Wang, R.; Fang, J. A novel microfluidic system for enrichment of functional circulating tumor cells in cancer patient blood samples by combining cell size and invasiveness. Biosens. Bioelectron. 2023, 227, 115159. [Google Scholar]
- Shen, C.; Rawal, S.; Brown, R.; Zhou, H.; Agarwal, A.; Watson, M.A.; Cote, R.J.; Yang, C. Automatic detection of circulating tumor cells and cancer associated fibroblasts using deep learning. Sci. Rep. 2023, 13, 5708. [Google Scholar]
- Bakhshi, M.S.; Rizwan, M.; Khan, G.J.; Duan, H.; Zhai, K. Design of a novel integrated microfluidic chip for continuous separation of circulating tumor cells from peripheral blood cells. Sci. Rep. 2022, 12, 17016. [Google Scholar]
- Edd, J.F.; Mishra, A.; Smith, K.C.; Kapur, R.; Maheswaran, S.; Haber, D.A.; Toner, M. Isolation of circulating tumor cells. Iscience 2022, 25, 104696. [Google Scholar]
- Kumar, S.; Wang, Y.; Zhan, H.; Gardner, K.; Thompson, T.; Li, W. Classification of Circulating Tumor Cells Using Machine Learning on Microfluidic Trajectory Data. J. ACM 2024, 37, 111. [Google Scholar]
- Zhou, M.; Zheng, H.; Wang, Z.; Li, R.; Liu, X.; Zhang, W.; Wang, Z.; Li, H.; Wei, Z.; Hu, Z. Precisely enumerating circulating tumor cells utilizing a multi-functional microfluidic chip and unique image interpretation algorithm. Theranostics 2017, 7, 4710. [Google Scholar]
- Pirone, D.; Montella, A.; Sirico, D.G.; Mugnano, M.; Villone, M.M.; Bianco, V.; Miccio, L.; Porcelli, A.M.; Kurelac, I.; Capasso, M. Label-free liquid biopsy through the identification of tumor cells by machine learning-powered tomographic phase imaging flow cytometry. Sci. Rep. 2023, 13, 6042. [Google Scholar] [CrossRef]
- Gangadhar, A.; Sari-Sarraf, H.; Vanapalli, S.A. Staining-free, in-flow enumeration of tumor cells in blood using digital holographic microscopy and deep learning. bioRxiv 2022. [Google Scholar] [CrossRef]
- Liang, N.; Li, B.; Jia, Z.; Wang, C.; Wu, P.; Zheng, T.; Wang, Y.; Qiu, F.; Wu, Y.; Su, J. Ultrasensitive detection of circulating tumour DNA via deep methylation sequencing aided by machine learning. Nat. Biomed. Eng. 2021, 5, 586–599. [Google Scholar]
- Muthamilselvan, S.; Ramasami Sundhar Baabu, P.; Palaniappan, A. Microfluidics for profiling miRNA biomarker panels in AI-assisted cancer diagnosis and prognosis. Technol. Cancer Res. Treat. 2023, 22, 15330338231185284. [Google Scholar]
- Kim, M.W.; Kim, J.Y.; Kim, Y.; Lee, S.; Moon, S.; Hyon, J.-y.; Hyun, K.-A.; Yang, Y.; Ha, S.; Park, S. Integrating machine learning with microfluidic technologies for proteomic profiling of extracellular vesicles in triple-negative breast cancer. Cancer Res. 2024, 84, 1843. [Google Scholar] [CrossRef]
- Wang, C.; Wang, C.; Wu, Y.; Gao, J.; Han, Y.; Chu, Y.; Qiang, L.; Qiu, J.; Gao, Y.; Wang, Y. High-Throughput, Living Single-Cell, Multiple Secreted Biomarker Profiling Using Microfluidic Chip and Machine Learning for Tumor Cell Classification. Adv. Healthc. Mater. 2022, 11, 2102800. [Google Scholar] [CrossRef] [PubMed]
- Noor, J.; Chaudhry, A.; Batool, S. Microfluidic technology, artificial intelligence, and biosensors as advanced technologies in cancer screening: A review article. Cureus 2023, 15, e39634. [Google Scholar] [CrossRef] [PubMed]
- Rejuan, R.; Aulisa, E.; Li, W.; Thompson, T.; Kumar, S.; Canic, S.; Wang, Y. Validation of a Microfluidic Device Prototype for Cancer Detection and Identification: Circulating Tumor Cells Classification Based on Cell Trajectory Analysis Leveraging Cell-Based Modeling and Machine Learning. bioRxiv 2024. [Google Scholar] [CrossRef]
- Wang, L.; Hong, R.; Shi, S.; Wang, S.; Chen, Y.; Han, C.; Li, M.; Ye, F. The prognostic significance of circulating tumor cell enumeration and HER2 expression by a novel automated microfluidic system in metastatic breast cancer. BMC Cancer 2024, 24, 1067. [Google Scholar] [CrossRef]
- Vora, N.; Shekar, P.; Hanulia, T.; Esmail, M.; Patra, A.; Georgakoudi, I. Deep learning-enabled detection of rare circulating tumor cell clusters in whole blood using label-free, flow cytometry. Lab Chip 2024, 24, 2237–2252. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, Y.; Zhang, W.; Tang, D.; Zhang, S.; Wang, L.; Zou, X.; Ni, Z.; Zhang, S.; Lv, Y. High-throughput enrichment of portal venous circulating tumor cells for highly sensitive diagnosis of CA19-9-negative pancreatic cancer patients using inertial microfluidics. Biosens. Bioelectron. 2024, 259, 116411. [Google Scholar] [CrossRef]
- Hua, H.; Zhou, Y.; Li, W.; Zhang, J.; Deng, Y.; Khoo, B.L. Microfluidics-based patient-derived disease detection tool for deep learning-assisted precision medicine. Biomicrofluidics 2024, 18, 14101. [Google Scholar] [CrossRef]
- Pirone, D.; Montella, A.; Cavina, B.; Giugliano, G.; Schiavo, M.; Mugnano, M.; Cerbone, V.; Scalia, G.; Porcelli, A.M.; Gasparre, G. Towards label-free liquid biopsy: Combining machine learning and tomographic phase imaging flow cytometry for the identification of tumor cells. In Biomedical Spectroscopy, Microscopy, and Imaging III; SPIE: Bellingham, WA, USA, 2024; pp. 178–181. [Google Scholar]
- Wu, W.; Zhang, Y.; Tan, X.; Chen, Y.; Cao, Y.; Sahai, V.; Peterson, N.; Goo, L.; Fry, S.; Kathawate, V. Antigen-independent single-cell circulating tumor cell detection using deep-learning-assisted biolasers. Biosens. Bioelectron. 2025, 271, 116984. [Google Scholar] [CrossRef]
- Kouhkord, A.; Naserifar, N. Ultrasound-assisted microfluidic cell separation: A study on microparticles for enhanced cancer diagnosis. Phys. Fluids 2025, 37, 12028. [Google Scholar] [CrossRef]
- Liu, Z.; Zhou, Y.; Lu, J.; Gong, T.; Ibáñez, E.; Cifuentes, A.; Lu, W. Microfluidic biosensors for biomarker detection in body fluids: A key approach for early cancer diagnosis. Biomark. Res. 2024, 12, 153. [Google Scholar]
- Qi, X.; Lin, S.; Li, M. Atomic force microscopy combined with microfluidics for label-free sorting and automated nanomechanics of circulating tumor cells in liquid biopsy. Nanoscale 2025, 17, 4695–4712. [Google Scholar] [PubMed]



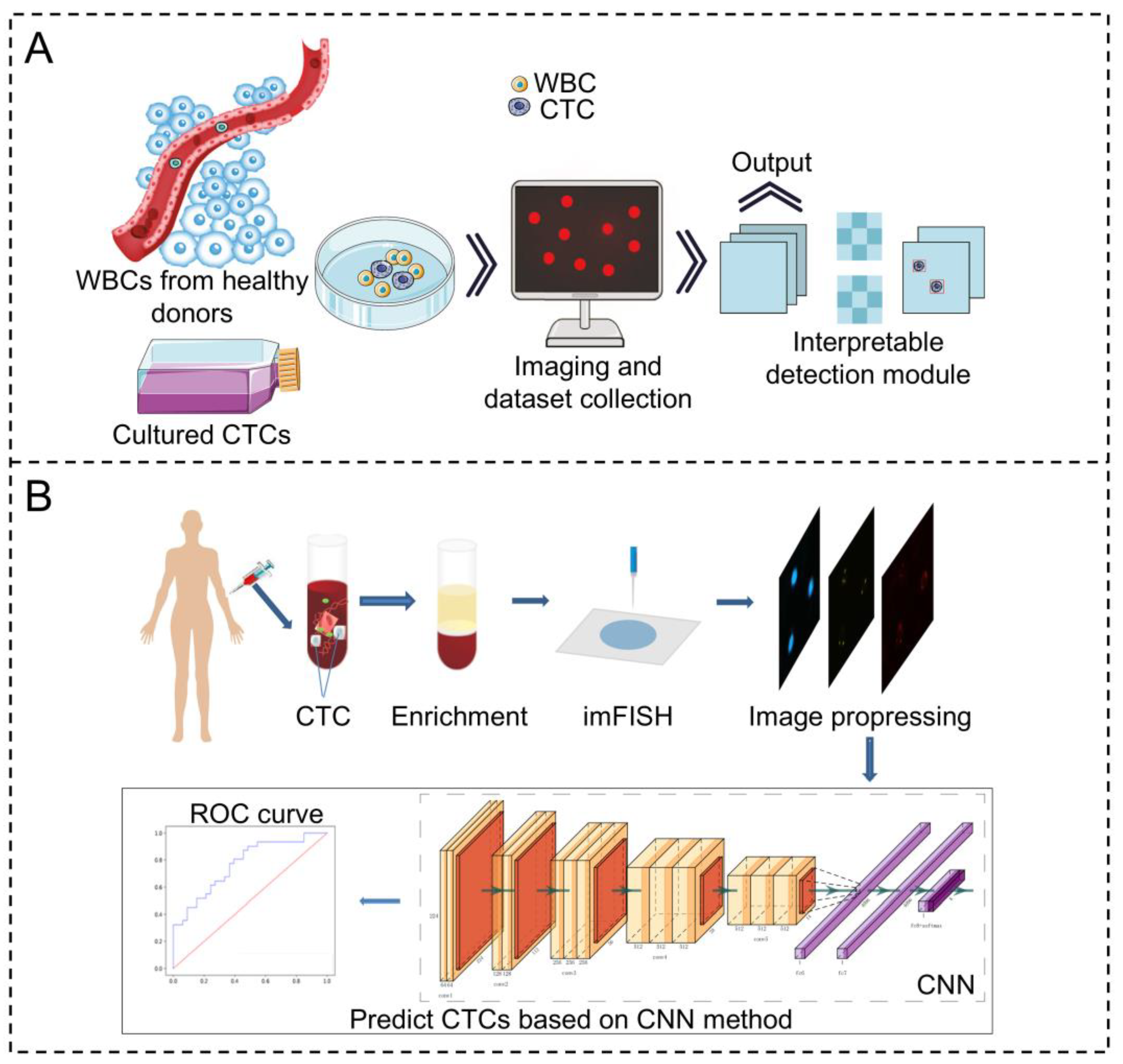
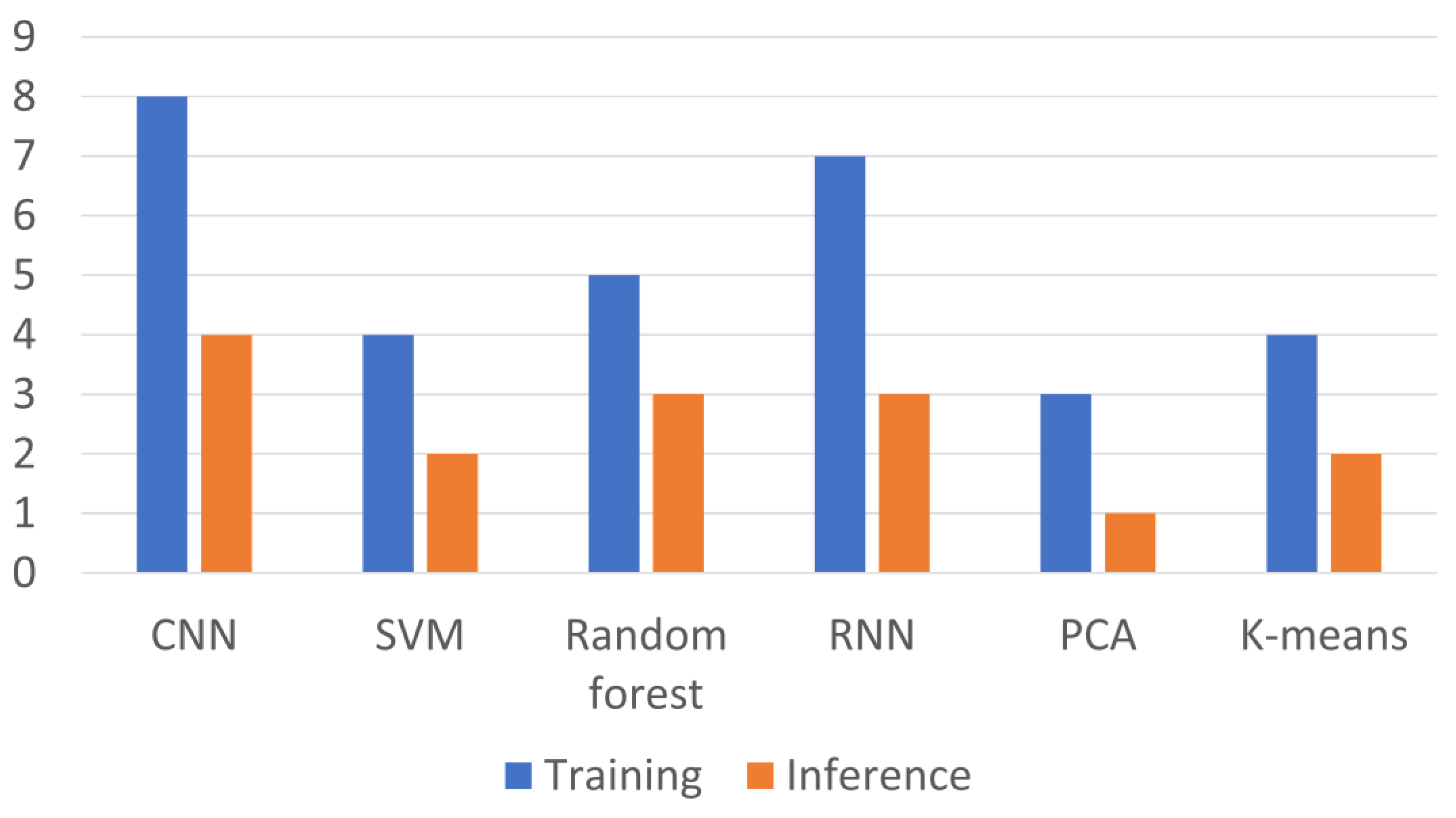
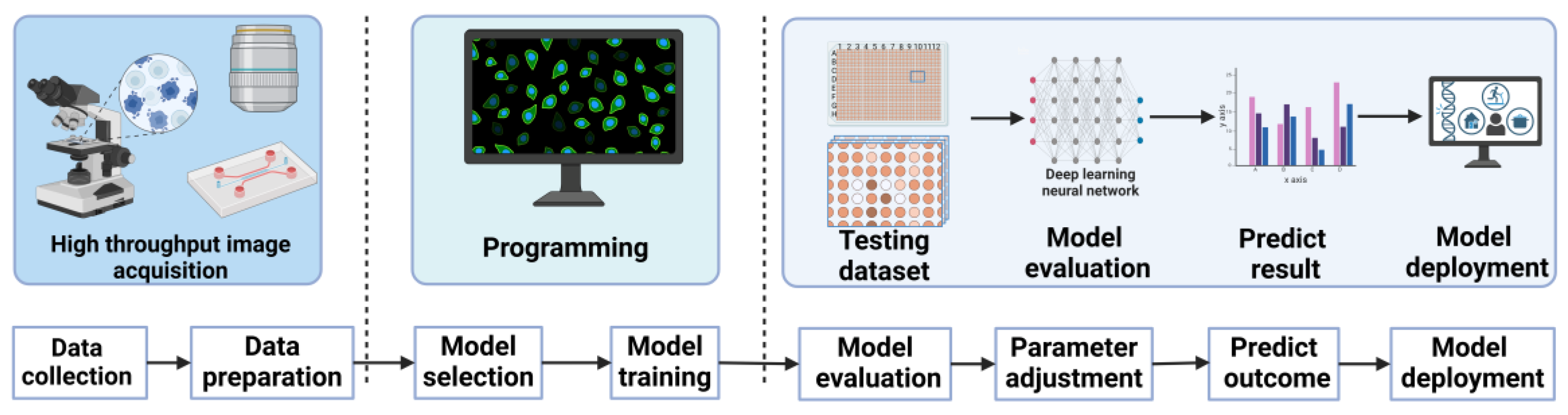
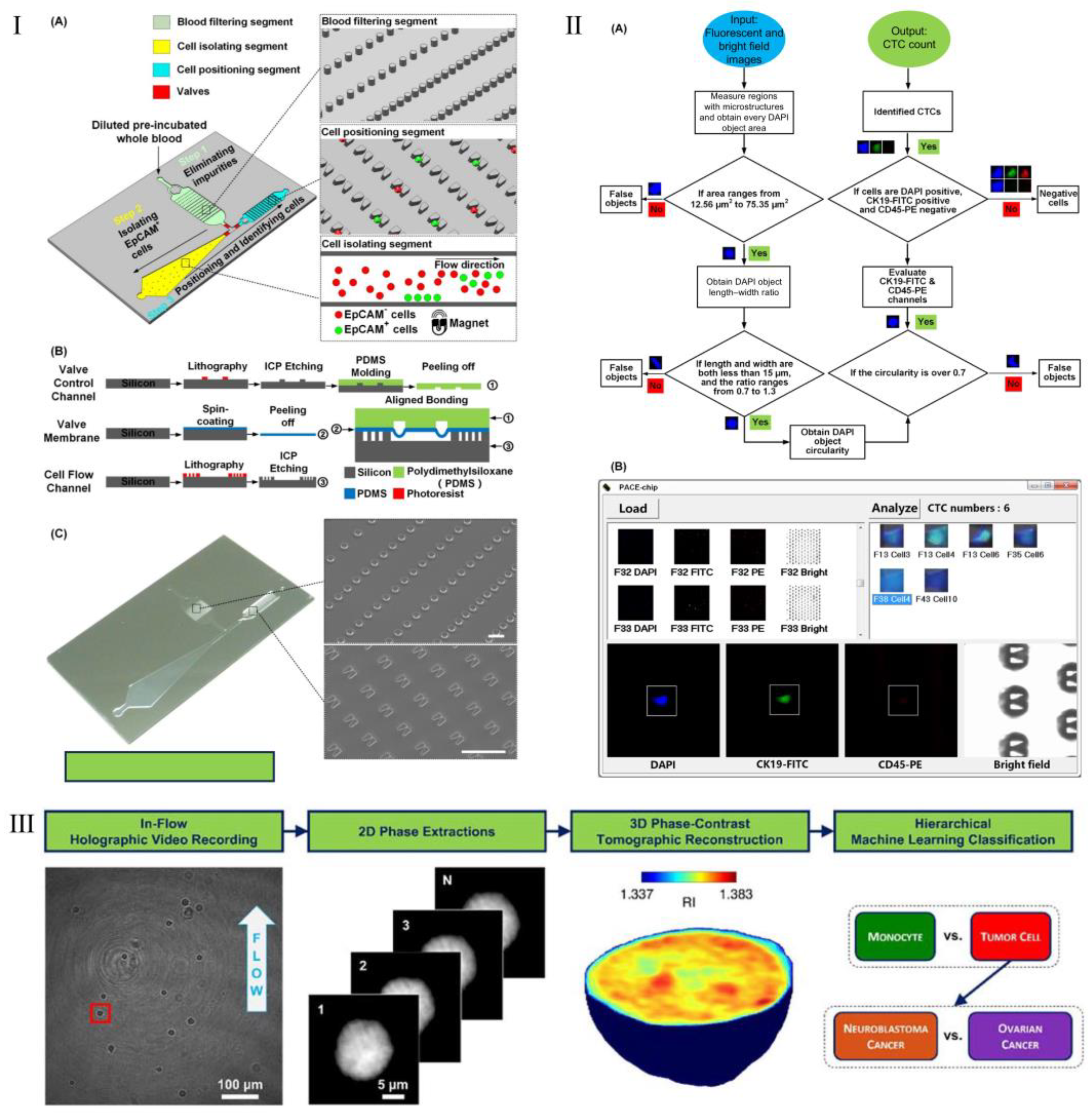
| Technology Category | Specific Technology | Advantages | Disadvantages | Application Scenarios | Efficiency | Sensitivity | Specificity | Throughput | Cell Viability |
|---|---|---|---|---|---|---|---|---|---|
| Physical (Size-Based) Methods | Microfiltration | High throughput, low cost, suitable for large sample volumes | Risk of clogging; may cause cell deformation | Pre-treatment of blood, bulk initial separation | High | Moderate | Moderate | High | Moderate |
| Inertial Focusing | Label-free, rapid processing | Lower separation purity; may require further refinement | Continuous flow separation, rapid screening | High | High | Moderate | High | High | |
| Affinity-Based Methods | Immunocapture | High specificity and selectivity; direct use for downstream analysis | Dependent on known CTC surface markers; lower throughput | Precise target cell capture, subsequent molecular analysis | Moderate | Very High | Very High | Moderate | Moderate |
| Magnetic Bead Separation | High specificity; adjustable binding efficiency | Requires magnetic labeling; involves multiple processing steps | Clinical diagnostics, targeted enrichment | Moderate | High | Very High | Low | Moderate | |
| Electrical-Based Methods | Dielectrophoresis | High precision, label-free, tunable | High equipment cost; complex operation and integration | Research applications, separation based on electrical properties | Moderate | High | High | Low | High |
| Integrated/Multi-Modal Platforms | CTC-iChip | Combines multiple separation principles; comprehensive performance; high automation | Complex design; higher initial cost | Precision medicine, comprehensive cell analysis | High | High | High | High | Moderate |
| Method | Function | Advantages | Limitations | Application Scenarios |
|---|---|---|---|---|
| CNN | Extracts hierarchical spatial features from image data, enabling automated feature learning for CTC recognition | Efficient image recognition and classification capability, suitable for identifying specific morphologies and markers of CTCs | Requires large, labeled datasets for training, computationally expensive | Recognition and classification of CTCs in microfluidic chip images |
| SVM | Maps data into a high-dimensional space to find the optimal hyperplane for classifying CTCs | Powerful classification performance, suitable for classification problems with clear boundaries | Sensitive to noisy data and outliers, less effective with unbalanced datasets | Differentiating CTCs from other blood cells, selective classification of cell features |
| Random Forest | Uses an ensemble of decision trees to improve classification reliability and handle complex, high-dimensional data | Improves accuracy and robustness by constructing multiple decision trees, suitable for handling a large number of features | Can overfit with noisy data, less interpretable than simpler models | Processing a large number of features, such as cell morphology and molecular markers, to improve classification accuracy |
| RNN | Captures temporal dependencies in sequential data, enabling dynamic tracking of CTC behaviors | Capability to process sequential data, useful for analyzing dynamic changes of CTCs | Struggles with very long sequences, requires large datasets for training | Monitoring the behavior of CTCs in microfluidic devices |
| PCA | Transforms high-dimensional data into a reduced set of principal components, preserving the most significant variance | Data dimensionality reduction, highlighting important features, simplifying the machine learning processing flow | May lose some important variance during dimensionality reduction | Reducing the dimensionality of CTC image data, highlighting the most important features |
| K-means | Partitions data into clusters based on similarity, helping to classify and differentiate CTC subpopulations | Unsupervised learning, can automatically discover different cell populations | Sensitive to the initial choice of K, ineffective with clusters of varying sizes | Automatically discovering different cell populations based on cell morphology and phenotypic features |
| Technology | Description | Cancer Type | Advantages | Reference |
|---|---|---|---|---|
| Trajectory-Based ML Classification in Microfluidics | Classifies CTCs by analyzing their movement trajectories in microchannels using ML trained on simulation-based datasets. | General (validation study) | Label-free, interpretable trajectory data, combines simulation and ML. | [103] |
| Automated Microfluidic HER2-CTC Analysis | Enumerates CTCs and evaluates HER2 expression via an automated microfluidic system for breast cancer prognosis. | Metastatic breast dancer | Automated, prognostic relevance, HER2 marker integration. | [104] |
| Deep Learning + Label-Free Flow Cytometry | Uses deep learning to detect rare CTC clusters in whole blood through label-free flow cytometry. | General | Label-free, detects rare clusters, high accuracy with DL. | [105] |
| Inertial Microfluidics for Pancreatic CTCs | Enriches portal vein CTCs in CA19-9-negative pancreatic cancer patients using inertial flow-based separation. | Pancreatic cancer | High throughput, useful for biomarker-negative cases. | [106] |
| Patient-Derived Microfluidic Disease Chips | Integrates microfluidic disease models with deep learning for precision diagnostics on patient-derived samples. | General (patient-specific) | Patient-specific, integrative ML platform, personalized. | [107] |
| Tomographic Phase Imaging + ML | Combines tomographic phase imaging with ML for label-free CTC detection in liquid biopsy. | General | Label-free, optical imaging + ML, early detection. | [108] |
| Biolaser + Deep Learning CTC Detection | Uses DL-enhanced biolaser readouts for antigen-independent single-cell CTC detection. | General | Antigen-independent, single-cell precision, DL-powered. | [109] |
| Ultrasound-Assisted Microfluidic Separation | Studies microparticle dynamics in ultrasound-enhanced microfluidics for potential cancer diagnostics. | General/exploratory | Non-contact, gentle separation, tunable system. | [110] |
| Microfluidic Biosensors for Body Fluid Biomarkers | Reviews microfluidic biosensors for detecting cancer biomarkers in fluids like blood, urine, and saliva. | Various (review) | Non-invasive, biosensor integration, early diagnosis. | [111] |
| AFM + Microfluidics for CTC Nano-Mechanics | Uses atomic force microscopy in microfluidic chips to sort and analyze nanomechanical properties of CTCs. | General | Label-free, mechanical profiling, single-cell level. | [112] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
An, L.; Liu, Y.; Liu, Y. Advancements in Circulating Tumor Cell Detection for Early Cancer Diagnosis: An Integration of Machine Learning Algorithms with Microfluidic Technologies. Biosensors 2025, 15, 220. https://doi.org/10.3390/bios15040220
An L, Liu Y, Liu Y. Advancements in Circulating Tumor Cell Detection for Early Cancer Diagnosis: An Integration of Machine Learning Algorithms with Microfluidic Technologies. Biosensors. 2025; 15(4):220. https://doi.org/10.3390/bios15040220
Chicago/Turabian StyleAn, Ling, Yi Liu, and Yaling Liu. 2025. "Advancements in Circulating Tumor Cell Detection for Early Cancer Diagnosis: An Integration of Machine Learning Algorithms with Microfluidic Technologies" Biosensors 15, no. 4: 220. https://doi.org/10.3390/bios15040220
APA StyleAn, L., Liu, Y., & Liu, Y. (2025). Advancements in Circulating Tumor Cell Detection for Early Cancer Diagnosis: An Integration of Machine Learning Algorithms with Microfluidic Technologies. Biosensors, 15(4), 220. https://doi.org/10.3390/bios15040220




