Nano- and Macroscale Imaging of Cholesterol Linoleate and Human Beta Defensin 2-Induced Changes in Pseudomonas aeruginosa Biofilms
Abstract
:1. Introduction
2. Results
2.1. Effects of CL-PL and CL-PL Combined with HBD2 on PA Surface at the Single Cell Level
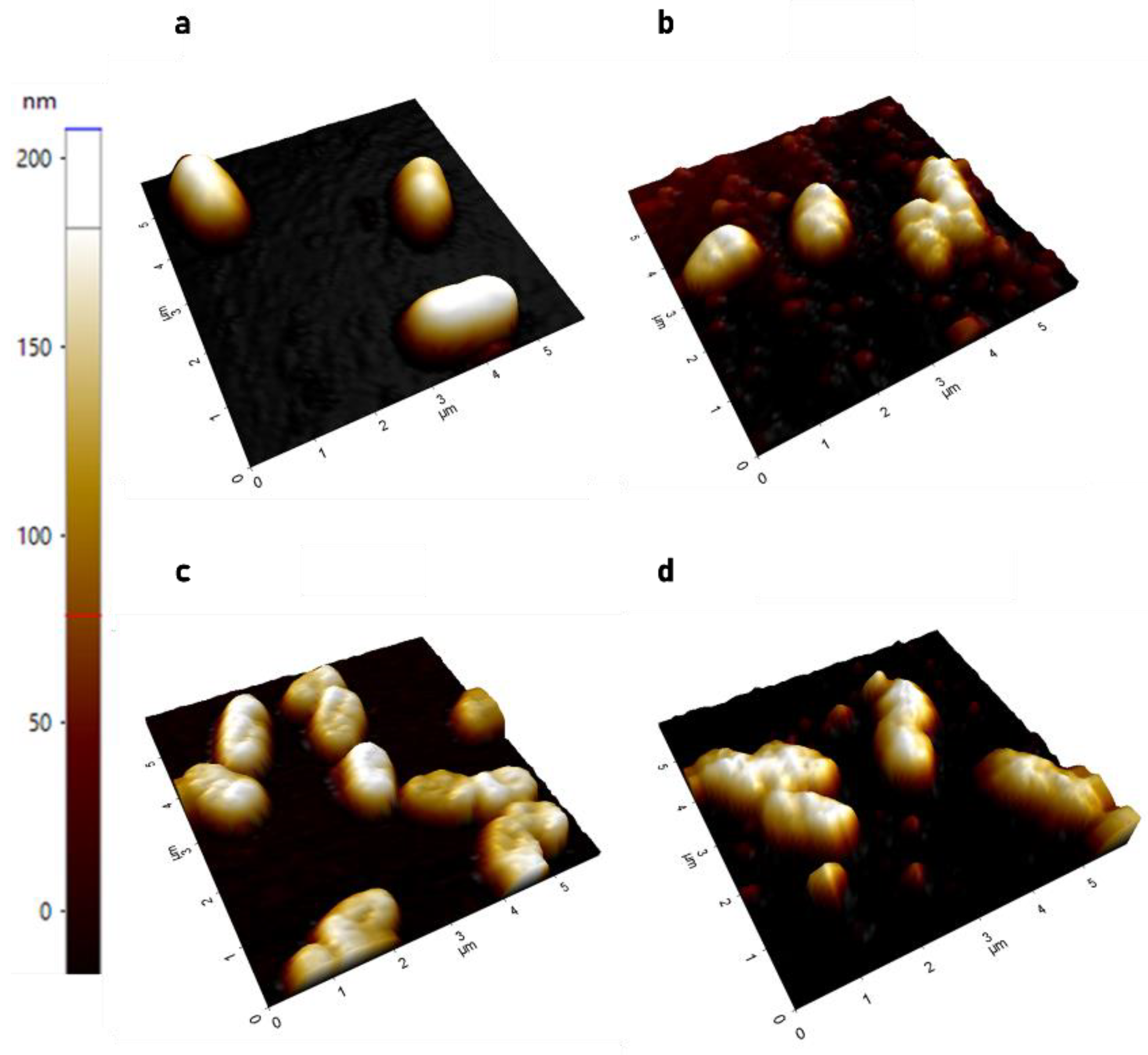
2.1.1. AFM Demonstrates Significant Increase in Surface Roughness in PACF after Treatment with CL-PL Alone and Combined with HBD2
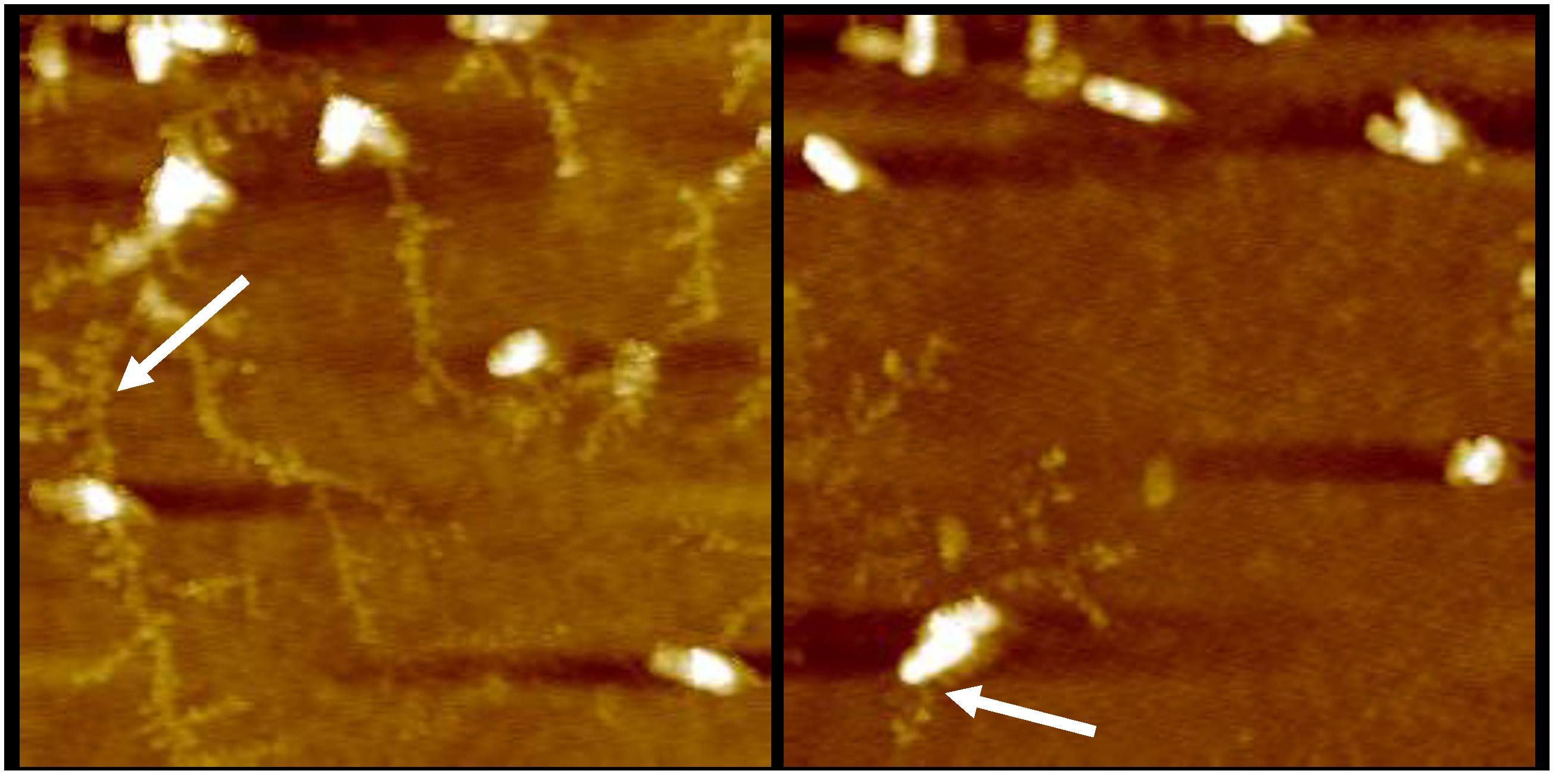
2.1.2. AFM Reveals Loss of Extracellular Network Surrounding PACF after Treatment with CL-PL and HBD2 Alone and Combined
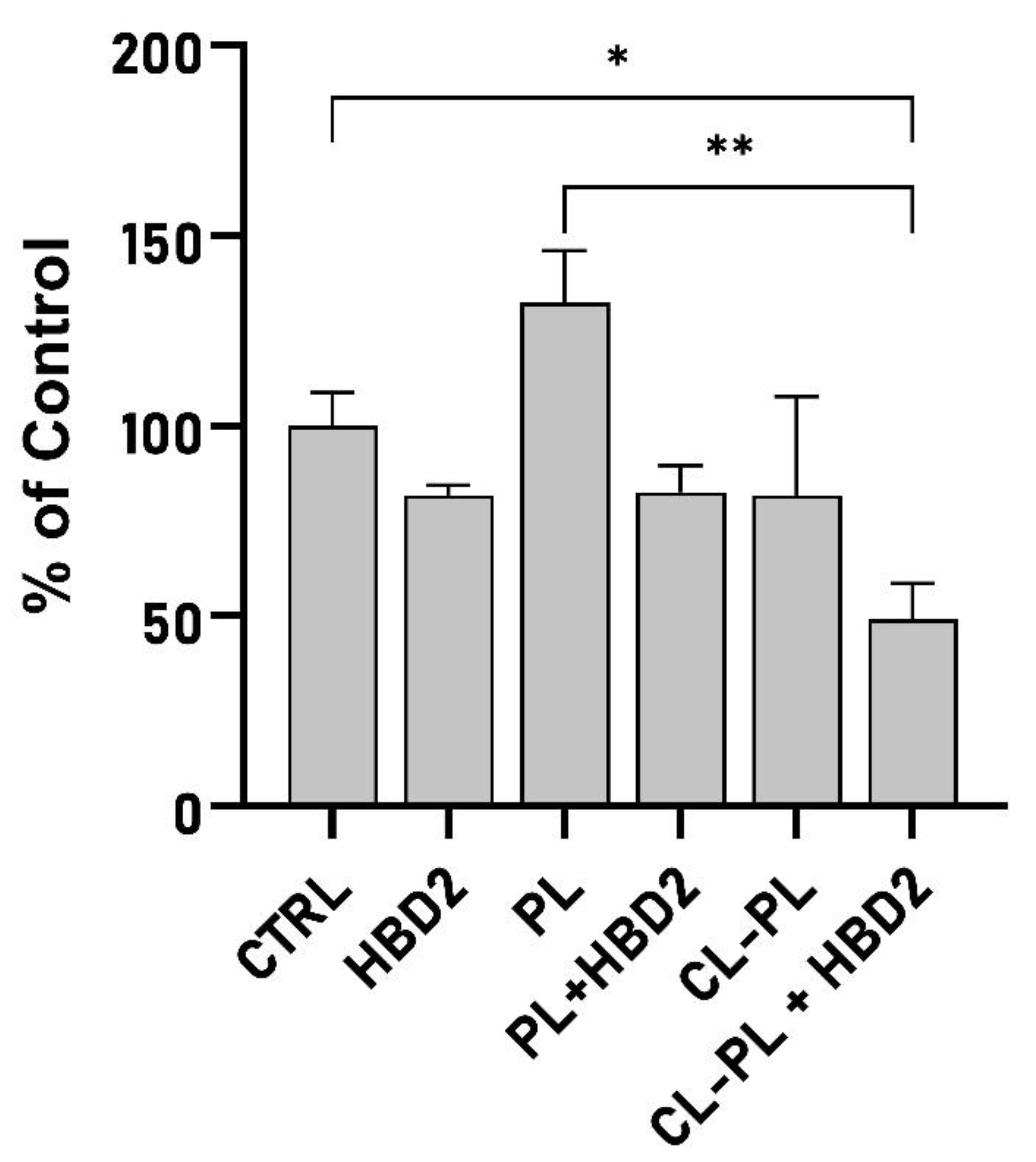
2.2. Effects of CL-PL Combined with HBD2 on PA Biofilm Production and Structure
3. Discussion
4. Materials and Methods
4.1. Antimicrobial Peptide Human Beta Defensin 2 (HBD2)
4.2. Liposome Preparation
4.3. Pseudomonas Aeruginosa
4.4. Atomic Force Microscopy
4.5. Biofilm Staining and Assessment
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

Appendix B
Appendix C

References
- Maunders, E.; Welch, M. Matrix exopolysaccharides; the sticky side of biofilm formation. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef] [Green Version]
- Jones, C.J.; Wozniak, D.J. Psl Produced by Mucoid Pseudomonas aeruginosa Contributes to the Establishment of Biofilms and Immune Evasion. mBio 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leid, J.G.; Willson, C.J.; Shirtliff, M.E.; Hassett, D.J.; Parsek, M.R.; Jeffers, A.K. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J. Immunol. 2005, 175, 7512–7518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otto, M. Bacterial evasion of antimicrobial peptides by biofilm formation. Curr. Top. Microbiol. Immunol. 2006, 306, 251–258. [Google Scholar] [CrossRef]
- Moradali, M.F.; Ghods, S.; Rehm, B.H. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front. Cell. Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Auvynet, C.; Rosenstein, Y. Multifunctional host defense peptides: Antimicrobial peptides, the small yet big players in innate and adaptive immunity. FEBS J. 2009, 276, 6497–6508. [Google Scholar] [CrossRef]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [Green Version]
- Ma, D.C.; Yoon, A.J.; Faull, K.F.; Desharnais, R.; Zemanick, E.T.; Porter, E. Cholesteryl esters are elevated in the lipid fraction of bronchoalveolar lavage fluid collected from pediatric cystic fibrosis patients. PLoS ONE 2015, 10, e0125326. [Google Scholar] [CrossRef]
- Fischer, C.L. Antimicrobial Activity of Host-Derived Lipids. Antibiotics 2020, 9, 75. [Google Scholar] [CrossRef]
- Cederlund, A.; Gudmundsson, G.H.; Agerberth, B. Antimicrobial peptides important in innate immunity. FEBS J. 2011, 278, 3942–3951. [Google Scholar] [CrossRef]
- Haney, E.F.; Mansour, S.C.; Hancock, R.E. Antimicrobial Peptides: An Introduction. Methods Mol. Biol. 2017, 1548, 3–22. [Google Scholar] [CrossRef]
- Fujii, G.; Selsted, M.E.; Eisenberg, D. Defensins promote fusion and lysis of negatively charged membranes. Protein Sci. A Publ. Protein Soc. 1993, 2, 1301–1312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Travkova, O.G.; Moehwald, H.; Brezesinski, G. The interaction of antimicrobial peptides with membranes. Adv. Colloid Interface Sci. 2017, 247, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, V.; Feio, M.J.; Bastos, M. Role of lipids in the interaction of antimicrobial peptides with membranes. Prog. Lipid Res. 2012, 51, 149–177. [Google Scholar] [CrossRef] [PubMed]
- Epand, R.M.; Vogel, H.J. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta 1999, 1462, 11–28. [Google Scholar] [CrossRef] [Green Version]
- Schneider, J.J.; Unholzer, A.; Schaller, M.; Schafer-Korting, M.; Korting, H.C. Human defensins. J. Mol. Med. 2005, 83, 587–595. [Google Scholar] [CrossRef]
- Ganz, T. Defensins: Antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003, 3, 710–720. [Google Scholar] [CrossRef]
- Hazlett, L.; Wu, M. Defensins in innate immunity. Cell Tissue Res. 2011, 343, 175–188. [Google Scholar] [CrossRef]
- Diamond, G.; Bevins, C.L. beta-Defensins: Endogenous antibiotics of the innate host defense response. Clin. Immunol. Immunopathol. 1998, 88, 221–225. [Google Scholar] [CrossRef]
- Schroder, J.M. Epithelial antimicrobial peptides: Innate local host response elements. Cell. Mol. Life Sci. CMLS 1999, 56, 32–46. [Google Scholar] [CrossRef]
- Hoover, D.M.; Rajashankar, K.R.; Blumenthal, R.; Puri, A.; Oppenheim, J.J.; Chertov, O.; Lubkowski, J. The structure of human beta-defensin-2 shows evidence of higher order oligomerization. J. Biol. Chem. 2000, 275, 32911–32918. [Google Scholar] [CrossRef] [Green Version]
- Rohrl, J.; Yang, D.; Oppenheim, J.J.; Hehlgans, T. Human beta-defensin 2 and 3 and their mouse orthologs induce chemotaxis through interaction with CCR2. J. Immunol. 2010, 184, 6688–6694. [Google Scholar] [CrossRef] [Green Version]
- Niyonsaba, F.; Ushio, H.; Nakano, N.; Ng, W.; Sayama, K.; Hashimoto, K.; Nagaoka, I.; Okumura, K.; Ogawa, H. Antimicrobial peptides human beta-defensins stimulate epidermal keratinocyte migration, proliferation and production of proinflammatory cytokines and chemokines. J. Investig. Dermatol. 2007, 127, 594–604. [Google Scholar] [CrossRef] [Green Version]
- Koeninger, L.; Armbruster, N.S.; Brinch, K.S.; Kjaerulf, S.; Andersen, B.; Langnau, C.; Autenrieth, S.E.; Schneidawind, D.; Stange, E.F.; Malek, N.P.; et al. Human β-Defensin 2 Mediated Immune Modulation as Treatment for Experimental Colitis. Front. Immunol. 2020, 11, 93. [Google Scholar] [CrossRef] [Green Version]
- Pinkerton, J.W.; Kim, R.Y.; Koeninger, L.; Armbruster, N.S.; Hansbro, N.G.; Brown, A.C.; Jayaraman, R.; Shen, S.; Malek, N.; Cooper, M.A.; et al. Human β-defensin-2 suppresses key features of asthma in murine models of allergic airways disease. Clin. Exp. Allergy 2021, 51, 120–131. [Google Scholar] [CrossRef]
- Parducho, K.R.; Beadell, B.; Ybarra, T.K.; Bush, M.; Escalera, E.; Trejos, A.T.; Chieng, A.; Mendez, M.; Anderson, C.; Park, H.; et al. The Antimicrobial Peptide Human Beta-Defensin 2 Inhibits Biofilm Production of Pseudomonas aeruginosa without Compromising Metabolic Activity. Front. Immunol. 2020, 11, 805. [Google Scholar] [CrossRef]
- Porter, E. Antimicrobial Lipids: Emerging Effector Molecules of Innate Host Defense. PLoS ONE 2015. [Google Scholar] [CrossRef]
- Thormar, H.; Hilmarsson, H. The role of microbicidal lipids in host defense against pathogens and their potential as therapeutic agents. Chem. Phys. Lipids 2007, 150, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wertz, P.W.; de Szalay, S. Innate Antimicrobial Defense of Skin and Oral Mucosa. Antibiotics 2020, 9, 159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Do, T.Q.; Moshkani, S.; Castillo, P.; Anunta, S.; Pogosyan, A.; Cheung, A.; Marbois, B.; Faull, K.F.; Ernst, W.; Chiang, S.M.; et al. Lipids including cholesteryl linoleate and cholesteryl arachidonate contribute to the inherent antibacterial activity of human nasal fluid. J. Immunol. 2008, 181, 4177–4187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheung Lam, A.H.; Sandoval, N.; Wadhwa, R.; Gilkes, J.; Do, T.Q.; Ernst, W.; Chiang, S.M.; Kosina, S.; Howard Xu, H.; Fujii, G.; et al. Assessment of free fatty acids and cholesteryl esters delivered in liposomes as novel class of antibiotic. BMC Res. Notes 2016, 9, 337. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.T.; Jansen, M.; Yilma, A.N.; Nguyen, A.; Desharnais, R.; Porter, E. Antimicrobial lipids: Novel innate defense molecules are elevated in sinus secretions of patients with chronic rhinosinusitis. Am. J. Rhinol. Allergy 2010, 24, 99–104. [Google Scholar] [CrossRef] [Green Version]
- Nocera, A.L.; Mueller, S.K.; Stephan, J.R.; Hing, L.; Seifert, P.; Han, X.; Lin, D.T.; Amiji, M.M.; Libermann, T.; Bleier, B.S. Exosome swarms eliminate airway pathogens and provide passive epithelial immunoprotection through nitric oxide. J. Allergy Clin. Immunol. 2019, 143, 1525–1535.e1521. [Google Scholar] [CrossRef]
- Martinez, J.G.; Waldon, M.; Huang, Q.; Alvarez, S.; Oren, A.; Sandoval, N.; Du, M.; Zhou, F.; Zenz, A.; Lohner, K.; et al. Membrane-targeted synergistic activity of docosahexaenoic acid and lysozyme against Pseudomonas aeruginosa. Biochem. J. 2009, 419, 193–200. [Google Scholar] [CrossRef] [Green Version]
- Tollin, M.; Bergsson, G.; Kai-Larsen, Y.; Lengqvist, J.; Sjovall, J.; Griffiths, W.; Skuladottir, G.V.; Haraldsson, A.; Jornvall, H.; Gudmundsson, G.H.; et al. Vernix caseosa as a multi-component defence system based on polypeptides, lipids and their interactions. Cell. Mol. Life Sci. CMLS 2005, 62, 2390–2399. [Google Scholar] [CrossRef] [Green Version]
- Akbarzadeh, A.; Rezaei-Sadabady, R.; Davaran, S.; Joo, S.W.; Zarghami, N.; Hanifehpour, Y.; Samiei, M.; Kouhi, M.; Nejati-Koshki, K. Liposome: Classification, preparation, and applications. Nanoscale Res. Lett. 2013, 8, 102. [Google Scholar] [CrossRef] [Green Version]
- Bangham, A.D.; Horne, R.W. Negative staining of phospholipids and their structural modification by surface-active agents as observed in the electron microscope. J. Mol. Biol. 1964, 8, 660–668. [Google Scholar] [CrossRef]
- Bangham, A.D.; Standish, M.M.; Watkins, J.C. Diffusion of univalent ions across the lamellae of swollen phospholipids. J. Mol. Biol. 1965, 13, 238–252. [Google Scholar] [CrossRef]
- Johnson, S.M.; Bangham, A.D. Potassium permeability of single compartment liposomes with and without valinomycin. Biochim. Biophys. Acta 1969, 193, 82–91. [Google Scholar] [CrossRef]
- Daraee, H.; Etemadi, A.; Kouhi, M.; Alimirzalu, S.; Akbarzadeh, A. Application of liposomes in medicine and drug delivery. Artif. Cells Nanomed. Biotechnol. 2016, 44, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Drulis-Kawa, Z.; Dorotkiewicz-Jach, A. Liposomes as delivery systems for antibiotics. Int. J. Pharm. 2010, 387, 187–198. [Google Scholar] [CrossRef]
- Adler-Moore, J.P.; Proffitt, R.T. Amphotericin B lipid preparations: What are the differences? Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2008, 14 (Suppl. S4), 25–36. [Google Scholar] [CrossRef] [Green Version]
- Bekersky, I.; Fielding, R.M.; Dressler, D.E.; Lee, J.W.; Buell, D.N.; Walsh, T.J. Pharmacokinetics, excretion, and mass balance of liposomal amphotericin B (AmBisome) and amphotericin B deoxycholate in humans. Antimicrob. Agents Chemother. 2002, 46, 828–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Wu, W.; Zhang, J.; Xia, X. Antimicrobial lipids in nano-carriers for antibacterial delivery. J. Drug Target. 2020, 28, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Forier, K.; Raemdonck, K.; De Smedt, S.C.; Demeester, J.; Coenye, T.; Braeckmans, K. Lipid and polymer nanoparticles for drug delivery to bacterial biofilms. J. Control. Release 2014, 190, 607–623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meers, P.; Neville, M.; Malinin, V.; Scotto, A.W.; Sardaryan, G.; Kurumunda, R.; Mackinson, C.; James, G.; Fisher, S.; Perkins, W.R. Biofilm penetration, triggered release and in vivo activity of inhaled liposomal amikacin in chronic Pseudomonas aeruginosa lung infections. J. Antimicrob. Chemother. 2008, 61, 859–868. [Google Scholar] [CrossRef]
- Alsteens, D.; Müller, D.J.; Dufrêne, Y.F. Multiparametric Atomic Force Microscopy Imaging of Biomolecular and Cellular Systems. ACC Chem. Res. 2017, 50, 924–931. [Google Scholar] [CrossRef] [PubMed]
- Cole, A.M.; Liao, H.I.; Stuchlik, O.; Tilan, J.; Pohl, J.; Ganz, T. Cationic polypeptides are required for antibacterial activity of human airway fluid. J. Immunol. 2002, 169, 6985–6991. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Parsek, M.R.; Wozniak, D.J.; Ma, L.Z. A spider web strategy of type IV pili-mediated migration to build a fibre-like Psl polysaccharide matrix in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 2013, 15, 2238–2253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Liu, X.; Liu, H.; Zhang, L.; Guo, Y.; Yu, S.; Wozniak, D.J.; Ma, L.Z. The exopolysaccharide Psl-eDNA interaction enables the formation of a biofilm skeleton in Pseudomonas aeruginosa. Environ. Microbiol. Rep. 2015, 7, 330–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drulis-Kawa, Z.; Dorotkiewicz-Jach, A.; Gubernator, J.; Gula, G.; Bocer, T.; Doroszkiewicz, W. The interaction between Pseudomonas aeruginosa cells and cationic PC:Chol:DOTAP liposomal vesicles versus outer-membrane structure and envelope properties of bacterial cell. Int. J. Pharm. 2009, 367, 211–219. [Google Scholar] [CrossRef]
- Wang, Z.; Ma, Y.; Khalil, H.; Wang, R.; Lu, T.; Zhao, W.; Zhang, Y.; Chen, J.; Chen, T. Fusion between fluid liposomes and intact bacteria: Study of driving parameters and in vitro bactericidal efficacy. Int. J. Nanomed. 2016, 11, 4025–4036. [Google Scholar] [CrossRef] [Green Version]
- Li, A.; Lee, P.Y.; Ho, B.; Ding, J.L.; Lim, C.T. Atomic force microscopy study of the antimicrobial action of Sushi peptides on Gram negative bacteria. Biochim. Biophys. Acta 2007, 1768, 411–418. [Google Scholar] [CrossRef] [Green Version]
- Meincken, M.; Holroyd, D.L.; Rautenbach, M. Atomic force microscopy study of the effect of antimicrobial peptides on the cell envelope of Escherichia coli. Antimicrob. Agents Chemother. 2005, 49, 4085–4092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossetto, G.; Bergese, P.; Colombi, P.; Depero, L.E.; Giuliani, A.; Nicoletto, S.F.; Pirri, G. Atomic force microscopy evaluation of the effects of a novel antimicrobial multimeric peptide on Pseudomonas aeruginosa. Nanomedicine 2007, 3, 198–207. [Google Scholar] [CrossRef] [PubMed]
- da Silva, A., Jr.; Teschke, O. Effects of the antimicrobial peptide PGLa on live Escherichia coli. Biochim. Biophys. Acta 2003, 1643, 95–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaske, A.M.; Danila, D.; Queen, M.C.; Golunski, E.; Conyers, J.L. Biological Atomic Force Microscopy for Imaging Gold-Labeled Liposomes on Human Coronary Artery Endothelial Cells. J. Pharm. 2013, 2013, 875906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barken, K.B.; Pamp, S.J.; Yang, L.; Gjermansen, M.; Bertrand, J.J.; Klausen, M.; Givskov, M.; Whitchurch, C.B.; Engel, J.N.; Tolker-Nielsen, T. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 2008, 10, 2331–2343. [Google Scholar] [CrossRef] [PubMed]
- Sheng, G.P.; Yu, H.Q.; Yu, Z. Extraction of extracellular polymeric substances from the photosynthetic bacterium Rhodopseudomonas acidophila. Appl. Microbiol. Biotechnol. 2005, 67, 125–130. [Google Scholar] [CrossRef]
- Cooke, A.C.; Florez, C.; Dunshee, E.B.; Lieber, A.D.; Terry, M.L.; Light, C.J.; Schertzer, J.W. Pseudomonas Quinolone Signal-Induced Outer Membrane Vesicles Enhance Biofilm Dispersion in Pseudomonas aeruginosa. mSphere 2020, 5. [Google Scholar] [CrossRef]
- James, S.A.; Hilal, N.; Wright, C.J. Atomic force microscopy studies of bioprocess engineering surfaces—imaging, interactions and mechanical properties mediating bacterial adhesion. Biotechnol. J. 2017, 12, 1600698. [Google Scholar] [CrossRef] [Green Version]
- Allen, A.; Habimana, O.; Casey, E. The effects of extrinsic factors on the structural and mechanical properties of Pseudomonas fluorescens biofilms: A combined study of nutrient concentrations and shear conditions. Colloids Surf. B Biointerfaces 2018, 165, 127–134. [Google Scholar] [CrossRef]
- Kart, D.; Reçber, T.; Nemutlu, E.; Sagiroglu, M. Sub-Inhibitory Concentrations of Ciprofloxacin Alone and Combinations with Plant-Derived Compounds against P. aeruginosa Biofilms and Their Effects on the Metabolomic Profile of P. aeruginosa Biofilms. Antibiotics 2021, 10, 414. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Lu, L.; Liao, L.; Zhang, B.; Zeng, M.; Zou, K.; Liu, X.; Zhang, M. Establishment of GC-MS method for the determination of Pseudomonas aeruginosa biofilm and its application in metabolite enrichment analysis. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2021, 1179, 122839. [Google Scholar] [CrossRef] [PubMed]
- Drotleff, B.; Roth, S.R.; Henkel, K.; Calderón, C.; Schlotterbeck, J.; Neukamm, M.A.; Lämmerhofer, M. Lipidomic profiling of non-mineralized dental plaque and biofilm by untargeted UHPLC-QTOF-MS/MS and SWATH acquisition. Anal. Bioanal. Chem. 2020, 412, 2303–2314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruhal, R.; Antti, H.; Rzhepishevska, O.; Boulanger, N.; Barbero, D.R.; Wai, S.N.; Uhlin, B.E.; Ramstedt, M. A multivariate approach to correlate bacterial surface properties to biofilm formation by lipopolysaccharide mutants of Pseudomonas aeruginosa. Colloids Surf. B Biointerfaces 2015, 127, 182–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, G.; de Leeuw, E.; Pazgier, M.; Yuan, W.; Zou, G.; Wang, J.; Ericksen, B.; Lu, W.Y.; Lehrer, R.I.; Lu, W. Through the looking glass, mechanistic insights from enantiomeric human defensins. J. Biol. Chem. 2009, 284, 29180–29192. [Google Scholar] [CrossRef] [Green Version]
- Merritt, J.H.; Kadouri, D.E.; O’Toole, G.A. Growing and analyzing static biofilms. Curr. Protoc. Microbiol. 2005, 22, 1B-1. [Google Scholar] [CrossRef] [Green Version]
- Adler, J.; Parmryd, I. Quantifying colocalization: Thresholding, void voxels and the H(coef). PLoS ONE 2014, 9, e111983. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beadell, B.A.; Chieng, A.; Parducho, K.R.; Dai, Z.; Ho, S.O.; Fujii, G.; Wang, Y.; Porter, E. Nano- and Macroscale Imaging of Cholesterol Linoleate and Human Beta Defensin 2-Induced Changes in Pseudomonas aeruginosa Biofilms. Antibiotics 2021, 10, 1279. https://doi.org/10.3390/antibiotics10111279
Beadell BA, Chieng A, Parducho KR, Dai Z, Ho SO, Fujii G, Wang Y, Porter E. Nano- and Macroscale Imaging of Cholesterol Linoleate and Human Beta Defensin 2-Induced Changes in Pseudomonas aeruginosa Biofilms. Antibiotics. 2021; 10(11):1279. https://doi.org/10.3390/antibiotics10111279
Chicago/Turabian StyleBeadell, Brent A., Andy Chieng, Kevin R. Parducho, Zhipeng Dai, Sam On Ho, Gary Fujii, Yixian Wang, and Edith Porter. 2021. "Nano- and Macroscale Imaging of Cholesterol Linoleate and Human Beta Defensin 2-Induced Changes in Pseudomonas aeruginosa Biofilms" Antibiotics 10, no. 11: 1279. https://doi.org/10.3390/antibiotics10111279
APA StyleBeadell, B. A., Chieng, A., Parducho, K. R., Dai, Z., Ho, S. O., Fujii, G., Wang, Y., & Porter, E. (2021). Nano- and Macroscale Imaging of Cholesterol Linoleate and Human Beta Defensin 2-Induced Changes in Pseudomonas aeruginosa Biofilms. Antibiotics, 10(11), 1279. https://doi.org/10.3390/antibiotics10111279





