FtsZ Interactions and Biomolecular Condensates as Potential Targets for New Antibiotics
Abstract
1. Introduction
2. Detection and Quantification of Direct FtsZ-Drug Binding in Solution
3. Methods to Identify Drugs Targeting FtsZ Polymerization and Their Mechanisms
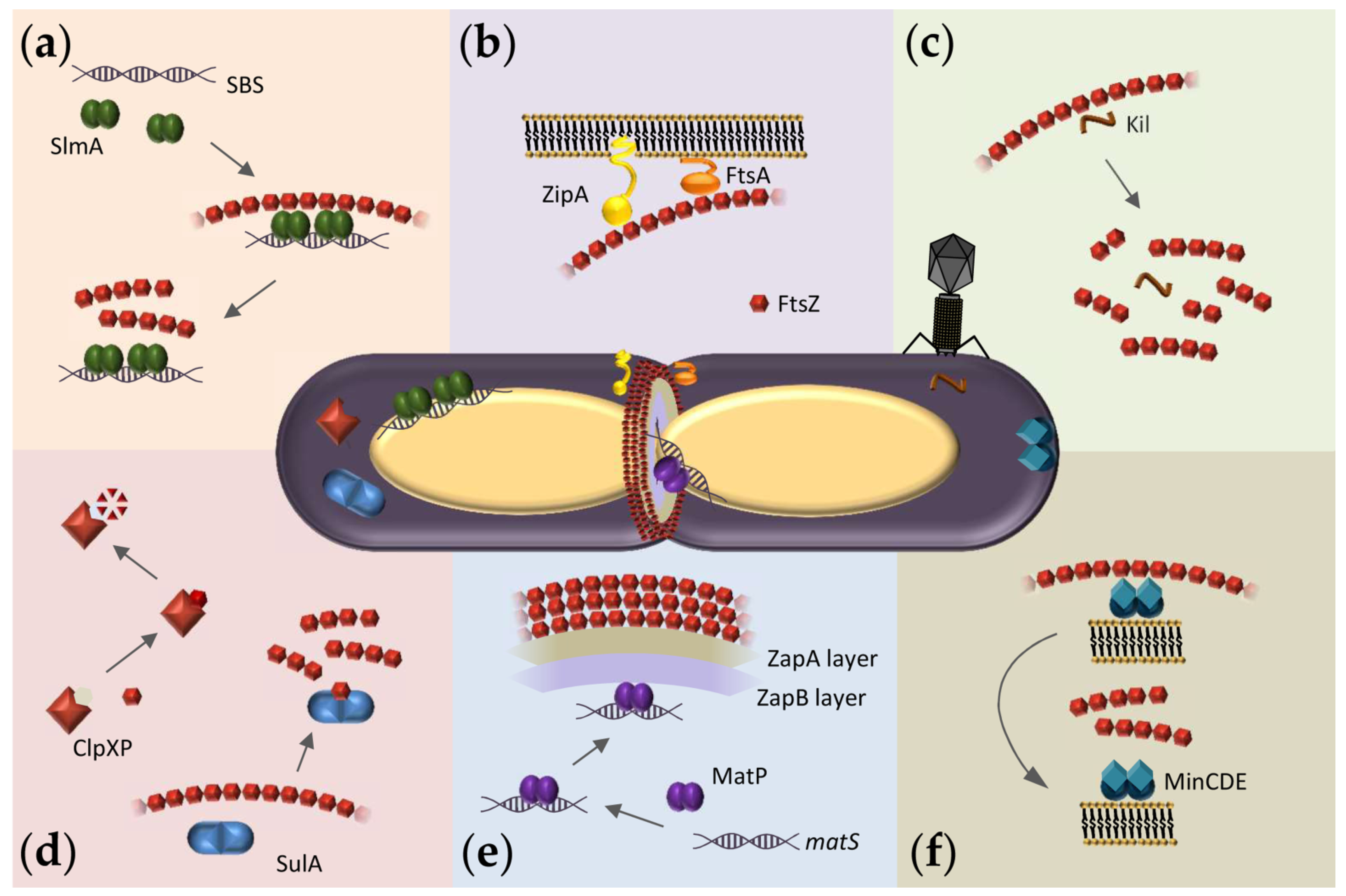
4. Exploiting the Interactions of FtsZ with Binding Partners in Solution to Discover New Antimicrobials
5. Reconstruction of Cellular FtsZ Subsystems
6. Could FtsZ Biomolecular Condensates Help Understand Persisters?
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Silber, N.; Matos de Opitz, C.L.; Mayer, C.; Sass, P. Cell division protein FtsZ: From structure and mechanism to antibiotic target. Future Microbiol. 2020, 15, 801–831. [Google Scholar] [CrossRef]
- Haeusser, D.P.; Margolin, W. Splitsville: Structural and functional insights into the dynamic bacterial Z ring. Nat. Rev. Microbiol. 2016, 14, 305–319. [Google Scholar] [CrossRef]
- Nogales, E.; Wolf, S.G.; Downing, K.H. Structure of the alpha beta tubulin dimer by electron crystallography. Nature 1998, 391, 199–203. [Google Scholar] [CrossRef]
- Kusuma, K.D.; Payne, M.; Ung, A.T.; Bottomley, A.L.; Harry, E.J. FtsZ as an Antibacterial Target: Status and Guidelines for Progressing This Avenue. Acs Infect Dis. 2019, 5, 1279–1294. [Google Scholar] [CrossRef]
- Andreu, J.M.; Schaffner-Barbero, C.; Huecas, S.; Alonso, D.; Lopez-Rodriguez, M.L.; Ruiz-Avila, L.B.; Núñez-Ramírez, R.; Llorca, O.; Martín-Galiano, A.J. The antibacterial cell division inhibitor PC190723 is an FtsZ polymer-stabilizing agent that induces filament assembly and condensation. J. Biol. Chem. 2010, 285, 14239–14246. [Google Scholar] [CrossRef]
- Vaughan, S.; Wickstead, B.; Gull, K.; Addinall, S.G. Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. J. Mol. Evol. 2004, 58, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Erickson, H.P.; Anderson, D.E.; Osawa, M. FtsZ in bacterial cytokinesis: Cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev. 2010, 74, 504–528. [Google Scholar] [CrossRef] [PubMed]
- Blasios, V.; Bisson-Filho, A.W.; Castellen, P.; Nogueira, M.L.; Bettini, J.; Portugal, R.V.; Zeri, A.C.; Gueiros-Filho, F.J. Genetic and biochemical characterization of the MinC-FtsZ interaction in Bacillus subtilis. PLoS ONE 2013, 8, e60690. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Mukherjee, A.; Pichoff, S.; Lutkenhaus, J. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. USA 1999, 96, 14819–14824. [Google Scholar] [CrossRef] [PubMed]
- Dajkovic, A.; Lan, G.; Sun, S.X.; Wirtz, D.; Lutkenhaus, J. MinC spatially controls bacterial cytokinesis by antagonizing the scaffolding function of FtsZ. Curr. Biol. 2008, 18, 235–244. [Google Scholar] [CrossRef]
- Bernhardt, T.G.; de Boer, P.A. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over Chromosomes in E. coli. Mol. Cell 2005, 18, 555–564. [Google Scholar] [CrossRef]
- Du, S.; Lutkenhaus, J. SlmA antagonism of FtsZ assembly employs a two-pronged mechanism like MinCD. PLoS Genet. 2014, 10, e1004460. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, M.A.; Zeng, W. Structures of the nucleoid occlusion protein SlmA bound to DNA and the C-terminal domain of the cytoskeletal protein FtsZ. Proc. Natl. Acad. Sci. USA 2016, 113, 4988–4993. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Margolin, W. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol. 1999, 181, 7531–7544. [Google Scholar] [CrossRef]
- Wang, X.; Huang, J.; Mukherjee, A.; Cao, C.; Lutkenhaus, J. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J. Bacteriol. 1997, 179, 5551–5559. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Pearce, K.H.; Payne, D.J. A conserved residue at the extreme C-terminus of FtsZ is critical for the FtsA-FtsZ interaction in Staphylococcus aureus. Biochem. Biophys. Res. Commun. 2000, 270, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Pazos, M.; Natale, P.; Vicente, M. A specific role for the ZipA protein in cell division: Stabilization of the FtsZ protein. J. Biol. Chem. 2013, 288, 3219–3226. [Google Scholar] [CrossRef]
- Hale, C.A.; de Boer, P.A. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 1997, 88, 175–185. [Google Scholar] [CrossRef]
- Mosyak, L.; Zhang, Y.; Glasfeld, E.; Haney, S.; Stahl, M.; Seehra, J.; Somers, W.S. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 2000, 19, 3179–3191. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, C.; Natale, P.; Cueto, L.; Vicente, M. The keepers of the ring: Regulators of FtsZ assembly. FEMS Microbiol. Rev. 2016, 40, 57–67. [Google Scholar] [CrossRef]
- Camberg, J.L.; Hoskins, J.R.; Wickner, S. ClpXP protease degrades the cytoskeletal protein, FtsZ, and modulates FtsZ polymer dynamics. Proc. Natl. Acad. Sci. USA 2009, 106, 10614–10619. [Google Scholar] [CrossRef]
- Mannik, J.; Bailey, M.W. Spatial coordination between chromosomes and cell division proteins in Escherichia coli. Front. Microbiol. 2015, 6, 306. [Google Scholar] [CrossRef] [PubMed]
- Mingorance, J.; Rivas, G.; Vélez, M.; Gómez-Puertas, P.; Vicente, M. Strong FtsZ is with the force: Mechanisms to constrict bacteria. Trends Microbiol. 2010, 18, 348–356. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Lutkenhaus, J. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 1998, 17, 462–469. [Google Scholar] [CrossRef]
- Du, S.; Lutkenhaus, J. At the Heart of Bacterial Cytokinesis: The Z Ring. Trends Microbiol. 2019, 27, 781–791. [Google Scholar] [CrossRef]
- Monterroso, B.; Ahijado-Guzman, R.; Reija, B.; Alfonso, C.; Zorrilla, S.; Minton, A.P.; Rivas, G. Mg(2+)-linked self-assembly of FtsZ in the presence of GTP or a GTP analogue involves the concerted formation of a narrow size distribution of oligomeric species. Biochemistry 2012, 51, 4541–4550. [Google Scholar] [CrossRef]
- Ahijado-Guzman, R.; Alfonso, C.; Reija, B.; Salvarelli, E.; Mingorance, J.; Zorrilla, S.; Monterroso, B.; Rivas, G. Control by potassium of the size distribution of Escherichia coli FtsZ polymers is independent of GTPase activity. J. Biol. Chem. 2013, 288, 27358–27365. [Google Scholar] [CrossRef] [PubMed]
- Huecas, S.; Llorca, O.; Boskovic, J.; Martin-Benito, J.; Valpuesta, J.M.; Andreu, J.M. Energetics and geometry of FtsZ polymers: Nucleated self-assembly of single protofilaments. Biophys. J. 2008, 94, 1796–1806. [Google Scholar] [CrossRef]
- Turner, D.J.; Portman, I.; Dafforn, T.R.; Rodger, A.; Roper, D.I.; Smith, C.J.; Turner, M.S. The mechanics of FtsZ fibers. Biophys. J. 2012, 102, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Erickson, H.P. Rapid in vitro assembly dynamics and subunit turnover of FtsZ demonstrated by fluorescence resonance energy transfer. J. Biol. Chem. 2005, 280, 22549–22554. [Google Scholar] [CrossRef] [PubMed]
- González, J.M.; Jiménez, M.; Vélez, M.; Mingorance, J.; Andreu, J.M.; Vicente, M.; Rivas, G. Essential cell division protein FtsZ assembles into one monomer-thick ribbons under conditions resembling the crowded intracellular environment. J. Biol. Chem. 2003, 278, 37664–37671. [Google Scholar] [CrossRef]
- Monterroso, B.; Reija, B.; Jimenez, M.; Zorrilla, S.; Rivas, G. Charged Molecules Modulate the Volume Exclusion Effects Exerted by Crowders on FtsZ Polymerization. PLoS ONE 2016, 11, e0149060. [Google Scholar] [CrossRef] [PubMed]
- Popp, D.; Iwasa, M.; Narita, A.; Erickson, H.P.; Maeda, Y. FtsZ condensates: An in vitro electron microscopy study. Biopolymers 2009, 91, 340–350. [Google Scholar] [CrossRef]
- Araujo-Bazan, L.; Ruiz-Avila, L.B.; Andreu, D.; Huecas, S.; Andreu, J.M. Cytological Profile of Antibacterial FtsZ Inhibitors and Synthetic Peptide MciZ. Front. Microbiol. 2016, 7, 1558. [Google Scholar] [CrossRef]
- Rivas, G.; López, A.; Mingorance, J.; Ferrándiz, M.J.; Zorrilla, S.; Minton, A.P.; Vicente, M.; Andreu, J.M. Magnesium-induced linear self-association of the FtsZ bacterial cell division protein monomer. The primary steps for FtsZ assembly. J. Biol. Chem. 2000, 275, 11740–11749. [Google Scholar] [CrossRef] [PubMed]
- Rivas, G.; Fernandez, J.A.; Minton, A.P. Direct observation of the enhancement of noncooperative protein self-assembly by macromolecular crowding: Indefinite linear self-association of bacterial cell division protein FtsZ. Proc. Natl. Acad. Sci. USA 2001, 98, 3150–3155. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Rocamora, V.M.; Reija, B.; Garcia, C.; Natale, P.; Alfonso, C.; Minton, A.P.; Zorrilla, S.; Rivas, G.; Vicente, M. Dynamic interaction of the Escherichia coli cell division ZipA and FtsZ proteins evidenced in nanodiscs. J. Biol. Chem. 2012, 287, 30097–30104. [Google Scholar] [CrossRef]
- Hernandez-Rocamora, V.M.; Garcia-Montanes, C.; Reija, B.; Monterroso, B.; Margolin, W.; Alfonso, C.; Zorrilla, S.; Rivas, G. MinC protein shortens FtsZ protofilaments by preferentially interacting with GDP-bound subunits. J. Biol. Chem. 2013, 288, 24625–24635. [Google Scholar] [CrossRef] [PubMed]
- Tonthat, N.K.; Arold, S.T.; Pickering, B.F.; Van Dyke, M.W.; Liang, S.; Lu, Y.; Beuria, T.K.; Margolin, W.; Schumacher, M.A. Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check. EMBO J. 2011, 30, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Milam, S.L.; Erickson, H.P. SulA inhibits assembly of FtsZ by a simple sequestration mechanism. Biochemistry 2012, 51, 3100–3109. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Rocamora, V.M.; Alfonso, C.; Margolin, W.; Zorrilla, S.; Rivas, G. Evidence That Bacteriophage lambda Kil Peptide Inhibits Bacterial Cell Division by Disrupting FtsZ Protofilaments and Sequestering Protein Subunits. J. Biol. Chem. 2015, 290, 20325–20335. [Google Scholar] [CrossRef]
- Monterroso, B.; Zorrilla, S.; Sobrinos-Sanguino, M.; Robles-Ramos, M.A.; Lopez-Alvarez, M.; Margolin, W.; Keating, C.D.; Rivas, G. Bacterial FtsZ protein forms phase-separated condensates with its nucleoid-associated inhibitor SlmA. EMBO Rep. 2019, 20, e45946. [Google Scholar] [CrossRef]
- Robles-Ramos, M.A.; Zorrilla, S.; Alfonso, C.; Margolin, W.; Rivas, G.; Monterroso, B. Assembly of bacterial cell division protein FtsZ into dynamic biomolecular condensates. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118986. [Google Scholar] [CrossRef]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Alberti, S.; Dormann, D. Liquid-Liquid Phase Separation in Disease. Annu. Rev. Genet. 2019, 53, 171–194. [Google Scholar] [CrossRef] [PubMed]
- Abbondanzieri, E.A.; Meyer, A.S. More than just a phase: The search for membraneless organelles in the bacterial cytoplasm. Curr. Genet. 2019, 65, 691–694. [Google Scholar] [CrossRef] [PubMed]
- Azaldegui, C.A.; Vecchiarelli, A.G.; Biteen, J.S. The emergence of phase separation as an organizing principle in bacteria. Biophys. J. 2020, 119, 1–16. [Google Scholar] [CrossRef]
- den Blaauwen, T.; Andreu, J.M.; Monasterio, O. Bacterial cell division proteins as antibiotic targets. Bioorganic Chem. 2014, 55, 27–38. [Google Scholar] [CrossRef]
- Vollmer, W. The prokaryotic cytoskeleton: A putative target for inhibitors and antibiotics? Appl. Microbiol. Biotechnol. 2006, 73, 37–47. [Google Scholar] [CrossRef]
- Lock, R.L.; Harry, E.J. Cell-division inhibitors: New insights for future antibiotics. Nat. Rev. Drug Discov. 2008, 7, 324–338. [Google Scholar] [CrossRef]
- Fisher, R.A.; Gollan, B.; Helaine, S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 2017, 15, 453–464. [Google Scholar] [CrossRef]
- US Food & Drug Administration. Available online: https://www.fda.gov/drugs/development-approval-process-drugs (accessed on 13 January 2021).
- European Medicines Agency. Available online: https://www.ema.europa.eu/en/about-us/what-we-do/authorisation-medicines/how-ema-evaluates-medicines (accessed on 13 January 2021).
- Monterroso, B.; Alfonso, C.; Zorrilla, S.; Rivas, G. Combined analytical ultracentrifugation, light scattering and fluorescence spectroscopy studies on the functional associations of the bacterial division FtsZ protein. Methods 2013, 59, 349–362. [Google Scholar] [CrossRef]
- Lagny, T.J.; Bassereau, P. Bioinspired membrane-based systems for a physical approach of cell organization and dynamics: Usefulness and limitations. Interface Focus 2015, 5, 20150038. [Google Scholar] [CrossRef]
- Rivas, G.; Vogel, S.K.; Schwille, P. Reconstitution of cytoskeletal protein assemblies for large-scale membrane transformation. Curr. Opin. Chem. Biol. 2014, 22, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Kretschmer, S.; Ganzinger, K.A.; Franquelim, H.G.; Schwille, P. Synthetic cell division via membrane-transforming molecular assemblies. BMC Biol. 2019, 17, 43. [Google Scholar] [CrossRef] [PubMed]
- Monterroso, B.; Robles-Ramos, M.A.; Zorrilla, S.; Rivas, G. Reconstituting bacterial cell division assemblies in crowded, phase-separated media. Methods Enzymol. 2021, 646, 19–49. [Google Scholar] [CrossRef] [PubMed]
- Ur Rahman, M.; Wang, P.; Wang, N.; Chen, Y. A key bacterial cytoskeletal cell division protein FtsZ as a novel therapeutic antibacterial drug target. Bosn. J. Basic Med. Sci. 2020, 20, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Casiraghi, A.; Suigo, L.; Valoti, E.; Straniero, V. Targeting Bacterial Cell Division: A Binding Site-Centered Approach to the Most Promising Inhibitors of the Essential Protein FtsZ. Antibiotics 2020, 9, 69. [Google Scholar] [CrossRef] [PubMed]
- Tripathy, S.; Sahu, S.K. FtsZ inhibitors as a new genera of antibacterial agents. Bioorganic Chem. 2019, 91, 103169. [Google Scholar] [CrossRef] [PubMed]
- Jameson, D.M.; Mocz, G. Fluorescence polarization/anisotropy approaches to study protein-ligand interactions: Effects of errors and uncertainties. Methods Mol. Biol. 2005, 305, 301–322. [Google Scholar] [CrossRef] [PubMed]
- Royer, C.A.; Scarlata, S.F. Fluorescence approaches to quantifying biomolecular interactions. Methods Enzymol. 2008, 450, 79–106. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, Q.; Berezin, M.Y. Fluorescence anisotropy (polarization): From drug screening to precision medicine. Expert Opin. Drug Discov. 2015, 10, 1145–1161. [Google Scholar] [CrossRef]
- Ferrer-Gonzalez, E.; Fujita, J.; Yoshizawa, T.; Nelson, J.M.; Pilch, A.J.; Hillman, E.; Ozawa, M.; Kuroda, N.; Al-Tameemi, H.M.; Boyd, J.M.; et al. Structure-Guided Design of a Fluorescent Probe for the Visualization of FtsZ in Clinically Important Gram-Positive and Gram-Negative Bacterial Pathogens. Sci. Rep. 2019, 9, 20092. [Google Scholar] [CrossRef] [PubMed]
- Nova, E.; Montecinos, F.; Brunet, J.E.; Lagos, R.; Monasterio, O. 4’,6-Diamidino-2-phenylindole (DAPI) induces bundling of Escherichia coli FtsZ polymers inhibiting the GTPase activity. Arch. Biochem. Biophys. 2007, 465, 315–319. [Google Scholar] [CrossRef]
- Kaul, M.; Parhi, A.K.; Zhang, Y.; LaVoie, E.J.; Tuske, S.; Arnold, E.; Kerrigan, J.E.; Pilch, D.S. A bactericidal guanidinomethyl biaryl that alters the dynamics of bacterial FtsZ polymerization. J. Med. Chem. 2012, 55, 10160–10176. [Google Scholar] [CrossRef]
- Huecas, S.; Schaffner-Barbero, C.; Garcia, W.; Yebenes, H.; Palacios, J.M.; Diaz, J.F.; Menendez, M.; Andreu, J.M. The interactions of cell division protein FtsZ with guanine nucleotides. J. Biol. Chem. 2007, 282, 37515–37528. [Google Scholar] [CrossRef]
- Ruiz-Avila, L.B.; Huecas, S.; Artola, M.; Vergonos, A.; Ramirez-Aportela, E.; Cercenado, E.; Barasoain, I.; Vazquez-Villa, H.; Martin-Fontecha, M.; Chacon, P.; et al. Synthetic inhibitors of bacterial cell division targeting the GTP-binding site of FtsZ. ACS Chem. Biol. 2013, 8, 2072–2083. [Google Scholar] [CrossRef] [PubMed]
- Freyer, M.W.; Lewis, E.A. Isothermal titration calorimetry: Experimental design, data analysis, and probing macromolecule/ligand binding and kinetic interactions. Methods Cell Biol. 2008, 84, 79–113. [Google Scholar] [CrossRef]
- Araujo-Bazan, L.; Huecas, S.; Valle, J.; Andreu, D.; Andreu, J.M. Synthetic developmental regulator MciZ targets FtsZ across Bacillus species and inhibits bacterial division. Mol. Microbiol. 2019, 111, 965–980. [Google Scholar] [CrossRef] [PubMed]
- Domadia, P.; Swarup, S.; Bhunia, A.; Sivaraman, J.; Dasgupta, D. Inhibition of bacterial cell division protein FtsZ by cinnamaldehyde. Biochem. Pharmacol. 2007, 74, 831–840. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Lutkenhaus, J. Analysis of FtsZ assembly by light scattering and determination of the role of divalent metal cations. J. Bacteriol. 1999, 181, 823–832. [Google Scholar] [CrossRef]
- Lui, H.K.; Gao, W.; Cheung, K.C.; Jin, W.B.; Sun, N.; Kan, J.W.Y.; Wong, I.L.K.; Chiou, J.; Lin, D.; Chan, E.W.C.; et al. Boosting the efficacy of anti-MRSA beta-lactam antibiotics via an easily accessible, non-cytotoxic and orally bioavailable FtsZ inhibitor. Eur. J. Med. Chem. 2019, 163, 95–115. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.W.; Wu, L.J.; Czaplewski, L.G.; Errington, J. Multiple effects of benzamide antibiotics on FtsZ function. Mol. Microbiol. 2011, 80, 68–84. [Google Scholar] [CrossRef] [PubMed]
- Haeusser, D.P.; Hoashi, M.; Weaver, A.; Brown, N.; Pan, J.; Sawitzke, J.A.; Thomason, L.C.; Court, D.L.; Margolin, W. The Kil peptide of bacteriophage lambda blocks Escherichia coli cytokinesis via ZipA-dependent inhibition of FtsZ assembly. PLoS Genet. 2014, 10, e1004217. [Google Scholar] [CrossRef]
- Anderson, D.E.; Kim, M.B.; Moore, J.T.; O’Brien, T.E.; Sorto, N.A.; Grove, C.I.; Lackner, L.L.; Ames, J.B.; Shaw, J.T. Comparison of small molecule inhibitors of the bacterial cell division protein FtsZ and identification of a reliable cross-species inhibitor. Acs Chem. Biol. 2012, 7, 1918–1928. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schmitz, K.S. An Introduction to Dynamic Light Scattering by Macromolecules; Academic Press: Boston, MA, USA, 1990. [Google Scholar]
- Hanlon, A.D.; Larkin, M.I.; Reddick, R.M. Free-solution, label-free protein-protein interactions characterized by dynamic light scattering. Biophys. J. 2010, 98, 297–304. [Google Scholar] [CrossRef]
- Hou, S.; Wieczorek, S.A.; Kaminski, T.S.; Ziebacz, N.; Tabaka, M.; Sorto, N.A.; Foss, M.H.; Shaw, J.T.; Thanbichler, M.; Weibel, D.B.; et al. Characterization of Caulobacter crescentus FtsZ protein using dynamic light scattering. J. Biol. Chem. 2012, 287, 23878–23886. [Google Scholar] [CrossRef]
- Haydon, D.J.; Stokes, N.R.; Ure, R.; Galbraith, G.; Bennett, J.M.; Brown, D.R.; Baker, P.J.; Barynin, V.V.; Rice, D.W.; Sedelnikova, S.E.; et al. An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science 2008, 321, 1673–1675. [Google Scholar] [CrossRef] [PubMed]
- Di Somma, A.; Avitabile, C.; Cirillo, A.; Moretta, A.; Merlino, A.; Paduano, L.; Duilio, A.; Romanelli, A. The antimicrobial peptide Temporin L impairs E. coli cell division by interacting with FtsZ and the divisome complex. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129606. [Google Scholar] [CrossRef]
- Caplan, M.R.; Erickson, H.P. Apparent cooperative assembly of the bacterial cell division protein FtsZ demonstrated by isothermal titration calorimetry. J. Biol. Chem. 2003, 278, 13784–13788. [Google Scholar] [CrossRef] [PubMed]
- iZon Homepage. Available online: https://izon.com/qnano-gold/ (accessed on 13 January 2021).
- Gross-Rother, J.; Blech, M.; Preis, E.; Bakowsky, U.; Garidel, P. Particle Detection and Characterization for Biopharmaceutical Applications: Current Principles of Established and Alternative Techniques. Pharmaceutics 2020, 12, 1112. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, A.; Kinjo, M. State-of-the-Art Fluorescence Fluctuation-Based Spectroscopic Techniques for the Study of Protein Aggregation. Int. J. Mol. Sci. 2018, 19, 964. [Google Scholar] [CrossRef]
- Wachsmuth, M.; Conrad, C.; Bulkescher, J.; Koch, B.; Mahen, R.; Isokane, M.; Pepperkok, R.; Ellenberg, J. High-throughput fluorescence correlation spectroscopy enables analysis of proteome dynamics in living cells. Nat. Biotechnol. 2015, 33, 384–389. [Google Scholar] [CrossRef] [PubMed]
- Bacia, K.; Schwille, P. Practical guidelines for dual-color fluorescence cross-correlation spectroscopy. Nat. Protoc. 2007, 2, 2842–2856. [Google Scholar] [CrossRef] [PubMed]
- Schwille, P.; Meyer-Almes, F.J.; Rigler, R. Dual-color fluorescence cross-correlation spectroscopy for multicomponent diffusional analysis in solution. Biophys. J. 1997, 72, 1878–1886. [Google Scholar] [CrossRef]
- Mikuni, S.; Kodama, K.; Sasaki, A.; Kohira, N.; Maki, H.; Munetomo, M.; Maenaka, K.; Kinjo, M. Screening for FtsZ Dimerization Inhibitors Using Fluorescence Cross-Correlation Spectroscopy and Surface Resonance Plasmon Analysis. PLoS ONE 2015, 10, e0130933. [Google Scholar] [CrossRef] [PubMed]
- Reija, B.; Monterroso, B.; Jimenez, M.; Vicente, M.; Rivas, G.; Zorrilla, S. Development of a homogeneous fluorescence anisotropy assay to monitor and measure FtsZ assembly in solution. Anal. Biochem. 2011, 418, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Kenny, C.H.; Ding, W.; Kelleher, K.; Benard, S.; Dushin, E.G.; Sutherland, A.G.; Mosyak, L.; Kriz, R.; Ellestad, G. Development of a fluorescence polarization assay to screen for inhibitors of the FtsZ/ZipA interaction. Anal. Biochem. 2003, 323, 224–233. [Google Scholar] [CrossRef] [PubMed]
- Cabre, E.J.; Monterroso, B.; Alfonso, C.; Sanchez-Gorostiaga, A.; Reija, B.; Jimenez, M.; Vicente, M.; Zorrilla, S.; Rivas, G. The Nucleoid Occlusion SlmA protein accelerates the disassembly of the FtsZ protein polymers without affecting their GTPase activity. PLoS ONE 2015, 10, e0126434. [Google Scholar] [CrossRef] [PubMed]
- Nag, D.; Chatterjee, A.; Chakrabarti, G. FtsA-FtsZ interaction in Vibrio cholerae causes conformational change of FtsA resulting in inhibition of ATP hydrolysis and polymerization. Int. J. Biol. Macromol. 2020, 142, 18–32. [Google Scholar] [CrossRef]
- Okuno, T.; Ogoh, M.; Tanina, H.; Funasaki, N.; Kogure, K. Direct monitoring of interaction between Escherichia coli proteins, MinC and monomeric FtsZ, in solution. Biol. Pharm. Bull. 2009, 32, 1473–1475. [Google Scholar] [CrossRef]
- Park, K.T.; Dajkovic, A.; Wissel, M.; Du, S.; Lutkenhaus, J. MinC and FtsZ mutant analysis provides insight into MinC/MinD-mediated Z ring disassembly. J. Biol. Chem. 2018, 293, 5834–5846. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, M.J.; Smith, I.; Parker, I.; Bootman, M.D. Fluorescence microscopy. Cold Spring Harb. Protoc. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Silber, N.; Mayer, C.; de Opitz, C.L.M.; Sass, P. Antibiotic-induced degradation of FtsZ reveals distinct stages of Bacillus subtilis FtsZ ring assembly and constriction. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ray, S.; Jindal, B.; Kunal, K.; Surolia, A.; Panda, D. BT-benzo-29 inhibits bacterial cell proliferation by perturbing FtsZ assembly. FEBS J. 2015, 282, 4015–4033. [Google Scholar] [CrossRef]
- Nath, A.; Atkins, W.M.; Sligar, S.G. Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry 2007, 46, 2059–2069. [Google Scholar] [CrossRef]
- Sobrinos-Sanguino, M.; Zorrilla, S.; Monterroso, B.; Minton, A.P.; Rivas, G. Nucleotide and receptor density modulate binding of bacterial division FtsZ protein to ZipA containing lipid-coated microbeads. Sci. Rep. 2017, 7, 13707. [Google Scholar] [CrossRef]
- Martos, A.; Monterroso, B.; Zorrilla, S.; Reija, B.; Alfonso, C.; Mingorance, J.; Rivas, G.; Jimenez, M. Isolation, characterization and lipid-binding properties of the recalcitrant FtsA division protein from Escherichia coli. PLoS ONE 2012, 7, e39829. [Google Scholar] [CrossRef]
- Robles-Ramos, M.A.; Margolin, W.; Sobrinos-Sanguino, M.; Alfonso, C.; Rivas, G.; Monterroso, B.; Zorrilla, S. The nucleoid occlusion protein SlmA binds to lipid membranes. mBio 2020, 11, e02094-20. [Google Scholar] [CrossRef]
- Arumugam, S.; Petrasek, Z.; Schwille, P. MinCDE exploits the dynamic nature of FtsZ filaments for its spatial regulation. Proc. Natl. Acad. Sci. USA 2014, 111, E1192–E1200. [Google Scholar] [CrossRef]
- Loose, M.; Mitchison, T.J. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat. Cell Biol. 2014, 16, 38–46. [Google Scholar] [CrossRef]
- Krupka, M.; Sobrinos-Sanguino, M.; Jimenez, M.; Rivas, G.; Margolin, W. Escherichia coli ZipA Organizes FtsZ Polymers into Dynamic Ring-Like Protofilament Structures. mBio 2018, 9. [Google Scholar] [CrossRef]
- Baranova, N.; Radler, P.; Hernandez-Rocamora, V.M.; Alfonso, C.; Lopez-Pelegrin, M.; Rivas, G.; Vollmer, W.; Loose, M. Diffusion and capture permits dynamic coupling between treadmilling FtsZ filaments and cell division proteins. Nat. Microbiol. 2020, 5, 407–417. [Google Scholar] [CrossRef]
- Godino, E.; Lopez, J.N.; Foschepoth, D.; Cleij, C.; Doerr, A.; Castella, C.F.; Danelon, C. De novo synthesized Min proteins drive oscillatory liposome deformation and regulate FtsA-FtsZ cytoskeletal patterns. Nat. Commun. 2019, 10, 4969. [Google Scholar] [CrossRef] [PubMed]
- Ahijado-Guzman, R.; Menten, J.; Prasad, J.; Lambertz, C.; Rivas, G.; Sonnichsen, C. Plasmonic Nanosensors for the Determination of Drug Effectiveness on Membrane Receptors. ACS Appl. Mater. Interfaces 2017, 9, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Ahijado-Guzman, R.; Gomez-Puertas, P.; Alvarez-Puebla, R.A.; Rivas, G.; Liz-Marzan, L.M. Surface-Enhanced Raman scattering-based detection of the interactions between the essential cell division FtsZ protein and bacterial membrane elements. ACS Nano 2012, 6, 7514–7520. [Google Scholar] [CrossRef]
- Lambertz, C.; Martos, A.; Henkel, A.; Neiser, A.; Kliesch, T.T.; Janshoff, A.; Schwille, P.; Sonnichsen, C. Single Particle Plasmon Sensors as Label-Free Technique To Monitor MinDE Protein Wave Propagation on Membranes. Nano Lett. 2016, 16, 3540–3544. [Google Scholar] [CrossRef] [PubMed]
- Theberge, A.B.; Courtois, F.; Schaerli, Y.; Fischlechner, M.; Abell, C.; Hollfelder, F.; Huck, W.T. Microdroplets in microfluidics: An evolving platform for discoveries in chemistry and biology. Angew. Chem. 2010, 49, 5846–5868. [Google Scholar] [CrossRef] [PubMed]
- Godino, E.; Lopez, J.N.; Zarguit, I.; Doerr, A.; Jimenez, M.; Rivas, G.; Danelon, C. Cell-free biogenesis of bacterial division proto-rings that can constrict liposomes. Commun. Biol. 2020, 3, 539. [Google Scholar] [CrossRef]
- Cabre, E.J.; Sanchez-Gorostiaga, A.; Carrara, P.; Ropero, N.; Casanova, M.; Palacios, P.; Stano, P.; Jimenez, M.; Rivas, G.; Vicente, M. Bacterial division proteins FtsZ and ZipA induce vesicle shrinkage and cell membrane invagination. J. Biol. Chem. 2013, 288, 26625–26634. [Google Scholar] [CrossRef]
- Furusato, T.; Horie, F.; Matsubayashi, H.T.; Amikura, K.; Kuruma, Y.; Ueda, T. De Novo Synthesis of Basal Bacterial Cell Division Proteins FtsZ, FtsA, and ZipA Inside Giant Vesicles. ACS Synth. Biol. 2018, 7, 953–961. [Google Scholar] [CrossRef]
- Fanalista, F.; Birnie, A.; Maan, R.; Burla, F.; Charles, K.; Pawlik, G.; Deshpande, S.; Koenderink, G.H.; Dogterom, M.; Dekker, C. Shape and Size Control of Artificial Cells for Bottom-Up Biology. ACS Nano 2019, 13, 5439–5450. [Google Scholar] [CrossRef]
- Mellouli, S.; Monterroso, B.; Vutukuri, H.R.; te Brinke, E.; Chokkalingam, V.; Rivas, G.; Huck, W.T.S. Self-organization of the bacterial cell-division protein FtsZ in confined environments. Soft Matter 2013, 9, 10493–10500. [Google Scholar] [CrossRef]
- Sobrinos-Sanguino, M.; Zorrilla, S.; Keating, C.D.; Monterroso, B.; Rivas, G. Encapsulation of a compartmentalized cytoplasm mimic within a lipid membrane by microfluidics. Chem. Commun. 2017, 53, 4775–4778. [Google Scholar] [CrossRef] [PubMed]
- Monterroso, B.; Zorrilla, S.; Sobrinos-Sanguino, M.; Keating, C.D.; Rivas, G. Microenvironments created by liquid-liquid phase transition control the dynamic distribution of bacterial division FtsZ protein. Sci. Rep. 2016, 6, 35140. [Google Scholar] [CrossRef]
- Tsao, D.H.; Sutherland, A.G.; Jennings, L.D.; Li, Y.; Rush, T.S., 3rd; Alvarez, J.C.; Ding, W.; Dushin, E.G.; Dushin, R.G.; Haney, S.A.; et al. Discovery of novel inhibitors of the ZipA/FtsZ complex by NMR fragment screening coupled with structure-based design. Bioorganic Med. Chem. 2006, 14, 7953–7961. [Google Scholar] [CrossRef]
- Silber, N.; Pan, S.; Schakermann, S.; Mayer, C.; Brotz-Oesterhelt, H.; Sass, P. Cell Division Protein FtsZ Is Unfolded for N-Terminal Degradation by Antibiotic-Activated ClpP. mBio 2020, 11. [Google Scholar] [CrossRef]
- Sass, P.; Josten, M.; Famulla, K.; Schiffer, G.; Sahl, H.G.; Hamoen, L.; Brotz-Oesterhelt, H. Antibiotic acyldepsipeptides activate ClpP peptidase to degrade the cell division protein FtsZ. Proc. Natl. Acad. Sci. USA 2011, 108, 17474–17479. [Google Scholar] [CrossRef] [PubMed]
- Kirstein, J.; Hoffmann, A.; Lilie, H.; Schmidt, R.; Rubsamen-Waigmann, H.; Brotz-Oesterhelt, H.; Mogk, A.; Turgay, K. The antibiotic ADEP reprogrammes ClpP, switching it from a regulated to an uncontrolled protease. EMBO Mol. Med. 2009, 1, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Pichoff, S.; Lutkenhaus, J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol. Microbiol. 2005, 55, 1722–1734. [Google Scholar] [CrossRef]
- Conti, J.; Viola, M.G.; Camberg, J.L. FtsA reshapes membrane architecture and remodels the Z-ring in Escherichia coli. Mol. Microbiol. 2018, 107, 558–576. [Google Scholar] [CrossRef]
- Schumacher, M.A. Bacterial Nucleoid Occlusion: Multiple Mechanisms for Preventing Chromosome Bisection During Cell Division. Subcell Biochem. 2017, 84, 267–298. [Google Scholar] [CrossRef]
- Cho, H.; McManus, H.R.; Dove, S.L.; Bernhardt, T.G. Nucleoid occlusion factor SlmA is a DNA-activated FtsZ polymerization antagonist. Proc. Natl. Acad. Sci. USA 2011, 108, 3773–3778. [Google Scholar] [CrossRef] [PubMed]
- Sobrinos-Sanguino, M. Actividad e Interacciones de la Proteína Esencial de División FtsZ: Ensayos Bioquímicos Con Potencial Aplicación en la Búsqueda de Nuevos Antimicrobianos. Undergraduate Thesis, Universidad Complutense de Madrid, Madrid, Spain, 2013. [Google Scholar]
- Ramirez-Diaz, D.A.; Garcia-Soriano, D.A.; Raso, A.; Mucksch, J.; Feingold, M.; Rivas, G.; Schwille, P. Treadmilling analysis reveals new insights into dynamic FtsZ ring architecture. PLoS Biol. 2018, 16, e2004845. [Google Scholar] [CrossRef]
- Shimizu, Y.; Inoue, A.; Tomari, Y.; Suzuki, T.; Yokogawa, T.; Nishikawa, K.; Ueda, T. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001, 19, 751–755. [Google Scholar] [CrossRef]
- Domanov, Y.A.; Kinnunen, P.K. Antimicrobial peptides temporins B and L induce formation of tubular lipid protrusions from supported phospholipid bilayers. Biophys J. 2006, 91, 4427–4439. [Google Scholar] [CrossRef]
- Rakowska, P.D.; Jiang, H.; Ray, S.; Pyne, A.; Lamarre, B.; Carr, M.; Judge, P.J.; Ravi, J.; Gerling, U.I.; Koksch, B.; et al. Nanoscale imaging reveals laterally expanding antimicrobial pores in lipid bilayers. Proc. Natl. Acad. Sci. USA 2013, 110, 8918–8923. [Google Scholar] [CrossRef]
- Minton, A.P. The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J. Biol. Chem. 2001, 276, 10577–10580. [Google Scholar] [CrossRef] [PubMed]
- Jiao, M.; Li, H.T.; Chen, J.; Minton, A.P.; Liang, Y. Attractive protein-polymer interactions markedly alter the effect of macromolecular crowding on protein association equilibria. Biophys. J. 2010, 99, 914–923. [Google Scholar] [CrossRef]
- Phillip, Y.; Schreiber, G. Formation of protein complexes in crowded environments--from in vitro to in vivo. FEBS Lett. 2013, 587, 1046–1052. [Google Scholar] [CrossRef] [PubMed]
- Maeda, Y.T.; Nakadai, T.; Shin, J.; Uryu, K.; Noireaux, V.; Libchaber, A. Assembly of MreB filaments on liposome membranes: A synthetic biology approach. ACS Synth. Biol. 2012, 1, 53–59. [Google Scholar] [CrossRef]
- Pinot, M.; Chesnel, F.; Kubiak, J.Z.; Arnal, I.; Nedelec, F.J.; Gueroui, Z. Effects of confinement on the self-organization of microtubules and motors. Curr. Biol. 2009, 19, 954–960. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.P.; Richmond, D.L.; Maibaum, L.; Pronk, S.; Geissler, P.L.; Fletcher, D.A. Membrane-induced bundling of actin filaments. Nat. Phys. 2008, 4, 789–793. [Google Scholar] [CrossRef] [PubMed]
- Nishigami, Y.; Ito, H.; Sonobe, S.; Ichikawa, M. Non-periodic oscillatory deformation of an actomyosin microdroplet encapsulated within a lipid interface. Sci. Rep. 2016, 6, 18964. [Google Scholar] [CrossRef] [PubMed]
- Osawa, M.; Anderson, D.E.; Erickson, H.P. Curved FtsZ protofilaments generate bending forces on liposome membranes. EMBO J. 2009, 28, 3476–3484. [Google Scholar] [CrossRef]
- Milam, S.L.; Osawa, M.; Erickson, H.P. Negative-stain electron microscopy of inside-out FtsZ rings reconstituted on artificial membrane tubules show ribbons of protofilaments. Biophys. J. 2012, 103, 59–68. [Google Scholar] [CrossRef][Green Version]
- Ganzinger, K.A.; Merino-Salomon, A.; Garcia-Soriano, D.A.; Butterfield, A.N.; Litschel, T.; Siedler, F.; Schwille, P. FtsZ Reorganization Facilitates Deformation of Giant Vesicles in Microfluidic Traps. Angew. Chem. 2020, 59, 21372–21376. [Google Scholar] [CrossRef] [PubMed]
- Arriaga, L.R.; Datta, S.S.; Kim, S.H.; Amstad, E.; Kodger, T.E.; Monroy, F.; Weitz, D.A. Ultrathin shell double emulsion templated giant unilamellar lipid vesicles with controlled microdomain formation. Small 2014, 10, 950–956. [Google Scholar] [CrossRef] [PubMed]
- Deng, N.N.; Yelleswarapu, M.; Huck, W.T. Monodisperse Uni- and Multicompartment Liposomes. J. Am. Chem. Soc. 2016, 138, 7584–7591. [Google Scholar] [CrossRef]
- Deshpande, S.; Brandenburg, F.; Lau, A.; Last, M.G.F.; Spoelstra, W.K.; Reese, L.; Wunnava, S.; Dogterom, M.; Dekker, C. Spatiotemporal control of coacervate formation within liposomes. Nat. Commun. 2019, 10, 1800. [Google Scholar] [CrossRef] [PubMed]
- Price, A.K.; Paegel, B.M. Discovery in Droplets. Anal. Chem. 2016, 88, 339–353. [Google Scholar] [CrossRef]
- Behera, B.; Anil Vishnu, G.K.; Chatterjee, S.; Sitaramgupta, V.V.; Sreekumar, N.; Nagabhushan, A.; Rajendran, N.; Prathik, B.H.; Pandya, H.J. Emerging technologies for antibiotic susceptibility testing. Biosens. Bioelectron. 2019, 142, 111552. [Google Scholar] [CrossRef]
- Balaban, N.Q.; Merrin, J.; Chait, R.; Kowalik, L.; Leibler, S. Bacterial persistence as a phenotypic switch. Science 2004, 305, 1622–1625. [Google Scholar] [CrossRef]
- Rabouille, C.; Alberti, S. Cell adaptation upon stress: The emerging role of membrane-less compartments. Curr. Opin. Cell Biol. 2017, 47, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Harms, A.; Maisonneuve, E.; Gerdes, K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 2016, 354. [Google Scholar] [CrossRef]
- Kriel, A.; Bittner, A.N.; Kim, S.H.; Liu, K.; Tehranchi, A.K.; Zou, W.Y.; Rendon, S.; Chen, R.; Tu, B.P.; Wang, J.D. Direct regulation of GTP homeostasis by (p)ppGpp: A critical component of viability and stress resistance. Mol. Cell 2012, 48, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zbornikova, E.; Rejman, D.; Gerdes, K. Novel (p)ppGpp Binding and Metabolizing Proteins of Escherichia coli. mBio 2018, 9. [Google Scholar] [CrossRef]
- Yu, J.; Liu, Y.; Yin, H.; Chang, Z. Regrowth-delay body as a bacterial subcellular structure marking multidrug-tolerant persisters. Cell Discov. 2019, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Conlon, B.P.; Nakayasu, E.S.; Fleck, L.E.; LaFleur, M.D.; Isabella, V.M.; Coleman, K.; Leonard, S.N.; Smith, R.D.; Adkins, J.N.; Lewis, K. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 2013, 503, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, R.J. Therapeutics-how to treat phase separation-associated diseases. Emerg. Top. Life Sci. 2020, 4, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Alberti, S.; Gladfelter, A.; Mittag, T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 2019, 176, 419–434. [Google Scholar] [CrossRef]
- Trivedi, P.; Palomba, F.; Niedzialkowska, E.; Digman, M.A.; Gratton, E.; Stukenberg, P.T. The inner centromere is a biomolecular condensate scaffolded by the chromosomal passenger complex. Nat. Cell Biol. 2019, 21, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Mitrea, D.M.; Chandra, B.; Ferrolino, M.C.; Gibbs, E.B.; Tolbert, M.; White, M.R.; Kriwacki, R.W. Methods for Physical Characterization of Phase-Separated Bodies and Membrane-less Organelles. J. Mol. Biol. 2018, 430, 4773–4805. [Google Scholar] [CrossRef] [PubMed]



| Method | Fundamentals Refs. | Information Obtained | Examples Refs. |
|---|---|---|---|
| Techniques in solution | |||
| 90° LS, Sedimentation, EM | [54] | Assessment of polymerization | [4,66,67,69,74,75,76] |
| Fluorescence anisotropy | [62,63] | Quantification of drug binding | [65,66,67,68,69] |
| Assessment of polymerization | [91] * | ||
| Interaction with Kil, ZipA, MinC, SlmA | [41,92], [38,39,93] * | ||
| FCS, FCCS | [63,88] | Assessment of polymerization | [41,90] |
| ITC | [70] | Quantification of drug binding | [71,72] |
| Assessment of polymerization | [83] * | ||
| Interaction with SulA | [40] * | ||
| DLS | [54,78] | Assessment of polymerization | [80,82] |
| SV | [54] | Assessment of polymerization | [41] |
| Interaction with SlmA | [93] * | ||
| FRET, intrinsic fluorescence | [63] | Assessment of polymerization | [30] * |
| Interaction with FtsA, MinC | [94,95] * | ||
| Biosensor | [56] | Interaction with SlmA, MinC | [12,96] * |
| Fluorescence microscopy | [97] | Assessment of polymerization | [98] |
| Interaction with ZapA | [75,99] | ||
| Reconstruction systems | |||
| Nanodiscs | [100] | Interaction with ZipA | [37] * |
| Microbeads | [56] | Interaction with ZipA | [101] * |
| Interaction of FtsA, SlmA with membrane | [102,103] * | ||
| SLBs | [55,57] | Interaction with MinCDE, FtsA, ZipA | [104,105,106,107,108] * |
| Biosensor, plasmonic sensor | [56] | Interaction with ZipA | [109,110] * |
| Interaction of SlmA, MinDE with membrane | [103,111] * | ||
| Microdroplets, liposomes | [55,56,57,58,112] | Interaction with FtsA, ZipA | [113,114,115,116] * |
| Interaction of MinCDE with membrane | [108]* | ||
| Arrangement & distribution of FtsZ species | [116,117,118,119] * | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zorrilla, S.; Monterroso, B.; Robles-Ramos, M.-Á.; Margolin, W.; Rivas, G. FtsZ Interactions and Biomolecular Condensates as Potential Targets for New Antibiotics. Antibiotics 2021, 10, 254. https://doi.org/10.3390/antibiotics10030254
Zorrilla S, Monterroso B, Robles-Ramos M-Á, Margolin W, Rivas G. FtsZ Interactions and Biomolecular Condensates as Potential Targets for New Antibiotics. Antibiotics. 2021; 10(3):254. https://doi.org/10.3390/antibiotics10030254
Chicago/Turabian StyleZorrilla, Silvia, Begoña Monterroso, Miguel-Ángel Robles-Ramos, William Margolin, and Germán Rivas. 2021. "FtsZ Interactions and Biomolecular Condensates as Potential Targets for New Antibiotics" Antibiotics 10, no. 3: 254. https://doi.org/10.3390/antibiotics10030254
APA StyleZorrilla, S., Monterroso, B., Robles-Ramos, M.-Á., Margolin, W., & Rivas, G. (2021). FtsZ Interactions and Biomolecular Condensates as Potential Targets for New Antibiotics. Antibiotics, 10(3), 254. https://doi.org/10.3390/antibiotics10030254







