5-Benzyliden-2-(5-methylthiazol-2-ylimino)thiazolidin-4-ones as Antimicrobial Agents. Design, Synthesis, Biological Evaluation and Molecular Docking Studies
Abstract
:1. Introduction
2. Results and Discussion
2.1. Toxicity Prediction
2.2. Chemistry
2.3. Biological Evaluation
2.3.1. Antibacterial Action
2.3.2. Antifungal Activity
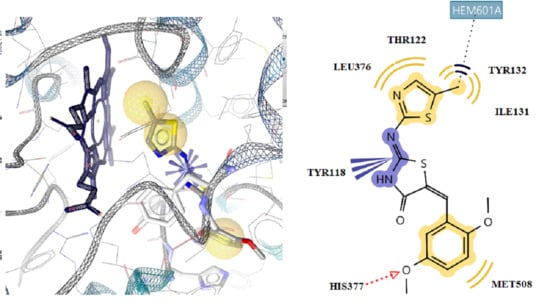
2.4. Docking Studies
2.4.1. Docking in E. coli-MurB
2.4.2. Docking in Antifungal Targets
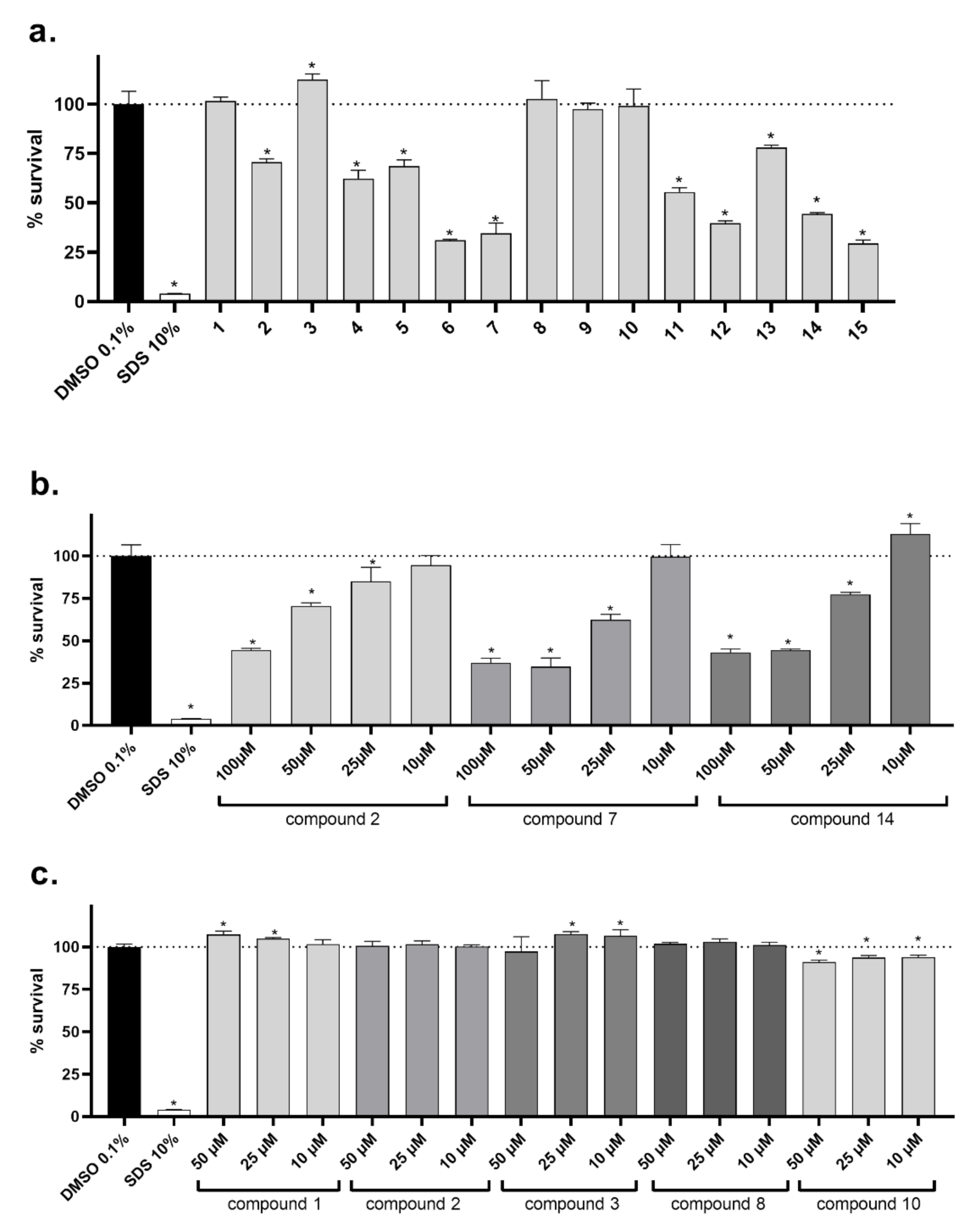
2.5. Cellular Toxicity
3. Materials and Methods
3.1. Chemistry
3.1.1. Synthesis of 5-Methylthiazol-2-yl chloroacetamide
3.1.2. Synthesis of 2-(5-Methylthiazol-2-yl) thiazolidin-4-one
3.1.3. General Procedure for the Synthesis of 5-Benzyliden-2-(5-methylthiazol-2-ylimino) thiazolidin-4-one
5-benzyliden -2-(5-methylthaizol-2-ylimino) thiazolidin-4-one (1)
2-(5-methylthiazol-2-ylimino)-5-(2-hydroxybenzyliden) thiazolidin-4-one (2)
2-(5-methylthiazol-2-ylimino)-5-(4-hydroxybenzyliden) thiazolidin-4-one (3)
2-(5-methylthiazol-2-ylimino)-5-(4-methoxybenzyliden) thiazolidin-4-one (4)
2-(5-methylthiazol-2-ylimino)-5-(2,5-dimethoxybenzyliden) thiazolidin-4-one (5)
2-(5-methylthiazol-2-ylimino)-5-(2-nitrobenzyliden) thiazolidin-4-one (6)
2-(5-methylthiazol-2-ylimino)-5-(3-nitrobenzyliden) thiazolidin-4-one (7)
2-(5-methylthiazol-2-ylimino)-5-(3-fluorobenzyliden) thiazolidin-4-one (8)
2-(5-methylthiazol-2-ylimino)-5-(4-fluorobenzyliden) thiazolidin-4-one (9)
2-(5-methylthiazol-2-ylimino)-5-(4-chlorobenzyliden) thiazolidin-4-one (10)
2-(5-methylthiazol-2-ylimino)-5-(2,3-dichlorobenzyliden) thiazolidin-4-one (11)
2-(5-methylthiazol-2-ylimino)-5-(2,4-dichlorobenzyliden) thiazolidin-4-one (12)
2-(5-methylthiazol-2-ylimino)-5-(2,6-dichlorobenzyliden) thiazolidin-4-one (13)
2-(5-methylthiazol-2-ylimino)-5-(3-bromobenzyliden) thiazolidin-4-one (14)
2-(5-methylthiazol-2-ylimino)-5-(4-bromobenzyliden) thiazolidin-4-one (15)
3.2. Biological Evaluation
3.2.1. Antibacterial Action
3.2.2. Methicillin-resistant Staphylococcus aureus (MRSA)
3.2.3. E. coli
3.2.4. Pseudomonas aeruginosa
3.2.5. Inhibition of Biofilm Formation
3.2.6. Antifungal Activity
3.3. Statistical Analysis
3.4. Cytotoxicity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus Biofilm: An Emerging Battleground in Microbial Communities. Antimicrob. Resist. Infect. Control 2019, 8, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as complex differentiated communities. Ann. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simoes, M.; Pereira, M.O.; Vieira, M.J. Effect of mechanical stress on biofilms challenged by different chemicals. Water Res. 2005, 39, 5142–5152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bjarnsholt, T. The role of bacterial biofilms in chronic infections. APMIS Suppl. 2013, 136, 1–51. [Google Scholar] [CrossRef]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G.J. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [Green Version]
- Otto, M. Staphylococcal infections: Mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu. Rev. Med. 2013, 64, 175–188. [Google Scholar] [CrossRef]
- Hacioglu, M.; Birteksor Tan, A.S.; Doser, S.; Inan, N.; Otuk, G. In vitro activities of antifungals alone and in combination with tigecycline against Candida albicans biofilm. Peer J. 2018, 6, e5263. [Google Scholar] [CrossRef] [Green Version]
- Holmes, A.H.; Moore, L.S.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karkey, A.; Guerin, P.J.; Piddock, L.J. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef]
- Winter, S.E.; Lopez, C.A.; Bäumler, A.J. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep. 2013, 14, 319–327. [Google Scholar] [CrossRef] [Green Version]
- Malhotra, S.; Hayes, D., Jr.; Wozniak, D.J. Cystic Fibrosis and Pseudomonas aeruginosa: The Host-Microbe Interface. Clin. Microbiol. Rev. 2019, 32, e00138-18. [Google Scholar] [CrossRef]
- Pfaller, M.A. Antifungal drug resistance: Mechanisms, epidemiology, and consequences for treatment. Am. J. Med. 2012, 125, S3–S13. [Google Scholar] [CrossRef]
- Munro, C.A. Echinocandin resistance in human pathogenic fungi. Expert Rev. Anti. Infect. Ther. 2012, 10, 115–116. [Google Scholar] [CrossRef]
- Bikobo, D.S.N.; Dan Vodnar, D.C.; Stana, A.; Tiperciuc, B.; Nastasă, C.; Douchet, M.; Oniga, O. Synthesis of 2-phenylamino-thiazole derivatives as antimicrobial agents. J. Saudi Chem. Soc. 2017, 21, 861–868. [Google Scholar] [CrossRef]
- Desaia, N.C.; Makwana, A.H.; Rajpara, K.M. Synthesis and study of 1,3,5-triazine based thiazole derivatives as antimicrobial agents. J. Saudi Chem. Soc. 2016, 20, S334–S341. [Google Scholar] [CrossRef] [Green Version]
- Mohammad, H.; Reddy, P.V.N.; Monteleone, D.; Mayhoub, A.S.; Cushman, M.; Hammac, G.K.; Seleem, M.N. Antibacterial Characterization of Novel Synthetic Thiazole Compounds against Methicillin-Resistant Staphylococcus pseudintermedius. PLoS ONE 2015, 10, e0130385. [Google Scholar] [CrossRef] [Green Version]
- Serpil, D. Synthesis of thiazole derivatives as antimicrobial agents by green chemistry technology. JOTCSA 2018, 5, 393–414. [Google Scholar]
- Althagafi, I.; El-Metwaly, N.; Farghaly, T.A. New Series of Thiazole Derivatives: Synthesis, Structural Elucidation, Antimicrobial Activity, Molecular Modeling and MOE Docking. Molecules 2019, 24, 1741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geronikaki, A.; Vicini, P.; Theophilidis, G.; Lagunin, A.; Poroikov, V.; Dabarakis, N.; Modarresi, H.; Dearden, J.C. Evaluation the local anaesthetic activity of derivatives of 3-amino-1,2- [d]benzoisothiazoles on sciatic nerve of rat. Eur. J. Med Chem. 2009, 44, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Kamble, R.D.; Meshram, R.J.; Hese, S.V.; More, R.A.; Kamble, S.S.; Gacche, R.N.; Dawane, B.S. Synthesis and in silico investigation of thiazoles bearing pyrazoles derivatives as anti-inflammatory agents. Comput. Biol. Chem. 2016, 61, 86–96. [Google Scholar] [CrossRef]
- Apostolidis, I.; Liaras, K.; Geronikaki, A.; Hadjipavlou-Litina, D.; Gavalas, A.; Sokovic, M.; Glamočlija, J.; Ciric, A. Synthesis and biological evaluation of some 5-arylidene-2-(1,3-thiazol-2-ylimino)-1,3-thiazolidin-4-ones as dual anti-inflamatory/antimicrobial agents. Bioorg. Med. Chem. 2013, 21, 532–539. [Google Scholar] [CrossRef]
- Mohareb, R.M.; Al-Omran, F.; Abdelaziz, M.A.; Ibrahim, R.A. Anti-inflammatory and Anti-ulcer Activities of New Fused Thiazole Derivatives Derived from 2-(2-Oxo-2H-chromen-3-yl)thiazol-4(5H)-one. Acta Chim. Slov. 2017, 64, 349–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.; Ba, M.; Zhou, H.; Cao, Y.; Tang, C.; Yang, Y.; He, R.; Liang, Y.; Zhang, X.; Li, Z.; et al. 2,4,5-Trisubstituted thiazole derivatives: A novel and potent class of non-nucleoside inhibitors of wild type and mutant HIV-1 reverse transcriptase. Eur. J. Med. Chem. 2014, 85, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Petrou, A.; Eleftheriou, P.; Geronikaki, A.; Akrivou, M.G.; Vizirianakis, I. Novel thiazolidin-4-ones as potential non-nucleoside inhibitors of HIV-1 reverse transcriptase. Molecules 2019, 24, 3821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saravanan, G.; Alagarsamy, V.; Prakash, C.R.; Kumar, P.D.; Selvam, T.P. Synthesis of Novel Thiazole Derivatives as Analgesic Agents. Asian J. Res. Pharm. Sci. 2011, 1, 134–138. [Google Scholar]
- Haroun, M. Novel hybrids of pyrazolidinedione and benzothiazole as tzd analogues. Rationale, design, synthesis and in vivo anti-diabetic evaluation. Med. Chem. 2019, 15, 622–631. [Google Scholar] [CrossRef]
- Khatik, G.L.; Datusalia, A.K.; Ahsan, W.; Kaur, P.; Vyas, M.; Amit Mittal, A.; Nayak, S.K.N. A Retrospect Study on Thiazole Derivatives as the Potential Antidiabetic Agents in Drug Discovery and Developments. Curr. Drug Discov. Technol. 2018, 15, 163–177. [Google Scholar] [CrossRef]
- Santana, T.I.; Barbosa, M.O.; Gomes, P.A.T.M.; da Cruz, A.C.N.; da Silva, T.G.; Leite, A.C.L. Synthesis, anticancer activity and mechanism of action of new thiazole derivatives. Eur. J. Med. Chem. 2018, 144, 874–886. [Google Scholar] [CrossRef]
- Jain, S.; Pattnaik, S.; Pathak, K.; Kumar, S.; Pathak, D.; Jain, S.; Vaidya, A. Anticancer Potential of Thiazole Derivatives: A Retrospective Review. Mini Rev. Med. Chem. 2018, 18, 640–655. [Google Scholar] [CrossRef]
- Grozav, A.; Porumb, I.-D.; Găină, L.A.; Filip, L.; Hanganu, D. Cytotoxicity and Antioxidant Potential of Novel2-(2-((1H-indol-5yl)methylene)-hydrazinyl)-thiazoleDerivatives. Molecules 2017, 22, 260. [Google Scholar] [CrossRef] [Green Version]
- Kenchappa, R.; Bodke, Y.D.; Telkar, S.; Aruna Sindhe, M. Antifungal and anthelmintic activity of novel benzofuran derivatives containing thiazolo benzimidazole nucleus: An in vitro evaluation. J. Chem. Biol. 2016, 10, 11–23. [Google Scholar] [CrossRef] [Green Version]
- Gupta, V. A Review on Biological Activity of Imidazole and thiazole moieties and their derivative. Sci. Int. 2013, 1, 253–260. [Google Scholar] [CrossRef] [Green Version]
- Bari, S.B.; Firake, S.D. Exploring Anti-inflammatory Potential of Thiazolidinone Derivatives of Benzenesulfonamide via Synthesis, Molecular Docking and Biological Evaluation. Anti-Inflamm. Anti-Allergy Agents Med. Chem. 2016, 15, 44–53. [Google Scholar] [CrossRef]
- Ali, Y.; Alam, M.S.; Hamid, N.; Husain, A.; Dhulap, A.; Bano, S.; Kharbanda, C. Design, synthesis and biological screening of Novel 2,4-dichlorophenoxy acetic acid substituted thiazolidin-4-ones as anti-inflammatory agents. Bioorg. Med. Chem. Lett. 2017, 27, 1017–1025. [Google Scholar] [CrossRef]
- Kouatly, O.; Eleftheriou, P.; Petrou, A.; Hadjipavlou-Litina, D.; Geronikaki, A. Docking assisted design of novel 4-adamantanyl-2-thiazolylimino-5-arylidene-4-thiazolidinones as potent NSAIDs. SAR QSAR Environ. Res. 2018, 29, 83–101. [Google Scholar] [CrossRef]
- Manjal, K.S.; Kaur, R.; Bhatia, R.; Kumar, K.; Singh, V.; Shankar, R.; Kaur, R.; Rawal, R. Synthetic and medicinal perspective of thiazolidinones: A review. Bioorg. Chem. 2017, 75, 406–423. [Google Scholar] [CrossRef]
- Djukic, M.; Fesatidou, M.; Xenikakis, I.; Geronikaki, A.; Stoyanova, V.; Savic, V.; Pasic, M.; Krilovic, B.; Djukic, D.; Gobeljic, B.; et al. In vitro antioxidant activity of thiazolidinone derivatives of 1,3-thiazole and 1,3,4-thiadiazole. Chem. Biol. Interact. 2018, 286, 119–131. [Google Scholar] [CrossRef]
- Szychowski, K.A.; Leja, M.L.; Kaminskyy, D.V.; Binduga, U.E.; Pinyazhko, O.R.; Lesyk, R.B.; Gmiński, J. Study of novel anticancer 4-thiazolidinone derivatives. Chem. Biol. Interact. 2017, 262, 46–56. [Google Scholar] [CrossRef]
- Kulabaş, N.; Bingöl Özakpınar, Ö.; Özsavcı, D.; Leyssen., P.; Neyts, J.; Küçükgüzel, İ. Synthesis, characterization and biological evaluation of thioureas, acylthioureas and 4-thiazolidinones as anticancer and antiviral agents. J. Res. Pharm. 2017, 21, 371–384. [Google Scholar]
- Gupta, A.; Singh, R.; Sonar, P.K.; Saraf, S.K. Novel 4-Thiazolidinone Derivatives as Anti-Infective Agents: Synthesis, Characterization, and Antimicrobial Evaluation. Biochem. Res. Int. 2016, 2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deep, A.; Narasimhan, B.; Lim, S.M.; Ramasamy, K.; Kumar, K.M. 4-Thiazolidinone derivatives: Synthesis, antimicrobial, anticancer evaluation and QSAR studies. RCS Adv. 2016, 111, 109485–109494. [Google Scholar]
- Gaikwad, S.; Bhake, A.; Bhandarkar, S. 4-thiazolidinone derivatives: Synthesis and biological study. J. Chem. Sci. 2015, 13, 1393–1400. [Google Scholar]
- Al-Ebaisat, H.; Ababne, T.; Shboul, T.; Jazzazi, T.M. Synthesis, Characterization and Antifungal Activity of Some Substituted 4-thiazolidinone Derivatives. J. Pure App. Chem. Res. 2016, 5, 125–130. [Google Scholar] [CrossRef] [Green Version]
- Vicini, P.; Geronikaki, A.; Anastasia, K.; Incerti, M.; Zani, F. Synthesis and antimicrobial activity of novel 2-thiazolylimino-5-arylidene-4-thiazolidinones. Bioorg. Med. Chem. 2006, 14, 3859–3864. [Google Scholar] [CrossRef] [PubMed]
- Hardy, B.; Douglas, N.; Helma, C.; Rautenberg, M.; Jeliazkova, N. Collaborative development of predictive toxicology applications. J. Chemin. Form. 2010, 2, 7–36. [Google Scholar] [CrossRef] [Green Version]
- OpenTox. Available online: http://www.opentox.org/toxicity-prediction (accessed on 5 May 2018).
- Bruno, G.; Costantino, L.; Curinga, C.; Maccari, R.; Monforte, F.; Nicolo, F.; Ottana, R.; Vigorita, M.G. Synthesis and aldose reductase inhibitory activity of 5-arylidene-2,4-thiazolidinediones. Bioorg. Med. Chem. 2002, 10, 1077–1084. [Google Scholar] [CrossRef]
- Ottana, R.; Maccari, R.; Barreca, M.L.; Bruno, G.; Rotondo, A.; Rossi, A.; Chiricosta, G.; Di Paola, R.; Sautebin, L.; Cuzzocrea, S.; et al. 5-Arylidene-2-imino-4-thiazolidinones: Design and synthesis of novel anti-inflammatory agents. Bioorg. Med. Chem. 2005, 13, 4243–4252. [Google Scholar] [CrossRef]
- Tratrat, C. Novel thiazole-based thiazolidinones as potent anti-infective Agents: In silico PASS and toxicity prediction, Synthesis, Biological Evaluation and Molecular Modelling. Comb. Chem. High Throughput Screen. 2020, 23, 126–140. [Google Scholar] [CrossRef]
- Ahmed, S.; Zayed, M.F.; El-Messery, S.M.; Al-Agamy, M.H.; Abdel-Rahman, H.M. Design, Synthesis, Antimicrobial Evaluation and Molecular Modeling Study of 1,2,4-Triazole-Based 4-Thiazolidinones. Molecules 2016, 21, 568. [Google Scholar] [CrossRef]
- Andres, C.J.; Bronson, J.J.; D’Andrea, S.V.; Deshpande, M.S.; Falk, P.J.; Grant-Young, K.A.; Harte, W.E.; Ho, H.T.; Misco, P.F.; Robertson, J.G.; et al. 4-Thiazolidinones: Novel inhibitors of the bacterial enzyme MurB. Bioorg. Med. Chem. Lett. 2000, 10, 715–717. [Google Scholar] [CrossRef]
- Benson, T.E.; Walsh, C.T.; Massey, V. Kinetic characterization of wild-type and S229A mutant MurB: Evidence for the role of Ser 229 as a general acid. Biochemistry 1997, 36, 796–805. [Google Scholar] [CrossRef]
- Nepali, K.; Lee, H.-Y.; Liou, J.-P. Nitro-Group-Containing Drugs. J. Med. Chem. 2019, 62, 2851–2893. [Google Scholar] [CrossRef]
- Chung, K.T.; Murdock, C.A.; Zhou, Y.; Stevens, S.E., Jr.; Li, Y.S.; Wei, C.I.; Fernando, S.Y.; Chou, M.W. Effects of the nitro-group on the mutagenicity and toxicity of some benzamines. Environ. Mol. Mutagen. 1996, 27, 67–74. [Google Scholar] [CrossRef]
- Moreno, S.N.; Docampo, R. Mechanism of toxicity of nitro compounds used in the chemotherapy of trichomoniasis. Environ. Health Perspect. 1985, 64, 199–208. [Google Scholar] [CrossRef]
- Hernandes, M.Z.; Cavalcanti, S.M.; Moreira, D.R.; de Azevedo Junior, W.F.; Leite, A.C. Halogen atoms in the modern medicinal chemistry: Hints for the drug design. Curr. Drug Targets. 2010, 11, 303–314. [Google Scholar] [CrossRef]
- Låg, M.; Søderlund, E.J.; Omichinski, J.G.; Brunborg, G.; Holme, J.A.; Dahl, J.E.; Nelson, S.D.; Dybing, E. Effect of bromine and chlorine positioning in the induction of renal and testicular toxicity by halogenated propanes. Chem. Res. Toxicol. 1991, 4, 528–534. [Google Scholar] [CrossRef]
- Kartsev, V.; Lichitsky, B.; Geronikaki, A.; Petrou, A.; Smiljkovic, M.; Kostic, M.; Radanovic, O.; Soković, M. Design, synthesis and antimicrobial activity of usnic acid derivatives. MedChemComm 2018, 9, 870–882. [Google Scholar] [CrossRef] [Green Version]
- 58. Kostić, M.; Smiljković, M..; Petrović, J.; Glamočilija, J.; Barros, L.; Ferreira, I.C.F.R.; Ćirić., A.; Soković, M. Chemical, nutritive composition and a wide range of bioactive properties of honey mushroom Armillariamellea (Vahl: Fr.) Kummer. Food Funct. 2017, 8, 3239–3249. [Google Scholar] [CrossRef] [Green Version]
- Stegger, M.; Andersen, P.S.; Kearns, A.; Pichon, B.; Holmes, M.A.; Edwards, G.; Laurent, F.; Teale, C.; Skov, R.; Larsen, A.R. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecA(LGA251). Clin. Microbiol. Infect. 2012, 18, 395–400. [Google Scholar] [CrossRef] [Green Version]
- Quinn, P.J.; Markey, B.K.; Leonard, F.C.; FitzPatrick, E.S.; Fanning, S.; Hartigan, P.J. Veterinary Microbiology and Microbial Disease; Wiley-Blackwell: Hoboken, NJ, USA, 2011. [Google Scholar]
- Cady, N.C.; McKean, K.A.; Behnke, J.; Kubec, R.; Mosier, A.P.; Kasper, S.H.; Burz, D.S.; Musah, R.A. Inhibition of biofilm formation, quorum sensing and infection in Pseudomonas aeruginosa by natural products-inspired organosulfur compounds. PLoS ONE 2012, 7, e38492. [Google Scholar] [CrossRef] [Green Version]
- Kritsi, E.; Matsoukas, M.T.; Potamitis, C.; Detsi, A.; Ivanov, M.; Sokovic, M.; Zoumpoulakis, P. Novel Hit Compounds as Putative Antifungals: The Case of Aspergillus fumigatus. Molecules 2019, 24, 3853. [Google Scholar] [CrossRef] [Green Version]
- Aleksić, M.; Stanisavljević, D.; Smiljković, M.; Vasiljević, P.; Stevanović, M.; Soković, M.; Stojković, D. Pyrimethanil: Between efficient fungicide against Aspergillus rot on cherry tomato and cytotoxic agent on human cell lines. Ann. Appl. Biol. 2019, 175, 228–235. [Google Scholar] [CrossRef]

| Compounds | R | MIC/MBC | B.c. | S.a. | M.f. | L.m. | P.a. | En.cl. | E.c. | S.t. |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | H | MIC | 33.2 ± 0.03 | 265.8 ± 0.03 | 265.8 ± 0.04 | 265.8 ± 0.06 | 265.8 ± 0.08 | 265.8 ± 0.03 | 33.2 ± 0.01 | 265.8 ± 0.05 |
| MBC | 66.4 ± 0.02 | 531.6 ± 0.4 | 531.6 ± 0.6 | 531.6 ± 0.3 | 531.6 ± 0.5 | 531.6 ± 0.5 | 66.4 ± 0.03 | 531.6 ± 0.3 | ||
| 2 | 2-OH | MIC | 94.6 ± 0.03 | 94.6 ± 0.05 | 94.6 ± 0.02 | 94.6 ± 0.01 | 94.6 ± 0.03 | 94.6 ± 0.03 | 94.6 ± 0.03 | 94.6 ± 0.02 |
| MBC | 189.2 ± 005 | 189.2 ± 0.08 | 189.2 ± 0.01 | 189.2 ± 0.05 | 189.2 ± 0.02 | 189.2 ± 0.04 | 189.2 ± 0.04 | 189.2 ± 0.03 | ||
| 3 | 4-OH | MIC | 63.0 ± 003 | 126.1 ± 0.06 | 189.2 ± 0.02 | 126.1 ± 0.04 | 378.5 ± 0.8 | 126.1 ± 0.05 | 63.0 ± 0.02 | 126.1 ± 0.02 |
| MBC | 126.1 ± 0.03 | 252.3 ± 0.05 | 378.5 ± 0.6 | 252.3 ± 0.03 | 757.0 ± 0.8 | 252.3 ± 0.05 | 126.1 ± 0.03 | 252.3 ± 0.03 | ||
| 4 | 4-OCH3 | MIC | 30.2 ± 0.04 | 120.8 ± 0.05 | 120.8 ± 0.03 | 120.8 ± 0.05 | 261.6 ± 0.05 | 120.8 ± 0.06 | 30.2 ± 0.01 | 120.8 ± 0.05 |
| MBC | 60.4 ± 006 | 241.6 ± 0.06 | 241.6 ± 0.02 | 241.6 ± 0.06 | 483.3 ± 0.6 | 241.6 ± 0.06 | 60.4 ± 0.04 | 241.6 ± 0.04 | ||
| 5 | 2,5-di-OCH3 | MIC | 96.9 ± 0.06 | 193.9 ± 0.05 | 193.9 ± 0.03 | 193.9 ± 0.04 | 193.9 ± 0.06 | 96.9 ± 0.04 | 193.9 ± 0.03 | 193.9 ± 0.03 |
| MBC | 193.9 ± 0.05 | 387.8 ± 0.3 | 387.8 ± 0.4 | 387.8 ± 0.3 | 387.8 ± 0.4 | 193.9 ± 0.08 | 387.8 ± 0.4 | 387.8 ± 0.4 | ||
| 6 | 2-NO2 | MIC | 57.8 ± 0.03 | 115.6 ± 0.04 | 231.2 ± 0.08 | 115.6 ± 0.03 | 115.6 ± 0.05 | 115.6 ± 0.04 | 115.6 ± 0.04 | 231.2 ± 0.06 |
| MBC | 115.6 ± 0.03 | 115.6 ± 0.03 | 462.4 ± 0.03 | 231.2 ± 0.02 | 231.2 ± 0.08 | 231.2 ± 0.04 | 231.2 ± 0.05 | 462.4 ± 0.4 | ||
| 7 | 3-NO2 | MIC | 86.7 ± 0.02 | 86.7 ± 0.02 | 86.7 ± 0.02 | 86.7 ± 0.02 | 86.7 ± 0.05 | 43.3 ± 0.03 | 43.3 ± 0.03 | 86.7 ± 0.05 |
| MBC | 173.4 ± 0.01 | 173.4 ± 0.06 | 173.4 ± 0.04 | 173.4 ± 0.03 | 173.4 ± 0.03 | 86.7 ± 0.02 | 86.7 ± 0.02 | 173.4 ± 0.06 | ||
| 8 | 3-F | MIC | 125.4 ± 0.03 | 125.4 ± 0.08 | 125.4 ± 0.04 | 125.4 ± 0.04 | 125.4 ± 0.04 | 62.7 ± 0.01 | 62.7 ± 0.01 | 125.4 ± 0.06 |
| MBC | 250.8 ± 0.06 | 250.8 ± 0.06 | 250.8 ± 0.06 | 250.8 ± 0.06 | 250.7 ± 0.05 | 125.4 ± 0.03 | 125.4 ± 0.03 | 250.8 ± 0.08 | ||
| 9 | 4-F | MIC | 344.8 ± 0.5 | 344.8 ± 0.3 | 344.8 ± 0.2 | 344.8 ± 0.3 | 344.8 ± 0.6 | 125.4 ± 0.05 | 125.4 ± 0.03 | 344.8 ± 0.4 |
| MBC | 689.6 ± 0.8 | 689.6 ± 0.6 | 689.6 ± 0.3 | 689.6 ± 0.8 | 689.6 ± 0.8 | 250.7 ± 0.07 | 250.7 ± 0.03 | 689.6 ± 0.6 | ||
| 10 | 4-Cl | MIC | 26.3 ± 0.01 | 89.4 ± 0.03 | 178.8 ± 0.04 | 89.4 ± 0.07 | 357.6 ± 0.4 | 178.8 ± 0.03 | 26.3 ± 0.02 | 357.6 ± 0.6 |
| MBC | 52.6 ± 0.01 | 178.8 ± 0.02 | 357.6 ± 0.3 | 178.8 ± 0.08 | 715.3 ± 0.8 | 357.6 ± 0.4 | 52.6 ± 0.03 | 715.3 ± 0.8 | ||
| 11 | 2,3-di-Cl | MIC | 189.2 ± 0.06 | 189.2 ± 0.06 | 189.2 ± 0.05 | 189.2 ± 0.06 | 189.2 ± 0.06 | 94.6 ± 0.02 | 94.6 ± 0.05 | 189.2 ± 0.05 |
| MBC | 378.4 ± 0.5 | 378.4 ± 0.6 | 378.4 ± 0.06 | 378.4 ± 0.8 | 378.4 ± 0.5 | 189.2 ± 0.03 | 189.2 ± 0.04 | 378.4 ± 0.3 | ||
| 12 | 2,4-di-Cl | MIC | 135.1 ± 0.03 | 135.1 ± 0.05 | 135.1 ± 0.04 | 270.2 ± 0.4 | 270.2 ± 0.3 | 270.2 ± 0.4 | 135.1 ± 0.02 | 270.2 ± 0.3 |
| MBC | 270.2 ± 0.3 | 270.2 ± 0.4 | 270.2 ± 0.4 | 540.5 ± 0.5 | 540.5 ± 0.4 | 540.5 ± 0.6 | 270.2 ± 0.3 | 540.5 ± 0.8 | ||
| 13 | 2,6-di-Cl | MIC | 40.5 ± 0,004 | 81.0 ± 0.06 | 81.0 ± 0.03 | 81.0 ± 0.05 | 162.1 ± 0.05 | 162.1 ± 0.05 | 81.0 ± 0.03 | 162.1 ± 0.05 |
| MBC | 81.0 ± 0.06 | 324.2 ± 0.4 | 162.1 ± 0.02 | 162.1 ± 0.06 | 324.2 ± 0.5 | 324.2 ± 0.3 | 162.1 ± 0.04 | 324.2 ± 0.5 | ||
| 14 | 3-Br | MIC | 78.9 ± 0.06 | 78.9 ± 0.05 | 78.9 ± 0.03 | 78.9 ± 0.06 | 157.9 ± 0.07 | 78.9 ± 0.03 | 78.9 ± 0.05 | 157.9 ± 0.05 |
| MBC | 157.9 ± 0.05 | 157.9 ± 0.04 | 157.9 ± 0.05 | 157.9 ± 0.05 | 315.8 ± 0.5 | 157.9 ± 0.04 | 157.9 ± 0.06 | 315.8 ± 0.3 | ||
| 15 | 4-Br | MIC | 78.9 ± 0.01 | 342.1 ± 0.5 | 342.1 ± 0.3 | 342.1 ± 0.5 | 342.1 ± 0.6 | 342.1 ± 0.6 | 78.9 ± 0.03 | 342.1 ± 0.4 |
| MBC | 157.9 ± 0.05 | 684.2 ± 0.8 | 684.2 ± 0.8 | 684.2 ± 0.8 | 684.2 ± 0.8 | 684.2 ± 0.8 | 157.9 ± 0.04 | 684.2 ± 0.9 | ||
| STM | MIC | 43.0 ± 0.04 | 172 ± 0.3 | 86 ± 0.02 | 258 ± 0.4 | 172 ± 0.3 | 43 ± 0.04 | 172 ± 0.3 | 172 ± 0.3 | |
| MBC | 86 ± 0.05 | 344 ± 0.5 | 172 ± 0.3 | 516 ± 0.5 | 344 ± 0.4 | 86 ± 0.05 | 344 ± 0.4 | 344 ± 0.4 | ||
| AMP | MIC | 248 ± 0.3 | 248 ± 0.4 | 248 ± 0.3 | 372 ± 0.3 | 744 ± 0.6 | 248 ± 0.6 | 372 ± 0.4 | 248 ± 0.4 | |
| MBC | 372 ± 0.4 | 372 ± 0.5 | 372 ± 0.6 | 744 ± 0.6 | 1240 ± 0.8 | 372 ± 0.4 | 492 ± 0.6 | 492 ± 0.3 |
| Νο. | MIC/MBC | MRSA | P.a. | E.c. |
|---|---|---|---|---|
| 1 | MIC | 255.7 ± 0.04 | 122.8 ± 0.1 | 33.2 ± 0.03 |
| MBC | 531.5 ± 0.08 | 265 ± 0.3 | 66.4 ± 0.02 | |
| 2 | MIC | 173.4 ± 0.05 | 173.4 ± 0.05 | 173.4 ± 0.04 |
| MBC | 346.8 ± 0.03 | 346.8 ± 0.4 | 346.8 ± 0.10 | |
| 3 | MIC | 189.2 ± 0.2 | 189.2 ± 0.4 | 31.5 ± 0.05 |
| MBC | 378.5 ± 0.03 | 378.5 ± 0.02 | 63.0 ± 0.04 | |
| 4 | MIC | 120.8 ± 0.08 | 120.8 ± 0.3 | 60.4 ± 0.04 |
| MBC | 241.6 ± 0.02 | 241.6 ± 0.03 | 120.8 ± 0.05 | |
| 5 | MIC | 193.9 ± 0.06 | 96.9 ± 0.10 | 96.9 ± 0.02 |
| MBC | 387.8 ± 0.03 | 193.9 ± 0.5 | 193.9 ± 0.04 | |
| 6 | MIC | 433.5 ± 0.06 | 433.5 ± 0.03 | 433.5 ± 0.4 |
| MBC | 867.0 ± 0.02 | 867.0 ± 0.3 | 867.0 ± 0.10 | |
| 7 | MIC | 289.0 ± 0.03 | 144.5 ± 0.06 | 144.5 ± 0.02 |
| MBC | 578.0 ± 0.04 | 289.0 ± 0.03 | 289.0 ± 0.4 | |
| 8 | MIC | 125.4 ± 0.02 | 125.4 ± 0.08 | 125.4 ± 0.05 |
| MBC | 250.8 ± 0.03 | 250.8 ± 0.01 | 250.8 ± 0.06 | |
| 9 | MIC | 219.4 ± 0.08 | 219.4 ± 0.3 | 219.4 ± 0.04 |
| MBC | 438.8 ± 0.03 | 438.8 ± 0.10 | 438.8 ± 0.02 | |
| 10 | MIC | 178.8 ± 0.06 | 89.4 ± 0.03 | 29.8 ± 0.02 |
| MBC | 357.6 ± 0.05 | 178.8 ± 0.02 | 59.6 ± 0.10 | |
| 11 | MIC | 189.2 ± 0.06 | 189.2 ± 0.5 | 189.2 ± 0.03 |
| MBC | 378.4 ± 0.04 | 378.4 ± 0.03 | 378.4 ± 0.02 | |
| 12 | MIC | 135.1 ± 0.08 | 67.5 ± 0.09 | 67.5 ± 0.06 |
| MBC | 270.2 ± 0.06 | 135.1 ± 0.3 | 135.1 ± 0.4 | |
| 13 | MIC | 270.2 ± 0.05 | 135.1 ± 0.3 | 135.1 ± 0.02 |
| MBC | 540.5 ± 0.06 | 270.2 ± 0.5 | 270.2 ± 0.03 | |
| 14 | MIC | 131.6 ± 0.04 | 131.6 ± 0.06 | 263.2 ± 0.06 |
| MBC | 263.2 ± 0.02 | 263.2 ± 0.04 | 526.4 ± 0.02 | |
| 15 | MIC | 157.9 ± 0.03 | 78.9 ± 0.02 | 78.9 ± 0.01 |
| MBC | 215.8 ± 0.04 | 157.9 ± 0.10 | 157.9 ± 0.05 | |
| Streptomycin | MIC | 171.9 ± 0.02 | 86 ± 0.3 | 171.9 ± 0.06 |
| MBC | / | 171.9 ± 0.03 | 343 ± 0.02 | |
| Ampicillin | MIC | / | 572 ± 0.6 | 572 ± 0.4 |
| MBC | / | / | / |
| Compound | MIC | 0.5 MIC |
|---|---|---|
| 2 | 9.89 | 15.33 |
| 7 | 36.64 | NE |
| 8 | 61.34 | 48.67 |
| 13 | 62.69 | 35.49 |
| 14 | 56.74 | 40.54 |
| Ampicillin | 71.94 | 55.42 |
| Streptomycin | 67.36 | 30.35 |
| Νο. | MIC/MFC | A.v. | T.v. | A.o. | A.n. | P.v.c. | P.f. | P.o. | A.fum. |
|---|---|---|---|---|---|---|---|---|---|
| 1 | MIC | 265.8 ± 0.2 | 132.9 ± 0.08 | 265.8 ± 0.2 | 265.8 ± 0.05 | 132.9 ± 0.2 | 265.8 ± 0.1 | 265.8 ± 0.2 | 265.8 ± 0.02 |
| MFC | 531.6 ± 0.3 | 265.8 ± 0.3 | 531.6 ± 0.4 | 531.6 ± 0.03 | 265.8 ± 0.3 | 531.6 ± 0.2 | 531.6 ± 0.3 | 531.6 ± 0.03 | |
| 2 | MIC | 441.6 ± 0.2 | 220.8 ± 0.06 | 220.8 ± 0.5 | 220.8 ± 0.08 | 220.8 ± 0.08 | 220.8 ± 0.03 | 630.9 ± 0.4 | 220.8 ± 0.05 |
| MFC | 883.2 ± 0.4 | 441.6 ± 0.2 | 441.6 ± 0.3 | 441.6 ± 0.06 | 441.6 ± 0.3 | 441.6 ± 0.2 | 883.2 ± 0.4 | 441.6 ± 0.06 | |
| 3 | MIC | 94.6 ± 0.4 | 94.6 ± 0.05 | 94.6 ± 0.1 | 189.2 ± 0.09 | 94.6 ± 0.4 | 189.2 ± 0.3 | 94.6 ± 0.4 | 94.6 ± 0.04 |
| MFC | 189.2 ± 0.2 | 189.2 ± 0.3 | 189.2 ± 0.4 | 378.4 ± 0.10 | 189.2 ± 0.5 | 378.4 ± 0.3 | 189.2 ± 0.3 | 94.6 ± 0.08 | |
| 4 | MIC | 120.8 ± 0.5 | 60.4 ± 0.02 | 120.8 ± 0.3 | 241.6 ± 0.03 | 120.8 ± 0.05 | 120.8 ± 0.06 | 60.4 ± 0.1 | 120.8 ± 0.05 |
| MFC | 241.6 ± 0.3 | 120.8 ± 0.4 | 241.6 ± 0.2 | 483.2 ± 0.06 | 241.6 ± 0.02 | 241.6 ± 0.06 | 120.8 ± 0.02 | 241.6 ± 0.03 | |
| 5 | MIC | 110.8 ± 0.4 | 27.7 ± 0.02 | 55.4 ± 0.3 | 110.8 ± 0.01 | 110.8 ± 0.05 | 110.8 ± 0.08 | 110.8 ± 0.04 | 110.8 ± 0.03 |
| MFC | 221.6 ± 0.5 | 55.4 ± 0.03 | 118 ± 0.4 | 221.6 ± 0.03 | 221.6 ± 0.06 | 221.6 ± 0.02 | 221.6 ± 0.04 | 221.6 ± 0.05 | |
| 6 | MIC | 289.0 ± 0.3 | 144.5 ± 0.05 | 289.0 ± 0.5 | 115.6 ± 0.03 | 115.6 ± 0.06 | 115.6 ± 0.05 | 289.0 ± 0.2 | 231.2 ± 0.002 |
| MFC | 578.0 ± 0.4 | 289.0 ± 0.3 | 578.0 ± 0.3 | 231.2 ± 0.05 | 231.2 ± 0.03 | 231.2 ± 0.06 | 289.0 ± 0.4 | 462.4 ± 0.03 | |
| 7 | MIC | 578.0 ± 0.3 | 289.0 ± 0.2 | 289.0 ± 0.5 | 72.2 ± 0.04 | 72.2 ± 0.6 | 34.6 ± 0.08 | 578.0 ± 0.2 | 72.2 ± 0.03 |
| MFC | 1156 ± 0.3 | 578.0 ± 0.3 | 578.0 ± 0.2 | 144.4 ± 0.06 | 144.4 ± 0.06 | 72.2 ± 0.06 | 1156.0 ± 0.5 | 144.4 ± 0.04 | |
| 8 | MIC | 156.7 ± 0.6 | 156.7 ± 0.08 | 313.4 ± 0.4 | 501.5 ± 0.08 | 501.5 ± 0.5 | 501.5 ± 0.2 | 313.4 ± 0.3 | 501.5 ± 0.06 |
| MFC | 313.4 ± 0.3 | 313.4 ± 0.3 | 626.9 ± 0.4 | 1003.0 ± 1 | 1003.0 ± 0.6 | 1003.0 ± 0.3 | 626.9 ± 0.3 | 1003.0 ± 1 | |
| 9 | MIC | 250.7 ± 0.5 | 141.0 ± 0.04 | 282.1 ± 0.4 | 250.8 ± 0.04 | 250.8 ± 0.08 | 250.8 ± 0.08 | 501.5 ± 0.3 | 250.8 ± 0.04 |
| MFC | 501.5 ± 0.3 | 282.1 ± 0.05 | 564.2 ± 0.1 | 501.6 ± 0.03 | 501.6 ± 0.3 | 501.6 ± 0.3 | 1003.1 ± 0.2 | 501.6 ± 0.08 | |
| 10 | MIC | 119.2 ± 0.2 | 59.6 ± 0.02 | 119.2 ± 0.6 | 119.2 ± 0.05 | 59.6 ± 0.02 | 59.6 ± 0.03 | 59.6 ± 0.05 | 59.6 ± 0.02 |
| MFC | 238.4 ± 0.2 | 119.2 ± 0.05 | 238.4 ± 0.5 | 238.4 ± 0.08 | 119.2 ± 0.03 | 119.2 ± 0.01 | 119.2 ± 0.03 | 119.2 ± 0.06 | |
| 11 | MIC | 432.4 ± 0.4 | 108.1 ± 0.06 | 216.2 ± 0.4 | 378.4 ± 0.04 | 189.2 ± 0.05 | 189.2 ± 0.06 | 432.4 ± 0.5 | 378.4 ± 0.06 |
| MFC | 864.8 ± 0.5 | 216.2 ± 0.05 | 432.4 ± 0.4 | 756.7 ± 0.06 | 378.4 ± 0.3 | 378.4 ± 0.08 | 864.8 ± 0.5 | 756.7 ± 0.08 | |
| 12 | MIC | 135.1 ± 0.3 | 135.1 ± 0.06 | 270.2 ± 0.3 | 135.1 ± 0.06 | 135.1 ± 0.06 | 270.2 ± 0.06 | 135.1 ± 0.3 | 270.2 ± 0.05 |
| MFC | 270.2 ± 0.2 | 270.2 ± 0.2 | 540.4 ± 0.2 | 270.2 ± 0.03 | 270.2 ± 0.3 | 540.4 ± 0.3 | 270.2 ± 0.2 | 540.4 ± 0.03 | |
| 13 | MIC | 81.0 ± 0.2 | 81.0 ± 0.06 | 162.0 ± 0.3 | 162.0 ± 0.06 | 81.0 ± 0.04 | 81.0 ± 0.04 | 81.0 ± 0.05 | 81.0 ± 0.01 |
| MFC | 162.0 ± 0.2 | 162.0 ± 0.04 | 324.0 ± 0.5 | 324.0 ± 0.08 | 162.0 ± 0.04 | 162.0 ± 0.04 | 162.0 ± 0.3 | 162.0 ± 0.05 | |
| 14 | MIC | 78.9 ± 0.4 | 78.9 ± 0.03 | 157.9 ± 0.2 | 157.9 ± 0.04 | 78.9 ± 0.02 | 78.9 ± 0.05 | 78.9 ± 0.3 | 157.9 ± 0.03 |
| MFC | 157.9 ± 0.3 | 157.9 ± 0.2 | 315.8 ± 0.1 | 315.8 ± 0.04 | 157.9 ± 0.08 | 157.9 ± 0.01 | 157.9 ± 0.4 | 315.8 ± 0.06 | |
| 15 | MIC | 342.2 ± 0.2 | 171.1 ± 0.4 | 342.2 ± 0.5 | 342.2 ± 0.08 | 342.2 ± 0.02 | 342.2 ± 0.03 | 342.2 ± 0.03 | 342.2 ± 0.04 |
| MFC | 684.4 ± 0.3 | 342.2 ± 0.5 | 684.4 ± 0.1 | 684.4 ± 0.03 | 684.4 ± 0.06 | 684.4 ± 0.02 | 684.4 ± 0.5 | 684.4 ± 0.05 | |
| Bifonazole | MIC | 480.0 ± 0.3 | 640.0 ± 0.1 | 480.0 ± 0.1 | 480.0 ± 0.10 | 480.0 ± 0.2 | 640 ± 0.2 | 480 ± 0.3 | 480 ± 0.06 |
| MFC | 640.0 ± 0.5 | 800 ± 0.3 | 800.0 ± 0.4 | 640.0 ± 0.08 | 640.0 ± 0.2 | 800 ± 0.2 | 640 ± 0.4 | 640 ± 0.03 | |
| Ketoconazole | MIC | 2850 ± 0.3 | 4750.0 ± 0.3 | 380.0 ± 0.2 | 380.0 ± 0.06 | 380.0 ± 0.3 | 380 ± 0.3 | 3800 ± 0.3 | 380 ± 0.06 |
| MFC | 3800 ± 0,06 | 5700.0 ± 0.2 | 950.0 ± 0.5 | 950.0 ± 0.10 | 950.0 ± 0.3 | 950 ± 0.4 | 3800 ± 0.5 | 950 ± 0.08 |
| No. | TPSA (A2) | Μ.Β. | nOHNH | nON | Violations | cLogP |
|---|---|---|---|---|---|---|
| 1 | 58.12 | 301.4 | 1 | 4 | 0 | 3.67 |
| 2 | 78.35 | 317.39 | 2 | 5 | 0 | 3.28 |
| 3 | 78.35 | 317.39 | 2 | 5 | 0 | 3.28 |
| 4 | 67.35 | 331.42 | 1 | 5 | 0 | 3.54 |
| 5 | 76.59 | 261.45 | 1 | 6 | 0 | 3.42 |
| 6 | 103.94 | 346.39 | 1 | 7 | 0 | 3.88 |
| 7 | 103.94 | 346.39 | 1 | 7 | 0 | 3.88 |
| 8 | 58.12 | 319.39 | 1 | 4 | 0 | 3.83 |
| 9 | 58.12 | 319.39 | 1 | 4 | 0 | 3.83 |
| 10 | 58.12 | 335.84 | 1 | 4 | 0 | 4.23 |
| 11 | 58.12 | 270.29 | 1 | 4 | 0 | 4.78 |
| 12 | 58.12 | 370.29 | 1 | 4 | 0 | 4.78 |
| 13 | 58.12 | 370.29 | 1 | 4 | 0 | 4.78 |
| 14 | 58.12 | 380.29 | 1 | 4 | 0 | 4.50 |
| 15 | 58.12 | 380.29 | 1 | 4 | 0 | 4.50 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haroun, M.; Tratrat, C.; Kolokotroni, A.; Petrou, A.; Geronikaki, A.; Ivanov, M.; Kostic, M.; Sokovic, M.; Carazo, A.; Mladěnka, P.; et al. 5-Benzyliden-2-(5-methylthiazol-2-ylimino)thiazolidin-4-ones as Antimicrobial Agents. Design, Synthesis, Biological Evaluation and Molecular Docking Studies. Antibiotics 2021, 10, 309. https://doi.org/10.3390/antibiotics10030309
Haroun M, Tratrat C, Kolokotroni A, Petrou A, Geronikaki A, Ivanov M, Kostic M, Sokovic M, Carazo A, Mladěnka P, et al. 5-Benzyliden-2-(5-methylthiazol-2-ylimino)thiazolidin-4-ones as Antimicrobial Agents. Design, Synthesis, Biological Evaluation and Molecular Docking Studies. Antibiotics. 2021; 10(3):309. https://doi.org/10.3390/antibiotics10030309
Chicago/Turabian StyleHaroun, Michelyne, Christophe Tratrat, Aggeliki Kolokotroni, Anthi Petrou, Athina Geronikaki, Marija Ivanov, Marina Kostic, Marina Sokovic, Alejandro Carazo, Přemysl Mladěnka, and et al. 2021. "5-Benzyliden-2-(5-methylthiazol-2-ylimino)thiazolidin-4-ones as Antimicrobial Agents. Design, Synthesis, Biological Evaluation and Molecular Docking Studies" Antibiotics 10, no. 3: 309. https://doi.org/10.3390/antibiotics10030309
APA StyleHaroun, M., Tratrat, C., Kolokotroni, A., Petrou, A., Geronikaki, A., Ivanov, M., Kostic, M., Sokovic, M., Carazo, A., Mladěnka, P., Sreeharsha, N., Venugopala, K. N., Nair, A. B., & Elsewedy, H. S. (2021). 5-Benzyliden-2-(5-methylthiazol-2-ylimino)thiazolidin-4-ones as Antimicrobial Agents. Design, Synthesis, Biological Evaluation and Molecular Docking Studies. Antibiotics, 10(3), 309. https://doi.org/10.3390/antibiotics10030309