Prevalence, Genetic Diversity, Antimicrobial Resistance, and Toxigenic Profile of Vibrio vulnificus Isolated from Aquatic Environments in Taiwan
Abstract
:1. Introduction
2. Results
2.1. Detection Rate of V. vulnificus from Water and Shellfish Samples Associated with River Basin and Fishing Harbors
2.2. Antimicrobial Susceptibility and Genotypic Profiling of Vibrio Vulnificus Isolates
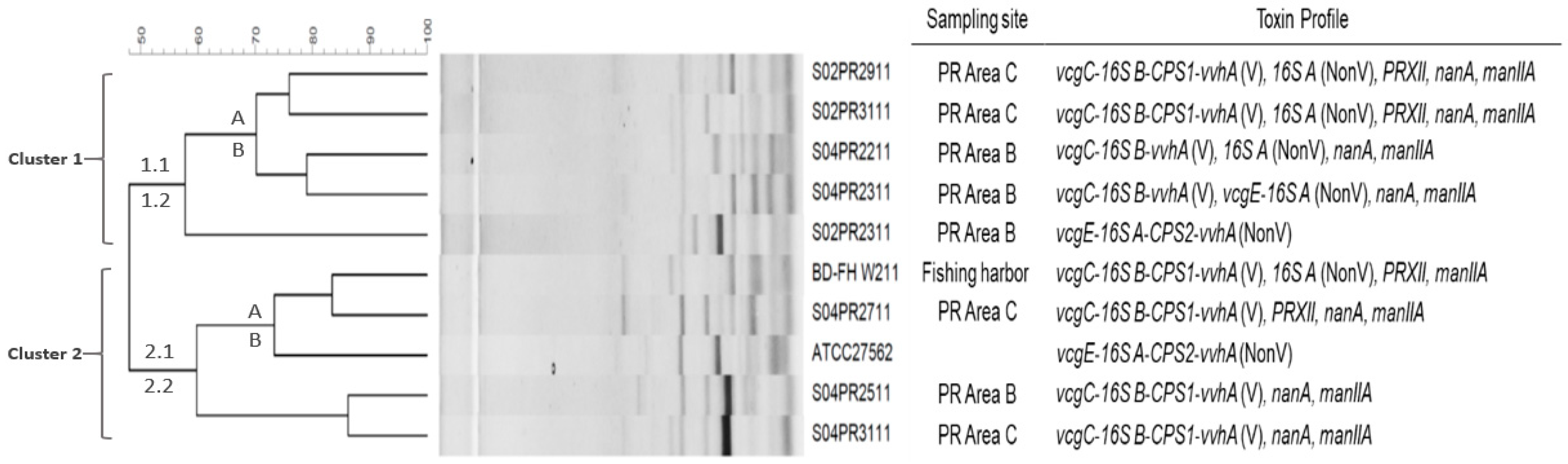
2.3. Genetic Analysis of V. vulnificus Strains by ERIC-PCR Fingerprinting Combined with Genotypic Profiling
3. Discussion
4. Materials and Methods
4.1. Sampling Information
4.2. Pre-Treatment of Water Samples
4.3. Enrichment, Cultivation, and Molecular Profiling of V. vulnificus
4.4. Antibiotic Susceptibility Testing and Multidrug Resistance Profiling of V. vulnificus Isolates
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bier, N.; Jäckel, C.; Dieckmann, R.; Brennholt, N.; Böer, S.I.; Strauch, E. Virulence profiles of Vibrio vulnificus in German coastal waters, a comparison of North sea and Baltic sea isolates. Int. J. Environ. Res. Public Health 2015, 12, 15943–15959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliver, J.D. The biology of Vibrio vulnificus. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef] [Green Version]
- Paydar, M.; Thong, K.L. Prevalence and genetic characterization of Vibrio vulnificus in raw seafood and seawater in Malaysia. J. Food Prot. 2013, 76, 1797–1800. [Google Scholar] [CrossRef]
- Oliver, J.D.; Hite, F.; McDougald, D.; Andon, N.L.; Simpson, L.M. Entry into, and resuscitation from, the viable but nonculturable state by Vibrio vulnificus in an estuarine environment. Appl. Environ. Microbiol. 1995, 61, 2624–2630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, N.V.; Shashidhar, R.; Bandekar, J.R. Induction, resuscitation and quantitative real-time polymerase chain reaction analyses of viable but nonculturable Vibrio vulnificus in artificial sea water. World J. Microbiol. Biotechnol. 2014, 30, 2205–2212. [Google Scholar] [CrossRef] [PubMed]
- DePaola, A.; Capers, G.M.; Alexander, D. Densities of Vibrio vulnificus in the intestines of fish from the US Gulf Coast. Appl. Environ. Microbiol. 1994, 60, 984–988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strom, M.S.; Paranjpye, R.N. Epidemiology and pathogenesis of Vibrio vulnificus. Microbes Infect. 2000, 2, 177–188. [Google Scholar] [CrossRef]
- Turner, J.W.; Good, B.; Cole, D.; Lipp, E.K. Plankton composition and environmental factors contribute to Vibrio seasonality. ISME J. 2009, 3, 1082–1092. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharyya, N.; Lemon, T.L.; Grove, A. A role for Vibrio vulnificus PecS during hypoxia. Sci. Rep. UK 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Heng, S.-P.; Letchumanan, V.; Deng, C.-Y.; Ab Mutalib, N.-S.; Khan, T.M.; Chuah, L.-H.; Chan, K.-G.; Goh, B.-H.; Pusparajah, P.; Lee, L.-H. Vibrio vulnificus: An Environmental and Clinical Burden. Front. Microbiol. 2017, 8, 997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.-W.; Lee, K.; Tang, H.-J.; Ko, W.-C.; Lee, H.-C.; Liu, Y.-C.; Hsueh, P.-R.; Chuang, Y.-C. Prognostic factors and antibiotics in Vibrio vulnificus septicemia. Arch. Intern. Med. 2006, 166, 2117–2123. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Trinanes, J.A.; Taylor, N.G.; Hartnell, R.; Siitonen, A.; Martinez-Urtaza, J. Emerging Vibrio risk at high latitudes in response to ocean warming. Nat. Clim. Chang. 2013, 3, 73–77. [Google Scholar] [CrossRef]
- Martinez-Urtaza, J.; Bowers, J.C.; Trinanes, J.; DePaola, A. Climate anomalies and the increasing risk of Vibrio parahaemolyticus and Vibrio vulnificus illnesses. Food Res. Int. 2010, 43, 1780–1790. [Google Scholar] [CrossRef]
- Paz, S.; Bisharat, N.; Paz, E.; Kidar, O.; Cohen, D. Climate change and the emergence of Vibrio vulnificus disease in Israel. Environ. Res 2007, 103, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Stockley, L.; Rangdale, R.; Martinez-Urtaza, J. Environmental occurrence and clinical impact of Vibrio vulnificus and Vibrio parahaemolyticus: A European perspective. Environ. Microbiol. Rep. 2010, 2, 7–18. [Google Scholar] [CrossRef]
- Reynaud, Y.; Pitchford, S.; De Decker, S.; Wikfors, G.H.; Brown, C.L. Molecular typing of environmental and clinical strains of Vibrio vulnificus isolated in the northeastern USA. PLoS ONE 2013, 8, e83357. [Google Scholar] [CrossRef] [Green Version]
- Baker-Austin, C.; Oliver, J.D. Vibrio vulnificus: New insights into a deadly opportunistic pathogen. Environ. Microbiol. 2018, 20, 423–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Wang, M.-Y. The role of Vibrio vulnificus virulence factors and regulators in its infection-induced sepsis. Folia Microbiol. 2020, 65, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Lee, S.-H.; Kim, M.; Moon, J.-S.; Kim, G.-W.; Jung, H.-G.; Kim, I.H.; Oh, J.E.; Jung, H.E.; Lee, H.K. Vibrio vulnificus quorum-sensing molecule cyclo (Phe-Pro) inhibits RIG-I-mediated antiviral innate immunity. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Hackbusch, S.; Wichels, A.; Gimenez, L.; Döpke, H.; Gerdts, G. Potentially human pathogenic Vibrio spp. in a coastal transect: Occurrence and multiple virulence factors. Sci. Total Environ. 2020, 707, 136113. [Google Scholar] [CrossRef]
- Han, F.; Ge, B. Quantitative detection of Vibrio vulnificus in raw oysters by real-time loop-mediated isothermal amplification. Int. J. Food Microbiol. 2010, 142, 60–66. [Google Scholar] [CrossRef]
- Senoh, M.; Miyoshi, S.I.; Okamoto, K.; Fouz, B.; Amaro, C.; Shinoda, S. The cytotoxin-hemolysin genes of human and eel pathogenic Vibrio vulnificus strains: Comparison of nucleotide sequences and application to the genetic grouping. Microbiol. Immunol. 2005, 49, 513–519. [Google Scholar] [CrossRef]
- Sanjuán, E.; Fouz, B.; Oliver, J.D.; Amaro, C. Evaluation of genotypic and phenotypic methods to distinguish clinical from environmental Vibrio vulnificus strains. Appl. Environ. Microbiol. 2009, 75, 1604–1613. [Google Scholar] [CrossRef] [Green Version]
- Cohen, A.L.V.; Oliver, J.D.; DePaola, A.; Feil, E.J.; Boyd, E.F. Emergence of a virulent clade of Vibrio vulnificus and correlation with the presence of a 33-kilobase genomic island. Appl. Environ. Microbiol. 2007, 73, 5553–5565. [Google Scholar] [CrossRef] [Green Version]
- Baker-Austin, C.; McArthur, J.V.; Lindell, A.H.; Wright, M.S.; Tuckfield, R.C.; Gooch, J.; Warner, L.; Oliver, J.; Stepanauskas, R. Multi-site analysis reveals widespread antibiotic resistance in the marine pathogen Vibrio vulnificus. Microb. Ecol. 2009, 57, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Kurdi Al-Dulaimi, M.M.; Ariffin, A.A. Multiple antibiotic resistance (MAR), plasmid profiles, and DNA polymorphisms among Vibrio vulnificus isolates. Antibiotics 2019, 8, 68. [Google Scholar] [CrossRef] [Green Version]
- Pan, J.; Zhang, Y.; Jin, D.; Ding, G.; Luo, Y.; Zhang, J.; Mei, L.; Zhu, M. Molecular characterization and antibiotic susceptibility of Vibrio vulnificus in retail shrimps in Hangzhou, People’s Republic of China. J. Food Prot. 2013, 76, 2063–2068. [Google Scholar] [CrossRef] [PubMed]
- Roig, F.J.; Llorens, A.; Fouz, B.; Amaro, C. Spontaneous quinolone resistance in the zoonotic serovar of Vibrio vulnificus. Appl. Environ. Microbiol. 2009, 75, 2577–2580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hor, L.-I.; Goo, C.-T.; Wan, L. Isolation and characterization ofVibrio vulnificus inhabiting the marine environment of the southwestern area of Taiwan. J. Biomed. Sci. 1995, 2, 384–389. [Google Scholar] [CrossRef]
- Hsueh, P.-R.; Lin, C.-Y.; Tang, H.-J.; Lee, H.-C.; Liu, J.-W.; Liu, Y.-C.; Chuang, Y.-C. Vibrio vulnificus in Taiwan. Emerg. Infect. Dis. 2004, 10, 1363. [Google Scholar] [CrossRef]
- Dafale, N.A.; Srivastava, S.; Purohit, H.J. Zoonosis: An Emerging Link to Antibiotic Resistance under “One Health Approach”. Indian J. Microbiol. 2020, 60, 139–152. [Google Scholar] [CrossRef]
- Tsai, H.-C.; Chou, M.-Y.; Shih, Y.-J.; Huang, T.-Y.; Yang, P.-Y.; Chiu, Y.-C.; Chen, J.-S.; Hsu, B.-M. Distribution and genotyping of aquatic Acinetobacter baumannii strains isolated from the Puzi River and its tributaries near areas of livestock farming. Water-Sui 2018, 10, 1374. [Google Scholar] [CrossRef] [Green Version]
- Ho, Y.-N.; Tsai, H.-C.; Hsu, B.-M.; Chiou, C.-S. The association of Salmonella enterica from aquatic environmental and clinical samples in Taiwan. Sci. Total Environ. 2018, 624, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.-H.; Hsu, B.-M.; Chou, M.-Y.; Tsai, H.-L.; Kao, P.-M.; Wang, H.-J.; Hsiao, H.-Y.; Su, M.-J.; Huang, Y.-L. Application of molecular biological techniques to analyze Salmonella seasonal distribution in stream water. FEMS Microbiol. Lett. 2014, 352, 87–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, H.-C.; Tao, C.-W.; Hsu, B.-M.; Yang, Y.-Y.; Tseng, Y.-C.; Huang, T.-Y.; Huang, S.-W.; Kuo, Y.-J.; Chen, J.-S. Multidrug-resistance in methicillin-resistant Staphylococcus aureus (MRSA) isolated from a subtropical river contaminated by nearby livestock industries. Ecotoxicol. Environ. Saf. 2020, 200, 110724. [Google Scholar] [CrossRef]
- Hsu, C.-Y.; Hsu, B.-M.; Ji, W.-T.; Chang, T.-Y.; Kao, P.-M.; Tseng, S.-F.; Shen, T.-Y.; Shih, F.-C.; Fan, C.-W.; Liu, J.-H. A Potential Association Between Antibiotic Abuse and Existence of Related Resistance Genes in Different Aquatic Environments. Water Air Soil Pollut. 2014, 226, 2235. [Google Scholar] [CrossRef]
- Kang, S.J.; Jung, S.I.; Peck, K.R. Historical and Clinical Perspective of Vibrio vulnificus Infections in Korea. Infect. Chemother. 2020, 52, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Prousalis, M. An Investigation of Vibrio vulnificus and the Influence of Environmental Factors on Bacterial Abundance and Activity in a Subtropical Coastal Estuary, Santa Rosa County, Florida, USA; The University of West Florida: Ann Arbor, FL, USA, 2020. [Google Scholar]
- Johnson, C.N.; Bowers, J.C.; Griffitt, K.J.; Molina, V.; Clostio, R.W.; Pei, S.; Laws, E.; Paranjpye, R.N.; Strom, M.S.; Chen, A.; et al. Ecology of Vibrio parahaemolyticus and Vibrio vulnificus in the Coastal and Estuarine Waters of Louisiana, Maryland, Mississippi, and Washington (United States). Appl. Environ. Microbiol. 2012, 78, 7249–7257. [Google Scholar] [CrossRef] [Green Version]
- Dalsgaard, A.; Høi, L. Prevalence and characterization of Vibrio vulnificus isolated from shrimp products imported into Denmark. J. Food Prot. 1997, 60, 1132–1134. [Google Scholar] [CrossRef]
- Lin, M.; Schwarz, J.R. Seasonal shifts in population structure of Vibrio vulnificus in an estuarine environment as revealed by partial 16S ribosomal DNA sequencing. FEMS Microbiol. Ecol. 2003, 45, 23–27. [Google Scholar] [CrossRef]
- Wang, Q.; Fu, S.; Yang, Q.; Hao, J.; Zhou, C.; Liu, Y. The Impact of Water Intrusion on Pathogenic Vibrio Species to Inland Brackish Waters of China. Int. J. Environ. Res. Public Health 2020, 17, 6781. [Google Scholar] [CrossRef] [PubMed]
- Di, D.Y.W.; Lee, A.; Jang, J.; Han, D.; Hur, H.-G. Season-Specific Occurrence of Potentially Pathogenic Vibrio spp. on the Southern Coast of South Korea. Appl. Environ. Microbiol. 2017, 83, e02680-16. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, M.; Fischer, M.; Ottesen, A.; McCarthy, P.J.; Lopez, J.V.; Brown, E.W.; Monday, S.R. Population dynamics of Vibrio spp. associated with marine sponge microcosms. ISME J. 2010, 4, 1608–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lipp, E.K.; Rodriguez-Palacios, C.; Rose, J.B. Occurrence and distribution of the human pathogen Vibrio vulnificus in a subtropical Gulf of Mexico estuary. In The Ecology and Etiology of Newly Emerging Marine Diseases; Springer: Berlin/Heidelberg, Germany, 2001; pp. 165–173. [Google Scholar]
- Yokochi, N.; Tanaka, S.; Matsumoto, K.; Oishi, H.; Tashiro, Y.; Yoshikane, Y.; Nakashima, M.; Kanda, K.; Kobayashi, G. Distribution of virulence markers among Vibrio vulnificus isolates of clinical and environmental origin and regional characteristics in Japan. PLoS ONE 2013, 8, e55219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.; Liu, D.; Hwang, C.; Chen, M.; Hwang, J.; Liu, Y.; Shankuan, L.; Lin, C.; Wu, T. Survey on the distribution of Vibrionaceae at the seaport areas in Taiwan, 1991–1994. Chin. J. Microbiol. Immunol. 1996, 29, 197–209. [Google Scholar]
- Sudha, S.; Mridula, C.; Silvester, R.; Hatha, A. Prevalence and antibiotic resistance of pathogenic Vibrios in shellfishes from Cochin market. Indian J. Geo-Mar. Sci. 2014, 43, 815–824. [Google Scholar]
- Wolny-Koładka, K.; Lenart-Boroń, A. Phenotypic and molecular assessment of drug resistance profile and genetic diversity of waterborne Escherichia coli. Water Air Soil Pollut. 2016, 227, 146. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.-C.; Hsu, B.-M.; Kao, P.-M.; Tao, C.-W.; Ho, Y.-N.; Kuo, C.-W.; Huang, Y.-L. Seasonal distribution and prevalence of diarrheagenic Escherichia coli in different aquatic environments in Taiwan. Ecotoxicol. Environ. Saf. 2016, 124, 37–41. [Google Scholar] [CrossRef]
- Yu, D.; Yi, X.; Ma, Y.; Yin, B.; Zhuo, H.; Li, J.; Huang, Y. Effects of administration mode of antibiotics on antibiotic resistance of Enterococcus faecalis in aquatic ecosystems. Chemosphere 2009, 76, 915–920. [Google Scholar] [CrossRef]
- Campagnolo, E.R.; Johnson, K.R.; Karpati, A.; Rubin, C.S.; Kolpin, D.W.; Meyer, M.T.; Esteban, J.E.; Currier, R.W.; Smith, K.; Thu, K.M. Antimicrobial residues in animal waste and water resources proximal to large-scale swine and poultry feeding operations. Sci. Total Environ. 2002, 299, 89–95. [Google Scholar] [CrossRef]
- Shaw, K.S.; Rosenberg Goldstein, R.E.; He, X.; Jacobs, J.M.; Crump, B.C.; Sapkota, A.R. Antimicrobial Susceptibility of Vibrio vulnificus and Vibrio parahaemolyticus Recovered from Recreational and Commercial Areas of Chesapeake Bay and Maryland Coastal Bays. PLoS ONE 2014, 9, e89616. [Google Scholar] [CrossRef]
- Biosca, E.G.; Amaro, C.; Larsen, J.L.; Pedersen, K. Phenotypic and genotypic characterization of Vibrio vulnificus: Proposal for the substitution of the subspecific taxon biotype for serovar. Appl. Environ. Microbiol. 1997, 63, 1460–1466. [Google Scholar] [CrossRef] [Green Version]
- Vickery, M.C.L.; Nilsson, W.B.; Strom, M.S.; Nordstrom, J.L.; DePaola, A. A real-time PCR assay for the rapid determination of 16S rRNA genotype in Vibrio vulnificus. J. Microbiol. Meth. 2007, 68, 376–384. [Google Scholar] [CrossRef]
- Panicker, G.; Call, D.R.; Krug, M.J.; Bej, A.K. Detection of pathogenic Vibrio spp. in shellfish by using multiplex PCR and DNA microarrays. Appl. Environ. Microbiol. 2004, 70, 7436–7444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, F.; Ge, B. Multiplex PCR assays for simultaneous detection and characterization of Vibrio vulnificus strains. Lett. Appl. Microbiol. 2010, 51, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Jeong, H.D. Development of 16S rRNA targeted PCR methods for the detection and differentiation of Vibrio vulnificus in marine environments. Aquaculture 2001, 193, 199–211. [Google Scholar] [CrossRef]
- Kim, H.-J.; Cho, J.-C. Genotypic diversity and population structure of Vibrio vulnificus strains isolated in Taiwan and Korea as determined by multilocus sequence typing. PLoS ONE 2015, 10, e0142657. [Google Scholar] [CrossRef] [Green Version]
- Han, F.; Pu, S.; Hou, A.; Ge, B. Characterization of clinical and environmental types of Vibrio vulnificus isolates from Louisiana oysters. Foodborne Pathog. Dis. 2009, 6, 1251–1258. [Google Scholar] [CrossRef] [PubMed]
- Çam, S.; Brinkmeyer, R.; Schwarz, J.R. Quantitative PCR enumeration of vcgC and 16S rRNA type A and B genes as virulence indicators for environmental and clinical strains of Vibrio vulnificus in Galveston Bay oysters. Can. J. Microbiol. 2019, 65, 613–621. [Google Scholar] [CrossRef]
- Chatzidaki-Livanis, M.; Hubbard, M.A.; Gordon, K.; Harwood, V.J.; Wright, A.C. Genetic distinctions among clinical and environmental strains of Vibrio vulnificus. Appl. Environ. Microbiol. 2006, 72, 6136–6141. [Google Scholar] [CrossRef] [Green Version]
- Chatzidaki-Livanis, M.; Jones, M.K.; Wright, A.C. Genetic variation in the Vibrio vulnificus group 1 capsular polysaccharide operon. J. Bacteriol. 2006, 188, 1987–1998. [Google Scholar] [CrossRef] [Green Version]
- Bier, N.; Bechlars, S.; Diescher, S.; Klein, F.; Hauk, G.; Duty, O.; Strauch, E.; Dieckmann, R. Genotypic diversity and virulence characteristics of clinical and environmental Vibrio vulnificus isolates from the Baltic Sea region. Appl. Environ. Microbiol. 2013, 79, 3570–3581. [Google Scholar] [CrossRef] [Green Version]
- Wong, H.-C.; You, W.-Y.; Chen, S.-Y. Detection of Toxigenic Vibrio cholerae, Vparahaemolyticus and Vvulnificus in Oyster by Multiplex-PCR with Internal Amplification Control. J. Food Drug Anal. 2012, 20, 48–58. [Google Scholar]
- Wei, S.; Zhao, H.; Xian, Y.; Hussain, M.A.; Wu, X. Multiplex PCR assays for the detection of Vibrio alginolyticus, Vibrio parahaemolyticus, Vibrio vulnificus, and Vibrio cholerae with an internal amplification control. Diagn. Microbiol. Infect. Dis. 2014, 79, 115–118. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.t.; Carmeli, Y.; Falagas, M.t.; Giske, C.t.; Harbarth, S.; Hindler, J.t.; Kahlmeter, G.; Olsson-Liljequist, B. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [Green Version]

| Seasons | DS & BD Fishing Harbor (HW) (Shellfish) | DS & BD Fishing Harbor (HW) (Water) | Area A of PR (Water) | Area B of PR (Water) | Area C of PR (Water) | Sum of PR (Water) |
|---|---|---|---|---|---|---|
| Spring | 0/10 (0%) | 0/6 (0%) | 0/8 (0%) | 0/8 (0%) | 0/8 (0%) | 0/24 (0%) |
| Summer | 0/10 (0%) | 0/6 (0%) | 0/8 (0%) | 1/8 (12.5%) | 2/8 (25%) | 3/24 (12.5%) |
| Autumn | 0/10 (0%) | 1/6 (16.7%) | 0/8 (0%) | 0/8 (0%) | 0/8 (0%) | 0/24 (0%) |
| Winter | 0/10 (0%) | 0/6 (0%) | 0/8 (0%) | 3/8 (37.5%) | 2/8 (25%) | 5/24 (20.8%) |
| Total | 0/40 (0%) | 1/24 (4.2%) | 0/32 (0%) | 4/32 (12.5%) | 4/32 (12.5%) | 8/96 (8.3%) |
| Strains | Zone of Inhibition (mm) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ampicillin | Amoxycillin-Clavulanic Acid | Ampicillin-Sulbactam | Cefepime | Chloramphenicol | Ciprofloxacin | Gentamicin | Imipenem | Tetracycline | Trimethoprim-Sulfamethoxazole | |
| S02PR2311 | 29 | 25 | 25 | 23 | 25 | 19 | 20 | 23 | 22 | 25 |
| S02PR2911 | 21 | 22 | 22 | 23 | 25 | 26 | 20 | 23 | 23 | 23 |
| S02PR3111 | 23 | 26 | 23 | 28 | 32 | 32 | 24 | 34 | 33 | 33 |
| S04PR2211 | 35 | 28 | 27 | 31 | 36 | 34 | 23 | 38 | 32 | 32 |
| S04PR2311 | 26 | 30 | 29 | 32 | 31 | 32 | 22 | 29 | 34 | 36 |
| S04PR2511 | 28 | 26 | 20 | 26 | 25 | 26 | 20 | 26 | 19 | 23 |
| S04PR2711 | 19 | 21 | 23 | 22 | 33 | 28 | 22 | 19 | 22 | 25 |
| BD-FH W211 | 23 | 22 | 23 | 31 | 34 | 37 | 23 | 27 | 26 | 28 |
| S04PR3111 | 19 | 19 | 19 | 25 | 26 | 31 | 25 | 22 | 28 | 24 |
| Resistant | ≤13 | ≤13 | ≤11 | ≤14 | ≤12 | ≤15 | ≤12 | ≤13 | ≤14 | ≤10 |
| Intermediate | 14–16 | 14–17 | 12–14 | 15–17 | 13–17 | 16–20 | 13–14 | 14–15 | 15–18 | 11–15 |
| Susceptible | ≥17 | ≥18 | ≥15 | ≥18 | ≥18 | ≥21 | ≥15 | ≥16 | ≥19 | ≥16 |
| Strain | Sampling Type | Virulent Type | Nonvirulent Type | PRXII | nanA | manIIA | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| vcgC | 16S B | CPS1 | vvhA | vcgE | 16S A | CPS2 | vvhA | |||||
| S02PR2911 | PR Area C | + | + | + | + | + | + | + | + | |||
| S02PR3111 | PR Area C | + | + | + | + | + | + | + | + | |||
| S04PR2211 | PR Area B | + | + | + | + | + | + | |||||
| S04PR2311 | PR Area B | + | + | + | + | + | + | + | ||||
| S02PR2311 | PR Area B | + | + | + | + | |||||||
| BD-FH W211 | Fishing harbor | + | + | + | + | + | + | + | ||||
| S04PR2711 | PR Area C | + | + | + | + | + | + | + | ||||
| S04PR2511 | PR Area B | + | + | + | + | + | + | |||||
| S04PR3111 | PR Area C | + | + | + | + | + | + | |||||
| Toal | 8/9 (88.9%) | 8/9 (88.9%) | 6/9 (66.7%) | 8/9 (88.9%) | 2/9 (22.2%) | 6/9 (66.7%) | 1/9 (11.1%) | 1/9 (11.1%) | 4/9 (44.4%) | 7/9 (77.8%) | 8/9 (88.9%) | |
| Target Gene | Size | Sequence (5′ to 3′) | Reaction Materials Final Volume: 25 μL | PCR Condition | Reference |
|---|---|---|---|---|---|
| vvhA | 505 | FDAvvhA-F: 5′-CCGCGGTACAGGTTGGCGCA-3′ FDAvvhA-R: 5′-CGCCACCCACTTTCGGGCC-3′ | DNA: 100–300 ng Primer: 300 nM Master mix: 5 μL | Pre-denaturation: 94 °C 3 min Denaturation: 94 °C 60 s Annealing: 60 °C 60 s Extension: 72 °C 60 s D.A.E. Cycles: 30 cycles Final extension: 72 °C 10 min | [65,66] |
| ERIC | - | ERIC1R: 5′-ATGTAAGCTCCTGGGGATTCAC-3′ ERIC2: 5′-AAGTAAGTGACTGGGGTGAGCG-3′ | DNA: 100–300 ng Primer: 5000 nM Master mix: 5 μL | Pre-denaturation: 95 °C 7 min Denaturation: 92 °C 45 s Annealing: 54 °C 60 s Extension: 70 °C 10 min D.A.E. Cycles: 35 cycles Final extension: 72 °C 20 min | [3] |
| Virulent type vcgC vcgC 16S B CPS1 | 99 278 839 342 | vcgC-F: 5′-AGCTGCCGATAGCGATCT-3′ vcgC-R: 5′-TGAGCTAACGCGAGTAGTGAG-3′ vcg-P1: 5′-AGCTGCCGATAGCGATCT-3′ vcg-P3: 5′-CGCTTAGGATGATCGGTG-3′ 16S B-F1: 5′-GCCTACGGGCCAAAGAGG-3′ 16S B-R1: 5′-CCTGCGTCTCCGCTGGCT-3′ CPS1HP-1F: 5′-TTTGGGATTTGAAAGGCTTG-3′ CPS1HP-1R: 5′-GTGCCTTTGCGAATTTTGAT-3′ | DNA: 100–300 ng Primer: 300 nM vcgC-FR, 200 nM vcg-P13, 200 nM 16S B-FR, 700 nM CPS1HP-FR Master mix: 5 μL | Pre-denaturation: 95 °C 5 min Denaturation: 94 °C 60 s Annealing: 56 °C 60 s Extension: 72 °C 60 s D.A.E. Cycles: 30 cycles Final extension: 72 °C 7 min | [21] |
| Nnvoirulent type vcgE 16S A CPS2 | 278 839 152 | vcg-P2: 5′-CTCAATTGACAATGATCT-3′ vcg-P3: 5′-CGCTTAGGATGATCGGTG-3′ 16S A-F2: 5′-AGCTTCGGCTCAAAGAGG-3′ 16S A-R2: 5′-CCAGCGTCTCCGCTAGAT-3′ CPS2HP-2F: 5′-TTCCATCAAACATCGCAGAA-3′ CPS2HP-2R: 5′-CTTTTGTCCGGCTTCTATCG-3′ | DNA: 100–300 ng Primer: 300 nM vcg-P23, 300 nM 16S A-FR, 200 nM CPS2HP-FR Master mix: 5 μL | Pre-denaturation: 95 °C 5 min Denaturation: 94 °C 60 s Annealing: 50 °C 60 s Extension: 72 °C 60 s D.A.E. Cycles: 30 cycles Final extension: 72 °C 7 min | [57] |
| Virulent type vvhA-1 | 814 | vvhA-1F: 5′-AGATTAAGTGTGTGTTGCACACAAGCGGTG-3′ vvhA-1R: 5′-ACCGAAAACAGCGCTGAAGGAAGAACGGTA-3′ | DNA: 100–300 ng Primer: 400 nM Master mix: 5 μL | Pre-denaturation: 95 °C 2 min Denaturation: 95 °C 30 s Annealing: 57 °C 30 s Extension: 72 °C 90 s D.A.E. Cycles: 30 cycles Final extension: 72 °C 3 min | [22] |
| Nnvoirulent type vvhA-2 | 814 | vvhA-2F: 5′-AAATTAAGTGCGTGCTACACACAAGTGGTG-3′ vvhA-2R: 5′-ACTGAGAAGAGTGCTGAAGGGATTACCGTA-3′ | DNA: 100–300 ng Primer: 400 nM Master mix: 5 μL | Pre-denaturation: 95 °C 2 min Denaturation: 95 °C 30 s Annealing: 57 °C 30 s Extension: 72 °C 90 s D.A.E. Cycles: 30 cycles Final extension: 72 °C 3 min | [22] |
| PRXII, nanA, manIIA | 2257 1299 243 | VVA1612F: 5′-ACCCTGATCGTTGGCTACTC-3′ VVA1613R: 5′-GGAGCGGTGTGATGGTGTTG-3′ rpiR-F: 5′-TACGCAAGCCCAGCGGCATG-3′ nanA-2R: 5′-TTGCCACTTCCGCGATCGGG-3′ ManIIA-F: 5′-GATGTTGGTGAACAACTTCTCTGC-3′ ManIIA-R: 5′-TCTGAAGCCTGTTGGATGCC-3′ | DNA: 100–300 ng Primer: 800 nM VVA-FR, 200 nM nanA-FR, 200 nM ManIIA-FR Master mix: 5 μL | Pre-denaturation: 94 °C 4 min Denaturation: 94 °C 30 s Annealing: 63 °C 30 s Extension: 72 °C 2.5 min D.A.E. Cycles: 30 cycles Final extension: 72 °C 10 min | [1] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, I.-C.; Hussain, B.; Hsu, B.-M.; Chen, J.-S.; Hsu, Y.-L.; Chiu, Y.-C.; Huang, S.-W.; Wang, J.-L. Prevalence, Genetic Diversity, Antimicrobial Resistance, and Toxigenic Profile of Vibrio vulnificus Isolated from Aquatic Environments in Taiwan. Antibiotics 2021, 10, 505. https://doi.org/10.3390/antibiotics10050505
Lin I-C, Hussain B, Hsu B-M, Chen J-S, Hsu Y-L, Chiu Y-C, Huang S-W, Wang J-L. Prevalence, Genetic Diversity, Antimicrobial Resistance, and Toxigenic Profile of Vibrio vulnificus Isolated from Aquatic Environments in Taiwan. Antibiotics. 2021; 10(5):505. https://doi.org/10.3390/antibiotics10050505
Chicago/Turabian StyleLin, I-Ching, Bashir Hussain, Bing-Mu Hsu, Jung-Sheng Chen, Yu-Ling Hsu, Yi-Chou Chiu, Shih-Wei Huang, and Jiun-Ling Wang. 2021. "Prevalence, Genetic Diversity, Antimicrobial Resistance, and Toxigenic Profile of Vibrio vulnificus Isolated from Aquatic Environments in Taiwan" Antibiotics 10, no. 5: 505. https://doi.org/10.3390/antibiotics10050505
APA StyleLin, I.-C., Hussain, B., Hsu, B.-M., Chen, J.-S., Hsu, Y.-L., Chiu, Y.-C., Huang, S.-W., & Wang, J.-L. (2021). Prevalence, Genetic Diversity, Antimicrobial Resistance, and Toxigenic Profile of Vibrio vulnificus Isolated from Aquatic Environments in Taiwan. Antibiotics, 10(5), 505. https://doi.org/10.3390/antibiotics10050505






