Comparison of Antibiotic Resistance Profile of Escherichia coli between Pristine and Human-Impacted Sites in a River
Abstract
:1. Introduction
2. Results and Discussion
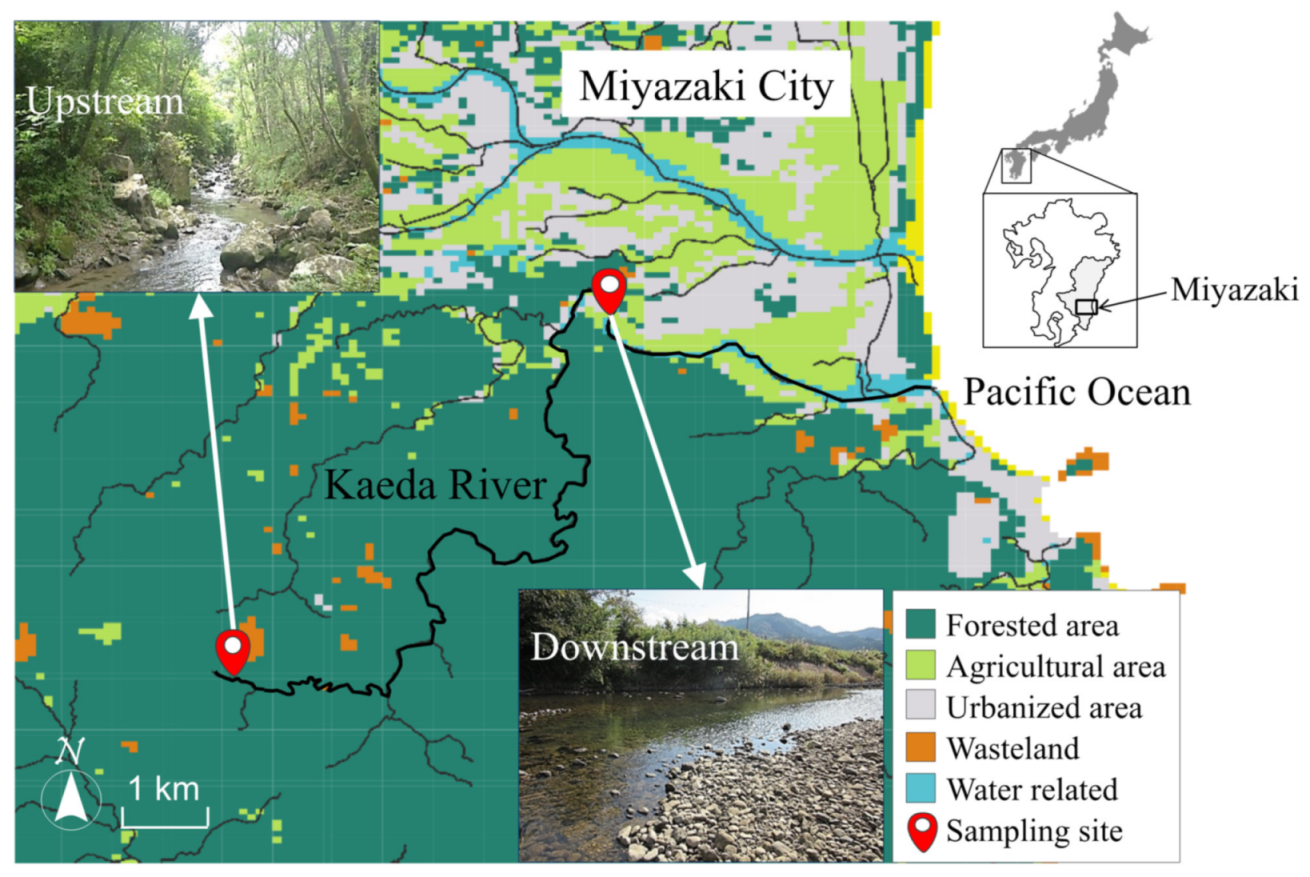
2.1. Water Qualities in the Kaeda River
2.2. Identification of E. coli Isolates from the Kaeda River
2.3. Seasonal Changes in the Antibiotic-Resistant Rate of E. coli
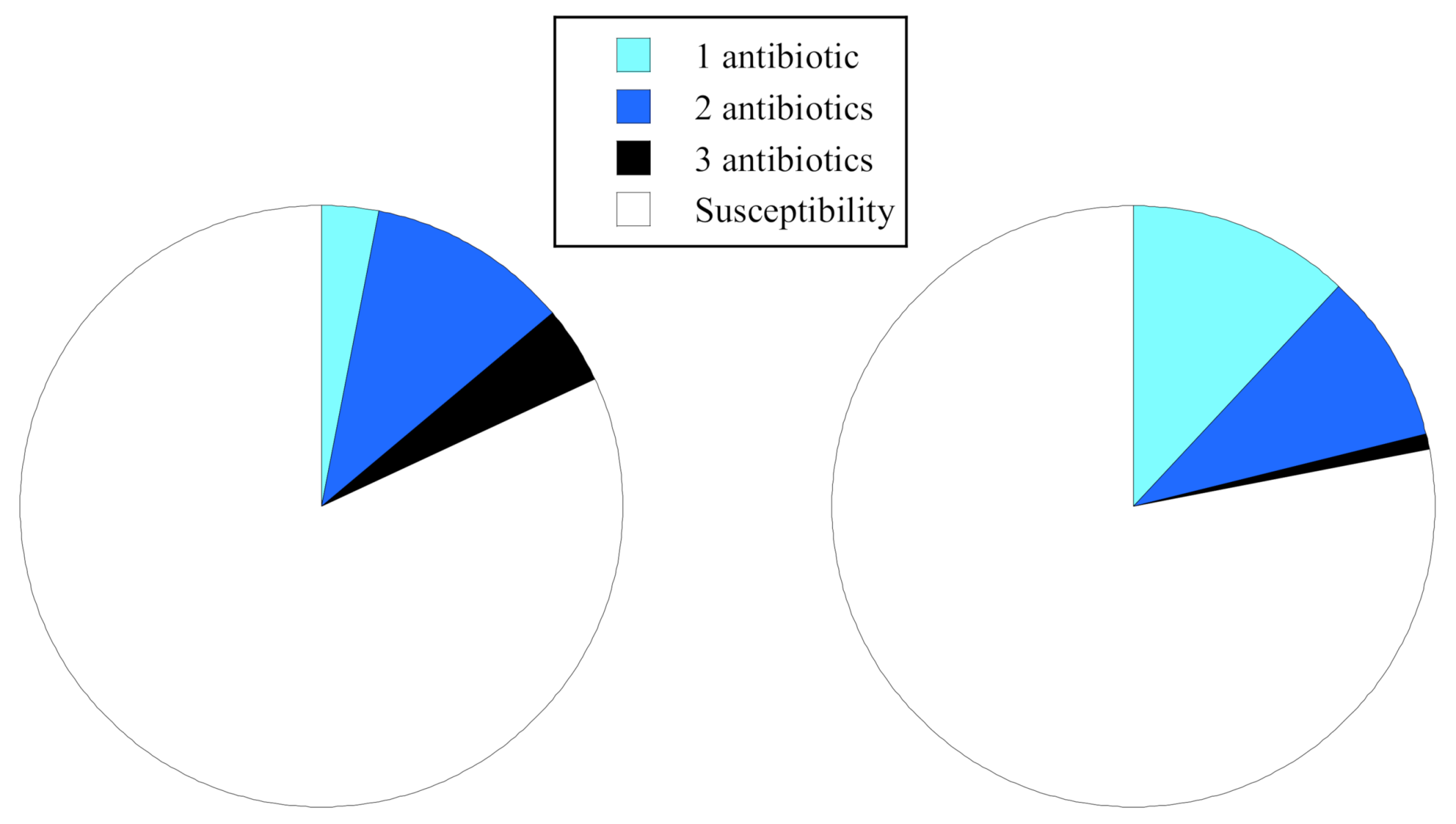
2.4. Comparisons of Antibiotic Resistance Rate and Resistance Patterns
2.5. Comparison of Antibiotic Resistance Profiles and PFGE Types
3. Materials and Methods
3.1. Sampling
3.2. Enumeration of Fecal Indicator Bacteria
3.3. Identification of E. coli by PCR Analysis
3.4. Determination of Minimum Inhibitory Concentration (MIC)
3.5. PFGE Typing
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- O’Neil, J. Review on antibiotic resistance. Antimicrobial resistance: Tackling a crisis for the health and wealth of nations. Health Wealth Nations 2014, December, 1–16. Available online: http://www.jpiamr.eu/wp-content/uploads/2014/12/AMR-Review-Paper-Tackling-a-crisis-for-the-health-and-wealth-of-nations_1-2.pdf (accessed on 12 May 2020).
- Tsuzuki, S.; Matsunaga, N.; Yahara, K.; Gu, Y.; Hayakawa, K.; Hirabayashi, A.; Kajihara, T.; Sugai, M.; Shibayama, K.; Ohmagari, N. National trend of blood-stream infection attributable deaths caused by Staphylococcus aureus and Escherichia coli in Japan. J. Infect. Chemother. 2020, 26, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Willyard, C. The drug-resistant bacteria that pose the greatest health threats. Nature 2017, 543, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Centers for Disease Control and Prevention (C.D.C.). Antibiotic Resistance Threats in the United States 2019; U.S. Department of Health and Human Services, C.D.C.: Atlanta, GA, USA, 2019; pp. 1–148. [Google Scholar]
- Müller, A.; Stephan, R.; Nüesch-Inderbinen, M. Distribution of virulence factors in ESBL-producing Escherichia coli isolated from the environment, livestock, food and humans. Sci. Total Environ. 2016, 541, 667–672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The AMR One Health Surveillance Committee. Nippon AMR One Health Report (NAOR) 2020; Infectious Diseases Control Division, Health Service Bureau, Ministry of Health, Labour and Welfare: Tokyo, Japan, 2020. [Google Scholar]
- Santos, T.; Silva, N.; Igrejas, G.; Rodrigues, P.; Micael, J.; Rodrigues, T.; Resendes, R.; Gonçalves, A.; Marinho, C.; Gonçalves, D.; et al. Dissemination of antibiotic resistant Enterococcus spp. and Escherichia coli from wild birds of Azores Archipelago. Anaerobe 2013, 24, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Raverty, S.A.; Rhodes, L.D.; Zabek, E.; Eshghi, A.; Cameron, C.E.; Hanson, M.B.; Schroeder, J.P. Respiratory microbiome of endangered southern resident killer whales and microbiota of surrounding sea surface microlayer in the eastern North Pacific. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Norman, S.A.; Lambourn, D.M.; Huggins, J.L.; Gaydos, J.K.; Dubpernell, S.; Berta, S.; Olson, J.K.; Souze, V.; Evans, A.; Carlson, B.; et al. Antibiotic resistance of bacteria in two marine mammal species, harbor seals and harbor porpoises, living in an urban marine ecosystem, the Salish Sea, Washington State, USA. Oceans 2021, 2, 86–104. [Google Scholar] [CrossRef]
- Bhullar, K.; Waglechner, N.; Pawlowski, A.; Koteva, K.; Banks, E.D.; Johnston, M.D.; Barton, H.A.; Wright, G.D. Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS ONE 2012, 7, e34953. [Google Scholar] [CrossRef] [PubMed]
- Segawa, T.; Takeuchi, N.; Rivera, A.; Yamada, A.; Yoshimura, Y.; Barcaza, G.; Shinbori, K.; Motoyama, H.; Kohshima, S.; Ushida, K. Distribution of antibiotic resistance genes in glacier environments. Environ. Microbiol. Rep. 2013, 5, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Urase, T.; Sato, T. Quantitative monitoring of resistance in Escherichia coli to clinically important antimicrobials in an urban watershed. J. Water Environ. Technol. 2016, 14, 341–349. [Google Scholar] [CrossRef] [Green Version]
- Swift, B.M.C.; Bennett, M.; Waller, K.; Dodd, C.; Murray, A.; Gomes, R.L.; Humphreys, B.; Hobman, J.L.; Jones, M.A.; Whitlock, S.E.; et al. Anthropogenic environmental drivers of antimicrobial resistance in wildlife. Sci. Total Environ. 2019, 649, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Ahlstrom, C.A.; van Toor, M.L.; Woksepp, H.; Chandler, J.C.; Reed, J.A.; Reeves, A.B.; Waldenström, J.; Franklin, A.B.; Douglas, D.C.; Bonnedahl, J.; et al. Evidence for continental-scale dispersal of antimicrobial resistant bacteria by landfill-foraging gulls. Sci. Total Environ. 2021, 764, 144551. [Google Scholar] [CrossRef] [PubMed]
- Ham, Y.S.; Kobori, H.; Kang, J.H.; Matsuzaki, T.; Iino, M.; Nomura, H. Distribution of antibiotic resistance in urban watershed in Japan. Environ. Pollut. 2012, 162, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Roth, N.; Käsbohrer, A.; Mayrhofer, S.; Zitz, U.; Hofacre, C.; Domig, K.J. The application of antibiotics in broiler production and the resulting antibiotic resistance in Escherichia coli: A global overview. Poult. Sci. 2019, 98, 1791–1804. [Google Scholar] [CrossRef] [PubMed]
- National Veterinary Assay Laboratory, Ministry of Agriculture, Forestry and Fishers. Report on the Japanese Veterinary Antimicrobial Resistance Monitoring System 2016–2017; AMR Clinical Reference Center: Tokyo, Japan, 2020. [Google Scholar]
- Nishiyama, M.; Ogura, Y.; Hayashi, T.; Suzuki, Y. Antibiotic resistance profiling and genotyping of vancomycin-resistant enterococci collected from an urban river basin in the Provincial City of Miyazaki, Japan. Water 2017, 9, 79. [Google Scholar] [CrossRef]
- Ogura, Y.; Ueda, T.; Nukazawa, K.; Hiroki, H.; Xie, H.; Arimizu, Y.; Hayashi, T.; Suzuki, Y. Antimicrobial resistance is more prevalent in various lineages of Escherichia coli strains isolated from sewage than in those from rivers. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Hashimoto, R.; Xie, H.; Nishimura, E.; Nishiyama, M.; Nukazawa, K.; Ishii, S. Growth and antibiotic resistance acquisition of Escherichia coli in a river that receives treated sewage effluent. Sci. Total Environ. 2019, 690, 696–704. [Google Scholar] [CrossRef] [PubMed]
- United States Environmental Protection Agency. Method 1600: Enterococci in Water by Membrane Filtration Using Membrane-Enterococcus Indoxyl-β-D-Glucoside Agar (mEI); U.S. Environmental Protection Agency: Washington, DC, USA, 2002. [Google Scholar]
- Osek, J.; Dacko, J. Development of a PCR-based method for specific identification of genotypic markers of shiga toxin-producing Escherichia coli strains. J. Vet. Med. B Infect. Dis. Vet. Public Health 2001, 48, 771–778. [Google Scholar] [CrossRef] [PubMed]
- Clinical Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Test for Bacteria That Grow Aerobically, 9th ed.; Clinical and Laboratory Standard Institute: Wayne, PA, USA, 2012; ISBN 1-56238-784-7. [Google Scholar]
- Clinical Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Second Informational Supplement; Clinical and Laboratory Standard Institute: Wayne, PA, USA, 2012; ISBN 1-56238-786-3. [Google Scholar]
- CDC PulseNet. Standard Operating Procedure for PulseNet PFGE of Escherichia coli O157: H7, Escherichia coli non-O157 (STEC), Salmonella Serotypes, Shigella Sonnei and Shigella Flexneri; PNL05 Centers for Disease Control and Prevention: Atlanta, GA, USA, 2017; Volume 157, pp. 1–16. [Google Scholar]

| Parameter | Pristine Upstream | Human-Impacted Downstream | ||||
|---|---|---|---|---|---|---|
| 29 Jul., 2016 | 26 Dec., 2016 | 6 Apr., 2017 | 29 Jul., 2016 | 26 Dec., 2016 | 6 Apr., 2017 | |
| Water temp (°C) | 21.2 | 11.2 | 12.6 | 28.3 | 12.4 | 14.2 |
| DO (mg/L) | 8.4 | 9.9 | 9.5 | 9.4 | 10 | 9.7 |
| pH (-) | 6.4 | 6.5 | 6.3 | 6.7 | 6.6 | 6.5 |
| EC (μS/cm) | 62 | 110 | 60 | 93 | 98 | 81 |
| Turbidity (kaolin unit) | 1.1 | 0.12 | 0.42 | 1.2 | 0.28 | 1.0 |
| TOC (mg-C/L) | 0.40 | 0.37 | 0.34 | 0.58 | 0.57 | 0.46 |
| Total coliform (CFU/100 mL) | (7.8 ± 1.4) * × 102 | (2.2 ± 0.5) × 102 | (3.0 ± 0.2) × 102 | (1.6 ± 0.3) × 103 | (7.2 ± 0.9) × 102 | (8.0 ± 0.4) × 102 |
| Escherichia coli (CFU/100 mL) | (1.8 ± 0.2) × 101 | 2 ± 2 | (1.3 ± 0.1) × 101 | (3.7 ± 1.7) × 101 | (8.0 ± 1.6) × 101 | (4.3 ± 1.9) × 101 |
| Enterococci (CFU/100 mL) | (3.6 ± 1.7) × 101 | 5 ± 0.1 | 5 ± 0.3 | (5.3 ± 0.4) × 101 | (1.5 ± 0.3) × 101 | 7 ± 4 |
| Sampling Point | Pristine Upstream (98 Isolates) | Human-Impacted Downstream (89 Isolates) | ||||||
|---|---|---|---|---|---|---|---|---|
| Date | 29 Jul., 2016 | 26 Dec., 2016 | 6 Apr., 2017 | Total | 29 Jul., 2016 | 26 Dec., 2016 | 6 Apr., 2017 | Total |
| Resistance rate (%) | 46 | 4 | 5 | 18 | 31 | 5 | 23 | 23 |
| (Isolates) | (15/33) | (1/22) | (2/41) | (18/98) | (12/39) | (1/19) | (7/31) | (20/89) |
| Group | Antimicrobial Agent | MIC Test Range (μg/mL) | Pristine Upstream (98 Isolates) | Human-Impacted Downstream (89 Isolates) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Susceptible | Intermediate | Rsistant | MIC50 (μg/mL) | MIC90 (μg/mL) | Susceptible | Intermediate | Rsistant | MIC50 (μg/mL) | MIC90 (μg/mL) | |||
| No. Isolates (% Isolates) | No. Isolates (% Isolates) | |||||||||||
| Penicillins | ABPC | 0.25–128 | 89 (91%) | 1 (1%) | 8 (8%) | 2 | 8 | 69 (78%) | 5 (6%) | 15 (16%) | 2 | 64 |
| Cephem | CEZ | 0.0625–32 | 67 (68%) | 14 (14%) | 17 (17%) | 1 | 32 | 58 (65%) | 17 (19%) | 14 (16%) | 1 | 16 |
| CTX | 0.03–16 | 98 (100%) | 0 (0%) | 0 (0%) | 0.03 | 0.25 | 89 (100%) | 0 (%) | 0 (%) | 0.03 | 0.125 | |
| Aminoglycosides | GM | 0.125–64 | 98 (100%) | 0 (0%) | 0 (0%) | 1 | 2 | 89 (100%) | 0 (%) | 0 (%) | 0.5 | 2 |
| TOB | 0.125–64 | 98 (100%) | 0 (0%) | 0 (0%) | 0.5 | 2 | 89 (100%) | 0 (%) | 0 (%) | 0.5 | 2 | |
| Carbapenems | IPM | 0.03–16 | 97 (99%) | 1 (1%) | 0 (0%) | 0.06 | 0.5 | 86 (97%) | 3 (3%) | 0 (%) | 0.06 | 0.25 |
| Fluoroquinolons | CPFX | 0.03–16 | 98 (100%) | 0 (0%) | 0 (0%) | — | 0.03 | 86 (97%) | 3 (3%) | 0 (%) | — | 0.03 |
| Tetracyclines | TC | 0.125–64 | 86 (88%) | 0 (0%) | 12 (12%) | 2 | 8 | 88 (99%) | 0 (%) | 1 (1%) | 4 | 8 |
| Chloramphenicols | CP | 0.25–128 | 96 (98%) | 2 (2%) | 0 (0%) | 1 | 32 | 89 (100%) | 0 (%) | 0 (%) | 1 | 4 |
| Year | Sampling Date | Sampling Point (n = Isolates) | Resistance Profile | Number of Isolates (%) |
|---|---|---|---|---|
| 2016 | 29 Jul. | Upstream | Susceptibility | 18 (55%) |
| (Summer) | (n = 33) | TC | 1 (3%) | |
| ABPC-CEZ | 3 (9%) | |||
| CEZ-TC | 7 (21%) | |||
| ABPC-CEZ-TC | 4 (12%) | |||
| Downstream | Susceptibility | 27 (69%) | ||
| (n = 39) | ABPC | 6 (15%) | ||
| CEZ | 1 (4%) | |||
| ABPC-CEZ | 4 (10%) | |||
| CEZ-TC | 1 (3%) | |||
| 2016 | 26 Dec. | Upstream | Susceptibility | 23 (96%) |
| (Winter) | (n = 24) | ABPC | 1 (4%) | |
| Downstream | Not examined | 18 (95%) | ||
| (n = 19) | CEZ | 1 (5%) | ||
| 2017 | 6 Apr. | Upstream | Susceptibility | 39 (95%) |
| (Spring) | (n = 41) | CEZ | 1 (2%) | |
| ABPC-CEZ | 1 (2%) | |||
| Down stream | Susceptibility | 24 (77%) | ||
| (n = 31) | ABPC | 1 (3%) | ||
| CEZ | 3 (10%) | |||
| ABPC-CEZ | 2 (7%) | |||
| ABPC-CEZ-TC | 1 (3%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nishimura, E.; Nishiyama, M.; Nukazawa, K.; Suzuki, Y. Comparison of Antibiotic Resistance Profile of Escherichia coli between Pristine and Human-Impacted Sites in a River. Antibiotics 2021, 10, 575. https://doi.org/10.3390/antibiotics10050575
Nishimura E, Nishiyama M, Nukazawa K, Suzuki Y. Comparison of Antibiotic Resistance Profile of Escherichia coli between Pristine and Human-Impacted Sites in a River. Antibiotics. 2021; 10(5):575. https://doi.org/10.3390/antibiotics10050575
Chicago/Turabian StyleNishimura, Emi, Masateru Nishiyama, Kei Nukazawa, and Yoshihiro Suzuki. 2021. "Comparison of Antibiotic Resistance Profile of Escherichia coli between Pristine and Human-Impacted Sites in a River" Antibiotics 10, no. 5: 575. https://doi.org/10.3390/antibiotics10050575
APA StyleNishimura, E., Nishiyama, M., Nukazawa, K., & Suzuki, Y. (2021). Comparison of Antibiotic Resistance Profile of Escherichia coli between Pristine and Human-Impacted Sites in a River. Antibiotics, 10(5), 575. https://doi.org/10.3390/antibiotics10050575






