Crosstalks between NOD1 and Histone H2A Contribute to Host Defense against Streptococcus agalactiae Infection in Zebrafish
Abstract
:1. Introduction
2. Results and Discussion
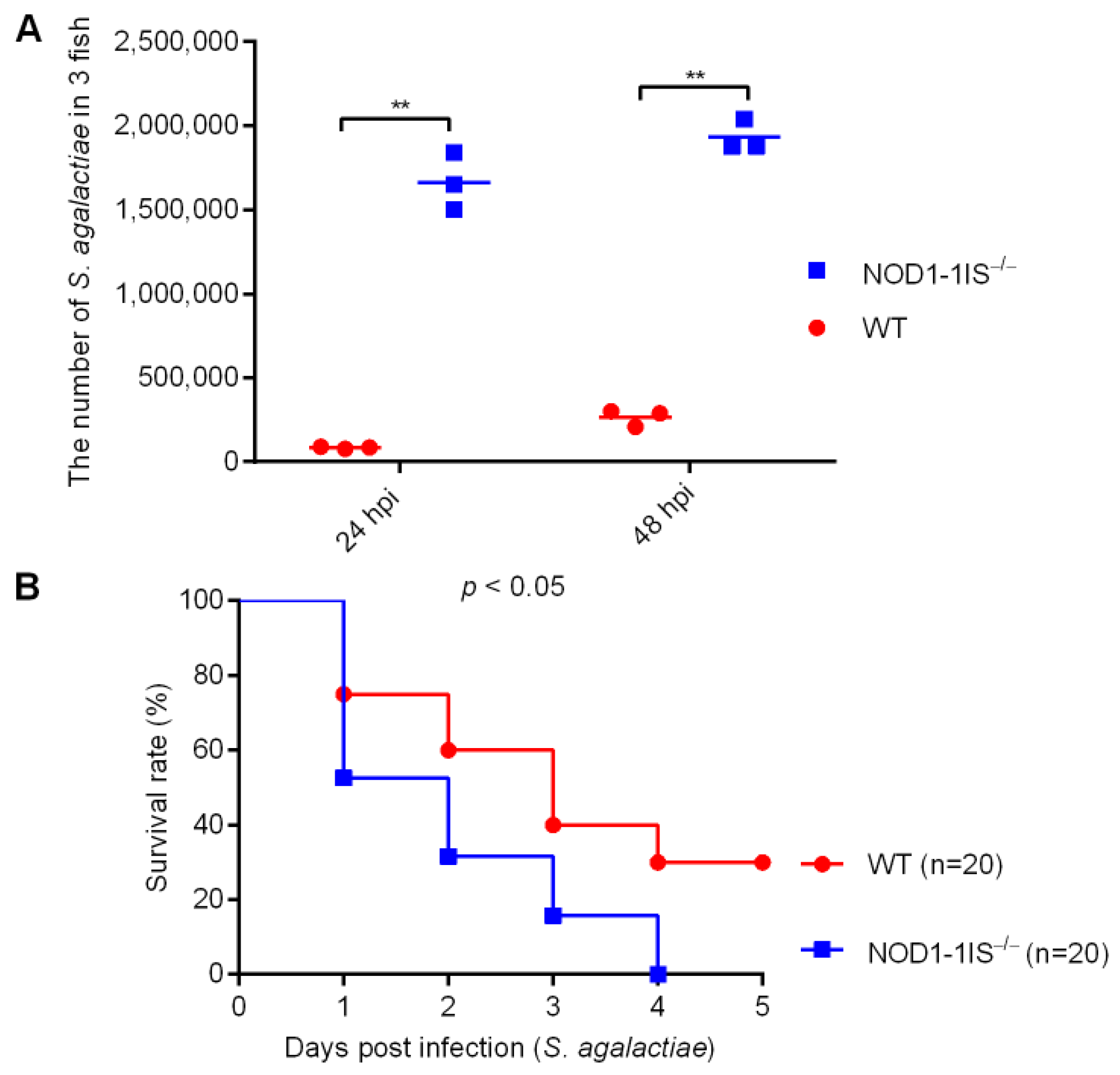
2.1. NOD1 Contributes to Host Defense against S. agalactiae Infection in Adult Zebrafish
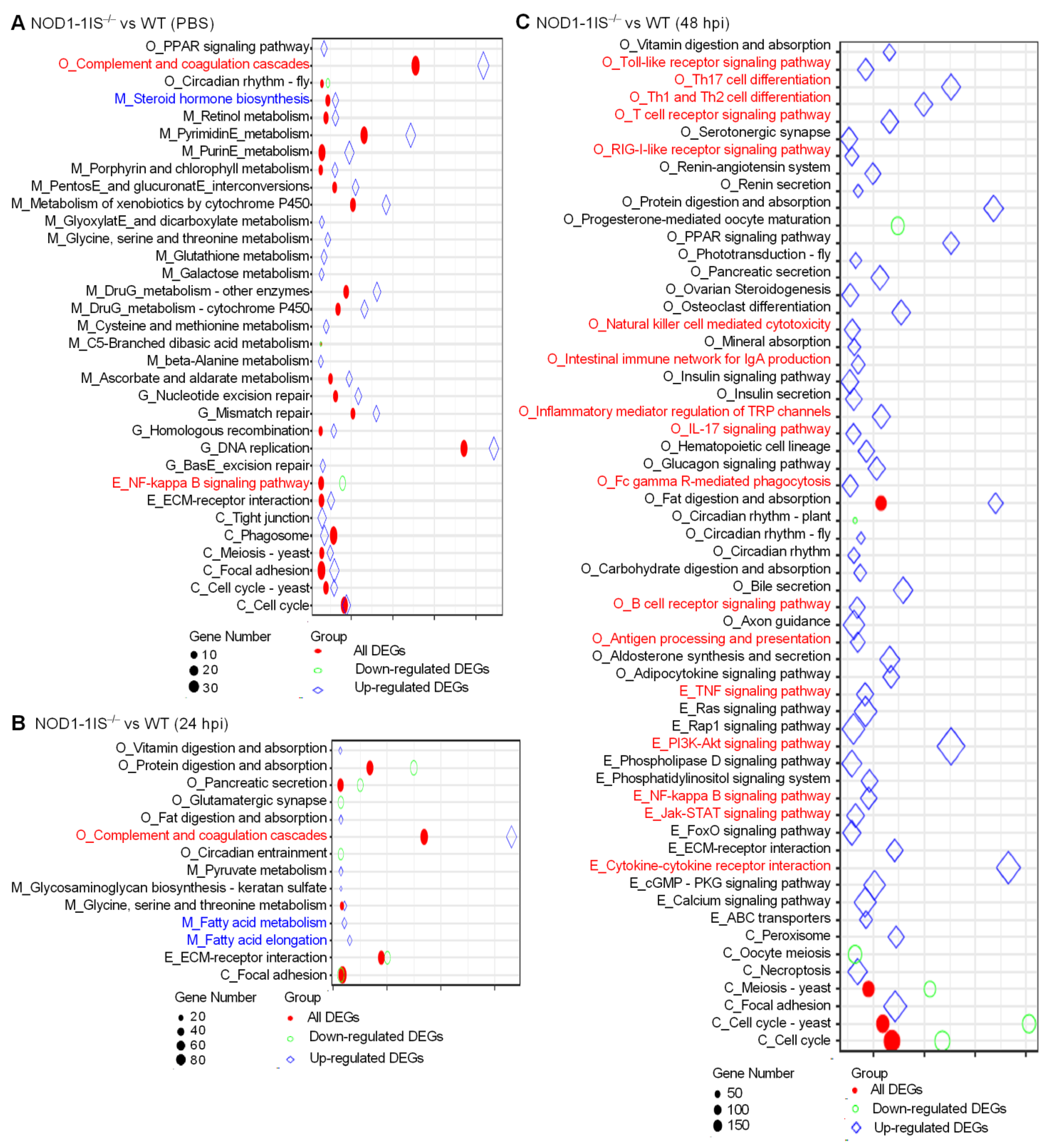
2.2. NOD1 Deficiency Mainly Affects Metabolism and Immune System Processes in Adult Zebrafish in Response to S. agalactiae Infection
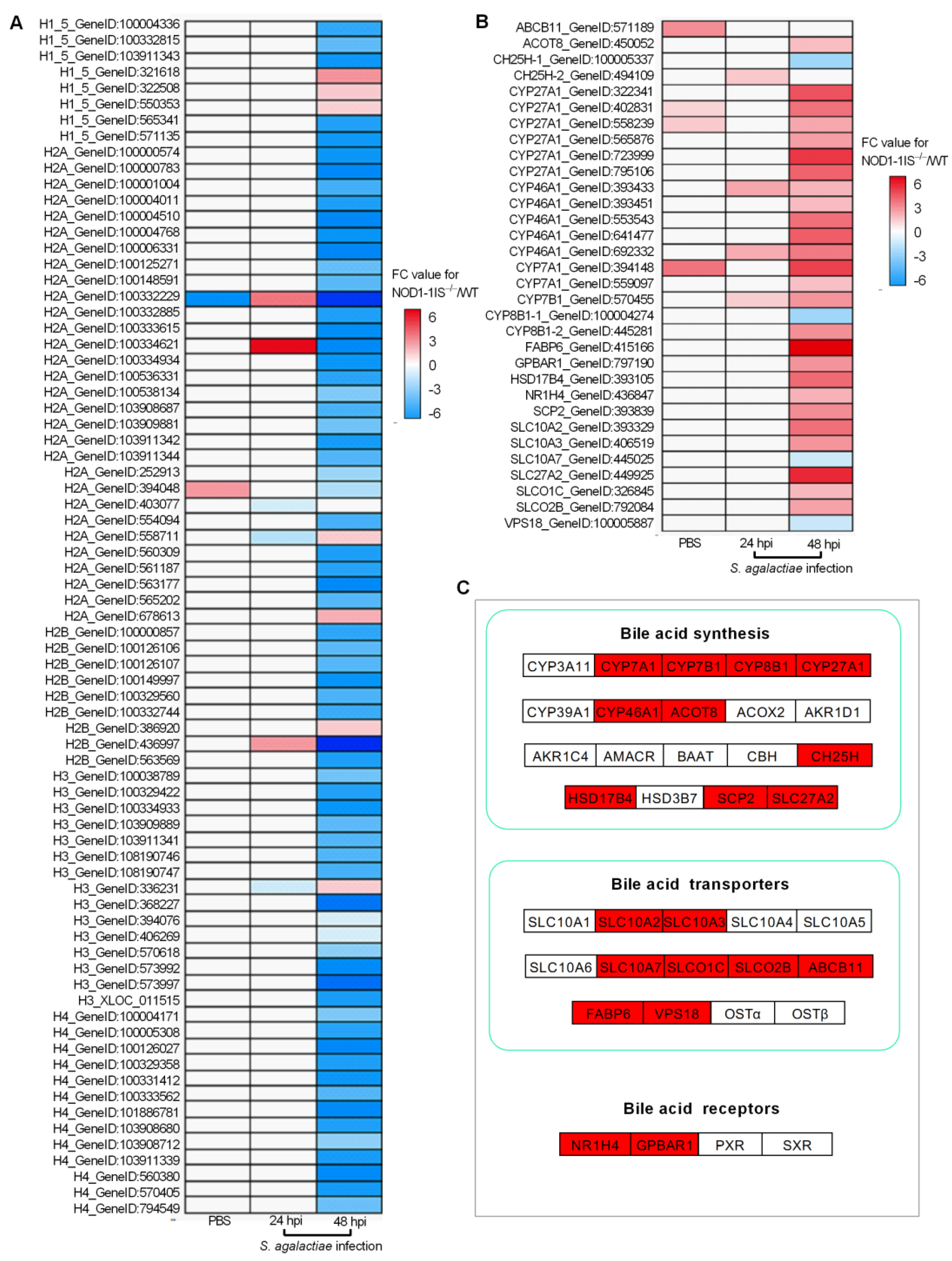
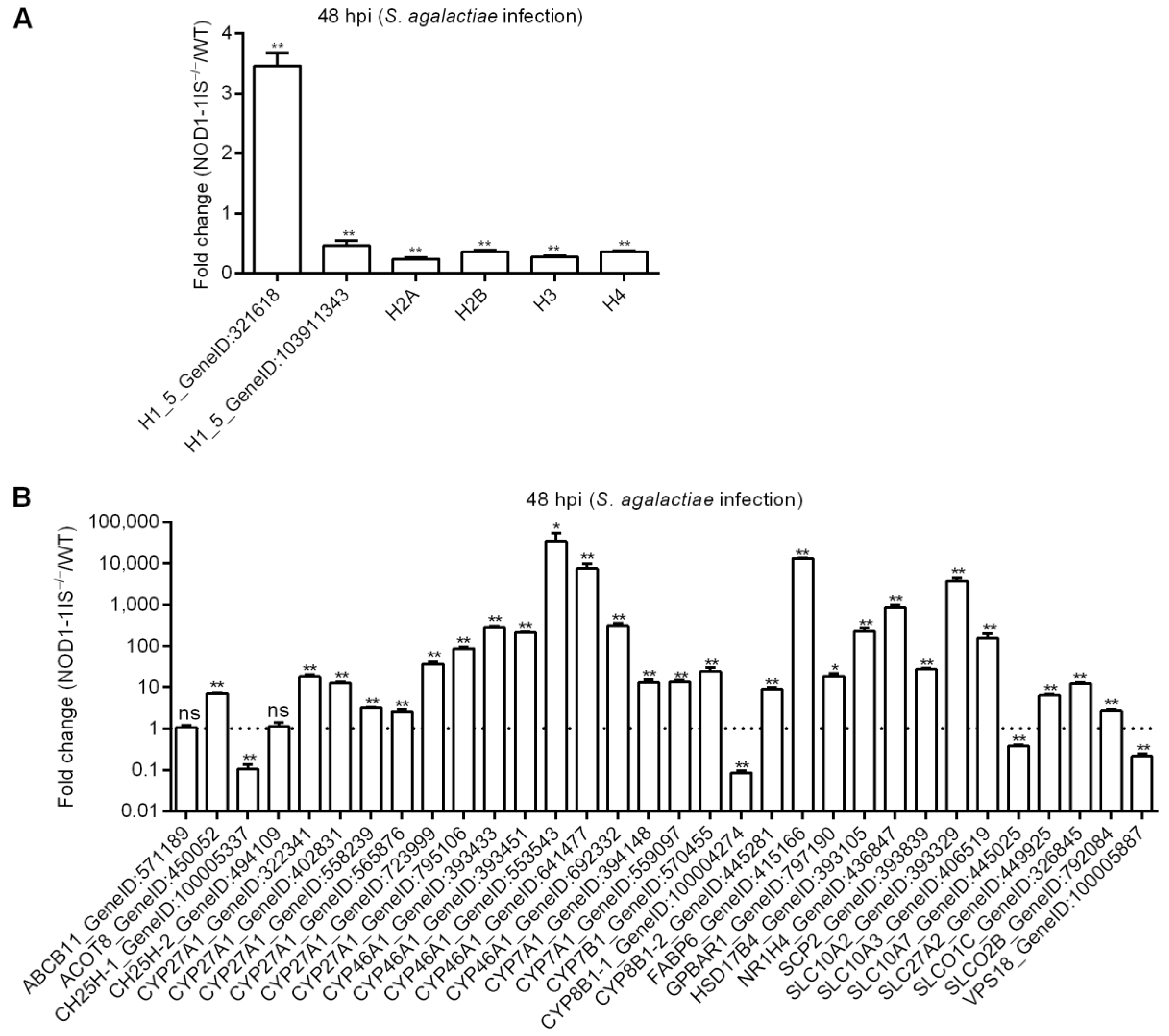
2.3. NOD1 Deficiency Regulated the Expression of Histone Variants and those Gene Variants Associated with Bile-Acid Signalling
2.4. Interaction Network Analysis of DEGs Related to Immunity and Metabolism Regulated by NOD1 Deficiency
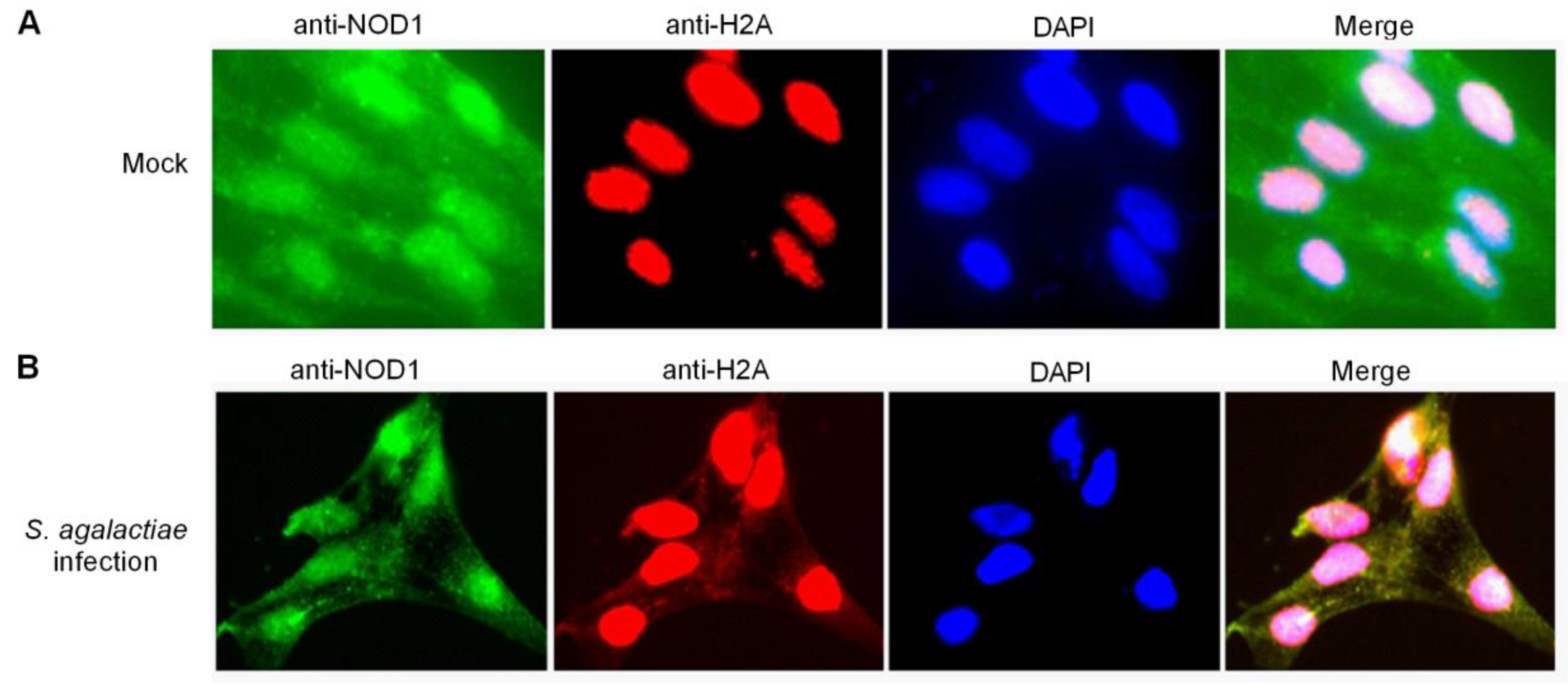
2.5. Histone H2A Is Translocated to the Cytoplasm and Colocalizes with NOD1 in the Case of S. agalactiae Infection
2.6. NOD1 Interacts and Cooperates with Histone H2A Variant to Protect against S. agalactiae Infection
3. Materials and Methods
3.1. Bacterial Infection for Adult Zebrafish and Sample Collection
3.2. Bacterial Infection for Zebrafish Larvae and In Vivo Antibacterial Analysis of zfH2A-6 and/or NOD1
3.3. Bacterial Infection for Zebrafish WT and NOD1−/− Cell Lines and Cell Counting kit-8 (CCK-8) Assay
3.4. cDNA Library Construction and Illumina Deep Sequencing
3.5. qRT-PCR Validation of DEGs
3.6. qRT-PCR Analysis of Antibacterial Genes Regulated by NOD1 and/or zfH2A-6
3.7. Fluorescence Microscopy
3.8. Co-Immunoprecipitation (Co-IP) and Western Blotting
3.9. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, S.L. Genomic Insights into the Distribution and Evolution of Group B Streptococcus. Front. Microbiol. 2019, 10, 1447. [Google Scholar] [CrossRef] [Green Version]
- Berardi, A.; Tzialla, C.; Riva, M.; Cerbo, R.M.; Creti, R. Group B streptococcus: Early- and late-onset infections. J. Chemother. 2007, 19 (Suppl. 2), 24–27. [Google Scholar] [CrossRef]
- Schuchat, A. Epidemiology of group B streptococcal disease in the United States: Shifting paradigms. Clin. Microbiol. Rev. 1998, 11, 497–513. [Google Scholar] [CrossRef] [Green Version]
- Farley, M.M. Group B streptococcal disease in nonpregnant adults. Clin. Infect. Dis. 2001, 33, 556–561. [Google Scholar] [CrossRef] [Green Version]
- Raabe, V.N.; Shane, A.L. Group B Streptococcus (Streptococcus agalactiae). Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klesius, P.H.; Shoemaker, C.A.; Evans, J.J. Streptococcus: A worldwide fish health problem. In Proceedings of the 8th International Symposium on Tilapia in Aquaculture; Ag. Press Unit Abbassa: Cairo, Egypt, 2008; pp. 83–107. [Google Scholar]
- Amal, M.N.A.; Zarif, S.T.; Suhaiba, M.S.; Aidil, M.R.M.; Shaqinah, N.N.; Zamri-Saad, M.; Ismail, A. The effects of fish gender on susceptibility to acute Streptococcus agalactiae infection in Javanese medaka Oryzias javanicus. Microb. Pathog. 2018, 114, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Bowater, R.O.; Forbes-Faulkner, J.; Anderson, I.G.; Condon, K.; Robinson, B.; Kong, F.; Gilbert, G.L.; Reynolds, A.; Hyland, S.; McPherson, G.; et al. Natural outbreak of Streptococcus agalactiae (GBS) infection in wild giant Queensland grouper, Epinephelus lanceolatus (Bloch), and other wild fish in northern Queensland, Australia. J. Fish. Dis. 2012, 35, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Chong, S.M.; Wong, W.K.; Lee, W.Y.; Tan, Z.B.; Tay, Y.H.; Teo, X.H.; Chee, L.D.; Fernandez, C.J. Streptococcus agalactiae outbreaks in cultured golden pomfret, Trachinotus blochii (Lacepede), in Singapore. J. Fish. Dis. 2017, 40, 971–974. [Google Scholar] [CrossRef]
- Baeck, G.W.; Kim, J.H.; Gomez, D.K.; Park, S.C. Isolation and characterization of Streptococcus sp. from diseased flounder (Paralichthys olivaceus) in Jeju Island. J. Vet. Sci. 2006, 7, 53–58. [Google Scholar] [CrossRef] [Green Version]
- Bergstrøm, B.; Aune, M.H.; Awuh, J.A.; Kojen, J.F.; Blix, K.J.; Ryan, L.; Flo, T.H.; Mollnes, T.E.; Espevik, T.; Stenvik, J. TLR8 Senses Staphylococcus aureus RNA in Human Primary Monocytes and Macrophages and Induces IFN-β Production via a TAK1-IKKβ-IRF5 Signaling Pathway. J. Immunol. 2015, 195, 1100–1111. [Google Scholar] [CrossRef] [Green Version]
- Mancuso, G.; Midiri, A.; Beninati, C.; Biondo, C.; Galbo, R.; Akira, S.; Henneke, P.; Golenbock, D.; Teti, G. Dual role of TLR2 and myeloid differentiation factor 88 in a mouse model of invasive group B streptococcal disease. J Immunol. 2004, 172, 6324–6329. [Google Scholar] [CrossRef] [Green Version]
- Mancuso, G.; Gambuzza, M.; Midiri, A.; Biondo, C.; Papasergi, S.; Akira, S.; Teti, G.; Beninati, C. Bacterial recognition by TLR7 in the lysosomes of conventional dendritic cells. Nat. Immunol. 2009, 10, 587–594. [Google Scholar] [CrossRef]
- Costa, A.; Gupta, R.; Signorino, G.; Malara, A.; Cardile, F.; Biondo, C.; Midiri, A.; Galbo, R.; Trieu-Cuot, P.; Papasergi, S.; et al. Activation of the NLRP3 inflammasome by group B streptococci. J. Immunol. 2012, 188, 1953–1960. [Google Scholar] [CrossRef]
- Santos-Sierra, S.; Golenbock, D.T.; Henneke, P. Toll-like receptor-dependent discrimination of streptococci. J. Endotoxin Res. 2006, 12, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.Y.; Pang, J.C.; Wang, M.; Lu, M.X.; Liu, Z.G.; Cao, J.M.; Ke, X.L.; Yi, M.M. Structurally diverse genes encode TLR13 in Nile tilapia: The two receptors can recognize Streptococcus 23S RNA and conduct signal transduction through MyD88. Mol. Immunol. 2021, 132, 60–78. [Google Scholar] [CrossRef] [PubMed]
- Lemire, P.; Calzas, C.; Segura, M. The NOD2 receptor does not play a major role in the pathogenesis of Group B Streptococcus in mice. Microb. Pathog. 2013, 65, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Wolffe, A.P. Transcription: In tune with the histones. Cell 1994, 77, 13–16. [Google Scholar] [CrossRef]
- Biterge, B.; Schneider, R. Histone variants: Key players of chromatin. Cell Tissue Res. 2014, 356, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Hamon, M.A.; Batsché, E.; Régnault, B.; Tham, T.N.; Seveau, S.; Muchardt, C.; Cossart, P. Histone modifications induced by a family of bacterial toxins. Proc. Natl. Acad. Sci. USA 2007, 104, 13467–13472. [Google Scholar] [CrossRef] [Green Version]
- Corr, S.C.; O’Neill, L.A. Listeria monocytogenes infection in the face of innate immunity. Cell Microbiol. 2009, 11, 703–709. [Google Scholar] [CrossRef]
- Hoeksema, M.; van Eijk, M.; Haagsman, H.P.; Hartshorn, K.L. Histones as mediators of host defense, inflammation and thrombosis. Future Microbiol. 2016, 11, 441–453. [Google Scholar] [CrossRef] [Green Version]
- Kawasaki, H.; Iwamuro, S. Potential roles of histones in host defense as antimicrobial agents. Infect. Disord. Drug Targets 2008, 8, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.M.; Kemp, G.D.; Molle, M.G.; Smith, V.J. Anti-microbial properties of histone H2A from skin secretions of rainbow trout, Oncorhynchus mykiss. Biochem. J. 2002, 368, 611–620. [Google Scholar] [CrossRef] [Green Version]
- Patat, S.A.; Carnegie, R.B.; Kingsbury, C.; Gross, P.S.; Chapman, R.; Schey, K.L. Antimicrobial activity of histones from hemocytes of the Pacific white shrimp. Eur. J. Biochem. 2004, 271, 4825–4833. [Google Scholar] [CrossRef]
- Doolin, T.; Gross, S.; Siryaporn, A. Physical Mechanisms of Bacterial Killing by Histones. Adv. Exp. Med. Biol. 2020, 1267, 117–133. [Google Scholar] [PubMed]
- Xu, J.; Zhang, X.; Monestier, M.; Esmon, N.L.; Esmon, C.T. Extracellular histones are mediators of death through TLR2 and TLR4 in mouse fatal liver injury. J. Immunol. 2011, 187, 2626–2631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.; Chen, H.W.; Evankovich, J.; Yan, W.; Rosborough, B.R.; Nace, G.W.; Ding, Q.; Loughran, P.; Beer-Stolz, D.; Billiar, T.R.; et al. Histones activate the NLRP3 inflammasome in Kupffer cells during sterile inflammatory liver injury. J. Immunol. 2013, 191, 2665–2679. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.W.; Wu, X.M.; Ren, S.S.; Cao, L.; Nie, P.; Chang, M.X. NOD1 deficiency impairs CD44a/Lck as well as PI3K/Akt pathway. Sci. Rep. 2017, 7, 2979. [Google Scholar] [CrossRef]
- Wu, X.M.; Chen, W.Q.; Hu, Y.W.; Cao, L.; Nie, P.; Chang, M.X. RIP2 Is a Critical Regulator for NLRs Signaling and MHC Antigen Presentation but Not for MAPK and PI3K/Akt Pathways. Front. Immunol. 2018, 9, 726. [Google Scholar] [CrossRef]
- Wu, X.M.; Cao, L.; Nie, P.; Chang, M.X. Histone H2A cooperates with RIP2 to induce the expression of antibacterial genes and MHC related genes. Dev. Comp. Immunol. 2019, 101, 103455. [Google Scholar] [CrossRef]
- Oehlers, S.H.; Flores, M.V.; Hall, C.J.; Swift, S.; Crosier, K.E.; Crosier, P.S. The inflammatory bowel disease (IBD) susceptibility genes NOD1 and NOD2 have conserved anti-bacterial roles in zebrafish. Dis. Model. Mech. 2011, 4, 832–841. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.M.; Zhang, J.; Li, P.W.; Hu, Y.W.; Cao, L.; Ouyang, S.; Bi, Y.H.; Nie, P.; Chang, M.X. NOD1 Promotes Antiviral Signaling by Binding Viral RNA and Regulating the Interaction of MDA5 and MAVS. J. Immunol. 2020, 204, 2216–2231. [Google Scholar] [CrossRef]
- Kim, B.J.; Hancock, B.M.; Del Cid, N.; Bermudez, A.; Traver, D.; Doran, K.S. Streptococcus agalactiae infection in zebrafish larvae. Microb. Pathog. 2015, 79, 57–60. [Google Scholar] [CrossRef] [Green Version]
- Saralahti, A.; Rämet, M. Zebrafish and Streptococcal Infections. Scand. J. Immunol. 2015, 82, 174–183. [Google Scholar] [CrossRef] [Green Version]
- Rubino, S.J.; Selvanantham, T.; Girardin, S.E.; Philpott, D.J. Nod-like receptors in the control of intestinal inflammation. Curr. Opin. Immunol. 2012, 24, 398–404. [Google Scholar] [CrossRef]
- Vasseur, F.; Sendid, B.; Jouault, T.; Standaert-Vitse, A.; Dubuquoy, L.; Francois, N.; Gower-Rousseau, C.; Desreumaux, P.; Broly, F.; Vermeire, S.; et al. Variants of NOD1 and NOD2 genes display opposite associations with familial risk of Crohn’s disease and anti-saccharomyces cerevisiae antibody levels. Inflamm. Bowel Dis. 2012, 18, 430–438. [Google Scholar] [CrossRef]
- Wu, X.M.; Cao, L.; Hu, Y.W.; Chang, M.X. Transcriptomic characterization of adult zebrafish infected with Streptococcus agalactiae. Fish Shellfish Immunol. 2019, 94, 355–372. [Google Scholar] [CrossRef]
- Clark, A.; Mach, N. The Crosstalk between the Gut Microbiota and Mitochondria during Exercise. Front. Physiol. 2017, 8, 319. [Google Scholar] [CrossRef]
- Noureldein, M.H.; Eid, A.A. Gut microbiota and mTOR signaling: Insight on a new pathophysiological interaction. Microb. Pathog. 2018, 118, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Magrone, T.; Jirillo, E. The interplay between the gut immune system and microbiota in health and disease: Nutraceutical intervention for restoring intestinal homeostasis. Curr. Pharm. Des. 2013, 19, 1329–1342. [Google Scholar] [PubMed]
- Inagaki, T.; Moschetta, A.; Lee, Y.K.; Peng, L.; Zhao, G.; Downes, M.; Yu, R.T.; Shelton, J.M.; Richardson, J.A.; Repa, J.J.; et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 3920–3925. [Google Scholar] [CrossRef] [Green Version]
- Travers, M.A.; Sow, C.; Zirah, S.; Deregnaucourt, C.; Chaouch, S.; Queiroz, R.M.; Charneau, S.; Allain, T.; Florent, I.; Grellier, P. Deconjugated Bile Salts Produced by Extracellular Bile-Salt Hydrolase-Like Activities from the Probiotic Lactobacillus johnsonii La1 Inhibit Giardia duodenalis In vitro Growth. Front. Microbiol. 2016, 7, 1453. [Google Scholar] [CrossRef] [Green Version]
- Hu, M.M.; He, W.R.; Gao, P.; Yang, Q.; He, K.; Cao, L.B.; Li, S.; Feng, Y.Q.; Shu, H.B. Virus-induced accumulation of intracellular bile acids activates the TGR5-β-arrestin-SRC axis to enable innate antiviral immunity. Cell Res. 2019, 29, 193–205. [Google Scholar] [CrossRef] [Green Version]
- Guo, C.; Xie, S.; Chi, Z.; Zhang, J.; Liu, Y.; Zhang, L.; Zheng, M.; Zhang, X.; Xia, D.; Ke, Y.; et al. Bile Acids Control Inflammation and Metabolic Disorder through Inhibition of NLRP3 Inflammasome. Immunity 2016, 45, 802–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Hartmann, P.; Haimerl, M.; Bathena, S.P.; Sjöwall, C.; Almer, S.; Alnouti, Y.; Hofmann, A.F.; Schnabl, B. Nod2 deficiency protects mice from cholestatic liver disease by increasing renal excretion of bile acids. J. Hepatol. 2014, 60, 1259–1267. [Google Scholar] [CrossRef] [Green Version]
- Wildenberg, M.E.; van den Brink, G.R. FXR activation inhibits inflammation and preserves the intestinal barrier in IBD. Gut 2011, 60, 432–433. [Google Scholar] [CrossRef]
- Alemi, F.; Poole, D.P.; Chiu, J.; Schoonjans, K.; Cattaruzza, F.; Grider, J.R.; Bunnett, N.W.; Corvera, C.U. The receptor TGR5 mediates the prokinetic actions of intestinal bile acids and is required for normal defecation in mice. Gastroenterology 2013, 144, 145–154. [Google Scholar] [CrossRef]
- Ding, L.; Yang, L.; Wang, Z.; Huang, W. Bile acid nuclear receptor FXR and digestive system diseases. Acta Pharm. Sin. B 2015, 5, 135–144. [Google Scholar] [CrossRef] [Green Version]
- Baars, A.; Oosting, A.; Knol, J.; Garssen, J.; van Bergenhenegouwen, J. The Gut Microbiota as a Therapeutic Target in IBD and Metabolic Disease: A Role for the Bile Acid Receptors FXR and TGR5. Microorganisms 2015, 3, 641–666. [Google Scholar] [CrossRef] [Green Version]
- Stepanov, V.; Stankov, K.; Mikov, M. The bile acid membrane receptor TGR5: A novel pharmacological target in metabolic, inflammatory and neoplastic disorders. J. Recept. Signal Transduct Res. 2013, 33, 213–223. [Google Scholar] [CrossRef]
- Xiong, F.; Cao, L.; Wu, X.M.; Chang, M.X. The function of zebrafish gpbar1 in antiviral response and lipid metabolism. Dev. Comp. Immunol. 2021, 116, 103955. [Google Scholar] [CrossRef]
- Mühlhäusser, P.; Müller, E.C.; Otto, A.; Kutay, U. Multiple pathways contribute to nuclear import of core histones. EMBO Rep. 2001, 2, 690–696. [Google Scholar] [CrossRef] [Green Version]
- Keck, K.M.; Pemberton, L.F. Histone chaperones link histone nuclear import and chromatin assembly. Biochim. Biophys. Acta 2013, 1819, 277–289. [Google Scholar] [CrossRef] [Green Version]
- Bernardes, N.E.; Chook, Y.M. Nuclear import of histones. Biochem. Soc. Trans. 2020, 48, 2753–2767. [Google Scholar] [CrossRef] [PubMed]
- Okamura, H.; Yoshida, K.; Amorim, B.R.; Haneji, T. Histone H1.2 is translocated to mitochondria and associates with Bak in bleomycin-induced apoptotic cells. J. Cell Biochem. 2008, 103, 1488–1496. [Google Scholar] [CrossRef] [PubMed]
- Klein, B.; Lütz-Meindl, U.; Kerschbaum, H.H. From the nucleus to the plasma membrane: Translocation of the nuclear proteins histone H3 and lamin B1 in apoptotic microglia. Apoptosis 2014, 19, 759–775. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.M.; Fang, H.; Nie, P.; Chang, M.X. Nucleotide polymorphism and function of piscine histone H2A in Edwardsiella piscicida infection. J. Fish. China 2020, 44, 1–14. [Google Scholar]
- Huang, J.; Xie, Y.; Sun, X.; Zeh, H.J., III; Kang, R.; Lotze, M.T.; Tang, D. DAMPs, ageing, and cancer: The ‘DAMP Hypothesis’. Ageing Res Rev. 2015, 24, 3–16. [Google Scholar] [CrossRef] [Green Version]
- Allam, R.; Scherbaum, C.R.; Darisipudi, M.N.; Mulay, S.R.; Hägele, H.; Lichtnekert, J.; Hagemann, J.H.; Rupanagudi, K.V.; Ryu, M.; Schwarzenberger, C.; et al. Histones from dying renal cells aggravate kidney injury via TLR2 and TLR4. J. Am. Soc. Nephrol. 2012, 23, 1375–1388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meissner, T.B.; Li, A.; Biswas, A.; Lee, K.H.; Liu, Y.J.; Bayir, E.; Iliopoulos, D.; van den Elsen, P.J.; Kobayashi, K.S. NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc. Natl. Acad. Sci. USA 2010, 107, 13794–13799. [Google Scholar] [CrossRef] [Green Version]
- Devaiah, B.N.; Singer, D.S. CIITA and Its Dual Roles in MHC Gene Transcription. Front. Immunol. 2013, 4, 476. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Wu, X.M.; Hu, Y.W.; Xue, N.N.; Nie, P.; Chang, M.X. The discrepancy function of NLRC5 isoforms in antiviral and antibacterial immune responses. Dev. Comp. Immunol. 2018, 84, 153–163. [Google Scholar] [CrossRef]

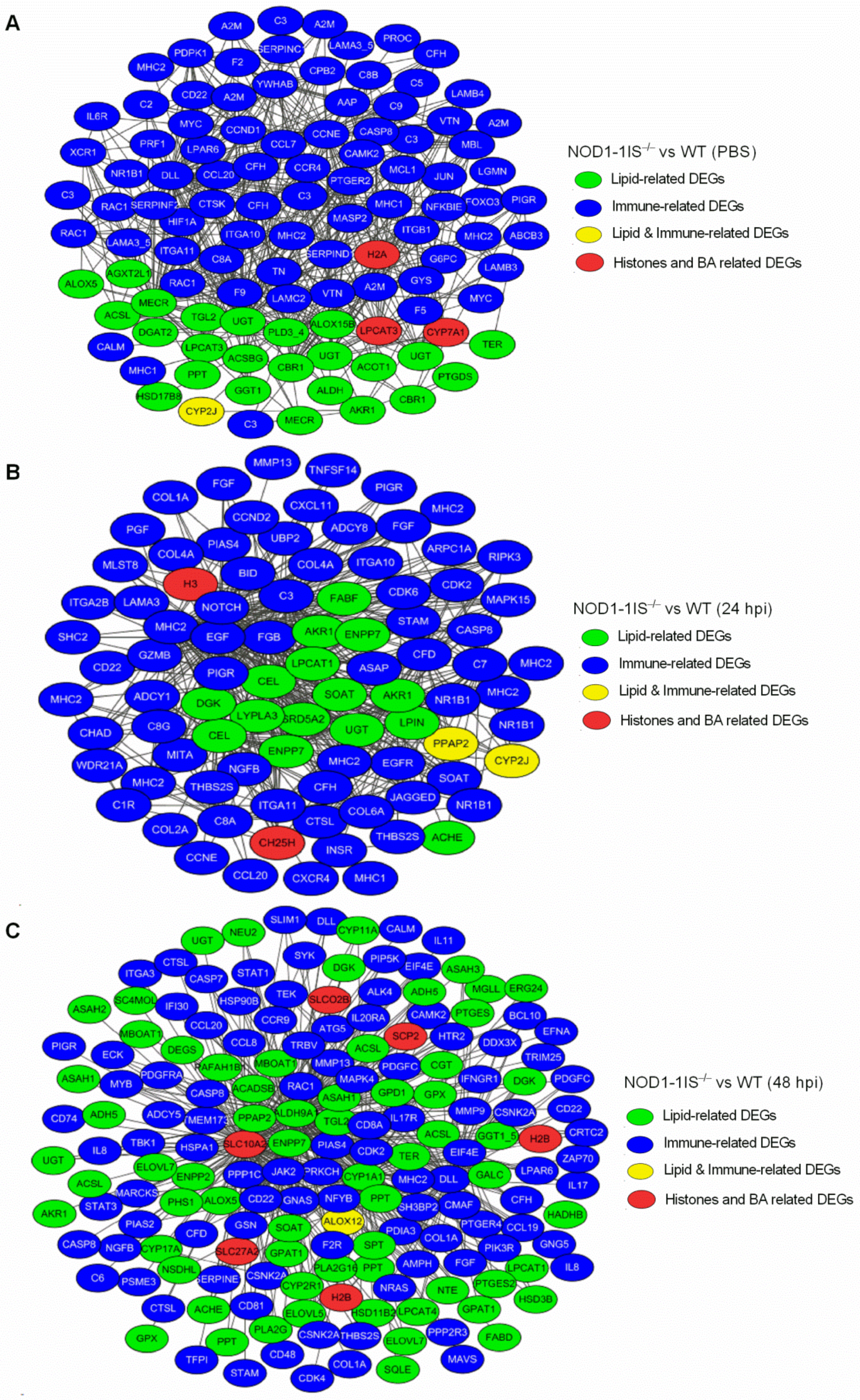

| Group | Total DEGs | Immune-Related DEGs | Lipid-Related DEGs | Histone Variants | Bile Acid-Related DEGs |
|---|---|---|---|---|---|
| NOD1-1IS−/− vs. WT (PBS) | 820 | 135 | 51 | 2 | 4 |
| NOD1-1IS−/− vs. WT (24 hpi) | 5407 | 340 | 117 | 6 | 4 |
| NOD1-1IS−/− vs. WT (48 hpi) | 12,240 | 719 | 282 | 74 | 30 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, X.; Xiong, F.; Fang, H.; Zhang, J.; Chang, M. Crosstalks between NOD1 and Histone H2A Contribute to Host Defense against Streptococcus agalactiae Infection in Zebrafish. Antibiotics 2021, 10, 861. https://doi.org/10.3390/antibiotics10070861
Wu X, Xiong F, Fang H, Zhang J, Chang M. Crosstalks between NOD1 and Histone H2A Contribute to Host Defense against Streptococcus agalactiae Infection in Zebrafish. Antibiotics. 2021; 10(7):861. https://doi.org/10.3390/antibiotics10070861
Chicago/Turabian StyleWu, Xiaoman, Fan Xiong, Hong Fang, Jie Zhang, and Mingxian Chang. 2021. "Crosstalks between NOD1 and Histone H2A Contribute to Host Defense against Streptococcus agalactiae Infection in Zebrafish" Antibiotics 10, no. 7: 861. https://doi.org/10.3390/antibiotics10070861
APA StyleWu, X., Xiong, F., Fang, H., Zhang, J., & Chang, M. (2021). Crosstalks between NOD1 and Histone H2A Contribute to Host Defense against Streptococcus agalactiae Infection in Zebrafish. Antibiotics, 10(7), 861. https://doi.org/10.3390/antibiotics10070861






