Effects of Tylosin, a Direct-Fed Microbial and Feedlot Pen Environment on Phenotypic Resistance among Enterococci Isolated from Beef Cattle Feces
Abstract
:1. Introduction
2. Materials and Methods
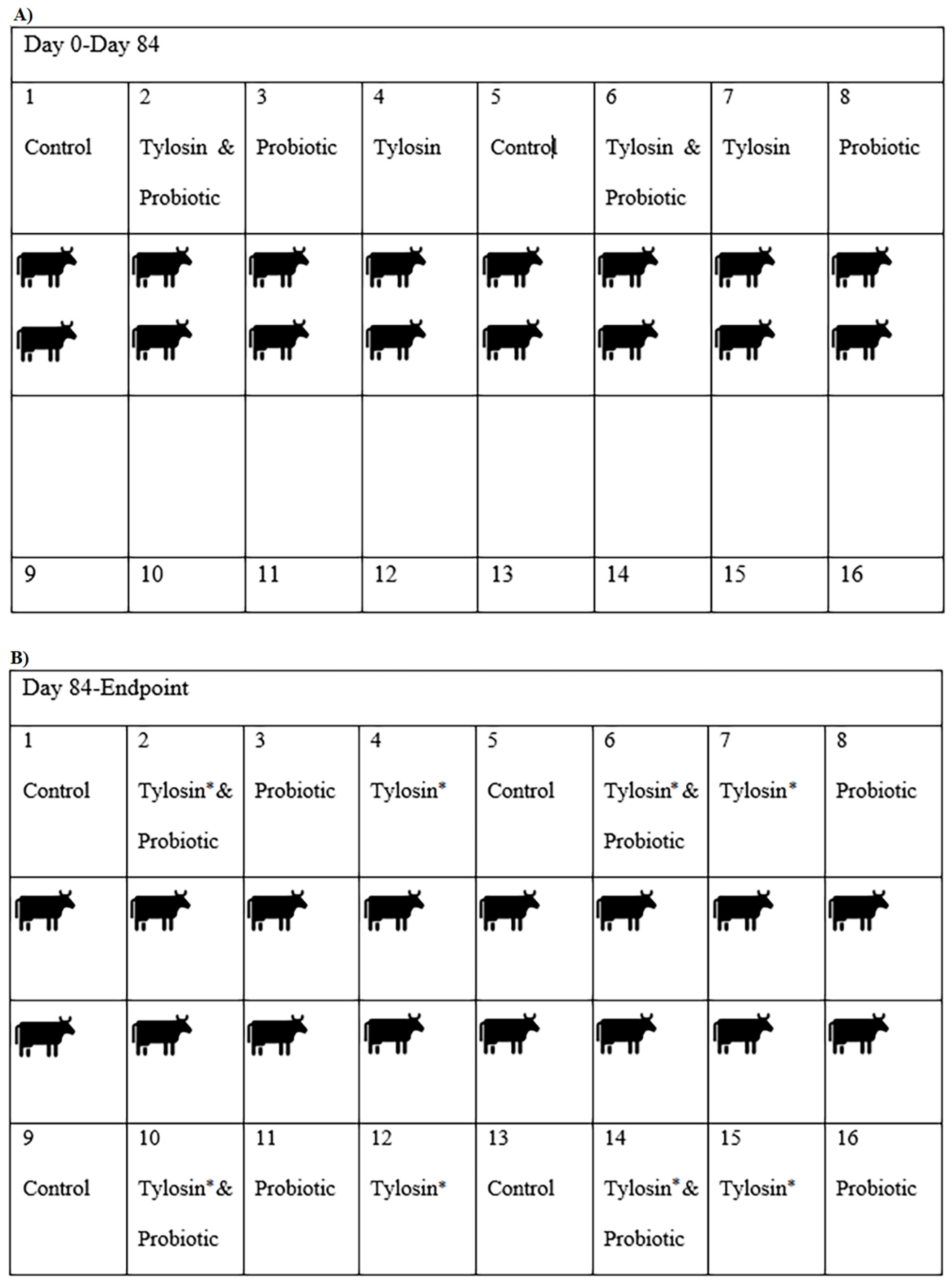
2.1. Experimental Design
2.2. Bacterial Enumeration, Isolation, and Speciation
2.3. Phenotypic Susceptibility Testing
2.4. Statistical Analysis
3. Results
3.1. Descriptive Statistics
3.2. Mixed Multivariable Regression Models
3.3. Descriptive Statistics of Phenotypic Resistance
3.4. Multi-Level Mixed Effects Logistic Regression Modeling of Antibiotic Resistance Phenotype
4. Discussion
4.1. Host Bacterial Response to Antibiotics and Direct-Fed Microbials
4.2. Role of the Pen and Ambient Environment
4.3. Assumptions and Future Potential
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Animal Care and Use Committee Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Amachawadi, R.; Nagaraja, T.G. Liver abscesses in cattle: A review of incidence in Holsteins and of bacteriology and vaccine approaches to control in feedlot cattle12. J. Anim. Sci. 2016, 94, 1620–1632. [Google Scholar] [CrossRef] [Green Version]
- Nagaraja, T.G.; Chengappa, M.M. Liver abscesses in feedlot cattle: A review. J. Anim. Sci. 1998, 76, 287–298. [Google Scholar] [CrossRef] [Green Version]
- Nagaraja, T.G.; Beharka, A.B.; Chengappa, M.M.; Carroll, L.H.; Raun, A.P.; Laudert, S.B.; Parrott, J.C. Bacterial flora of liver abscesses in feedlot cattle fed tylosin or no tylosin. J. Anim. Sci. 1999, 77, 973–978. [Google Scholar] [CrossRef]
- Amachawadi, R.; Scott, H.; Aperce, C.; Vinasco, J.; Drouillard, J.; Nagaraja, T. Effects of in-feed copper and tylosin supplementations on copper and antimicrobial resistance in faecal enterococci of feedlot cattle. J. Appl. Microbiol. 2015, 118, 1287–1297. [Google Scholar] [CrossRef]
- Brink, D.R.; Lowry, S.R.; Stock, R.A.; Parrott, J.C. Severity of liver abscesses and efficiency of feed utilization of feedlot cattle. J. Anim. Sci. 1990, 68, 1201–1207. [Google Scholar] [CrossRef]
- Brown, H.; Bing, R.F.; Grueter, H.P.; McAskill, J.W.; Cooley, C.O.; Rathmacher, R.P. Tylosin and Chlortetracycline for the Prevention of Liver Abscesses, Improved Weight Gains and Feed Efficiency in Feedlot Cattle. J. Anim. Sci. 1975, 40, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Brown, T.R.; Lawrence, T.E. Association of liver abnormalities with carcass grading performance and value. J. Anim. Sci. 2010, 88, 4037–4043. [Google Scholar] [CrossRef] [PubMed]
- Wileman, B.W.; Thomson, D.U.; Reinhardt, C.D.; Renter, D.G. Analysis of modern technologies commonly used in beef cattle production: Conventional beef production versus nonconventional production using meta-analysis. J. Anim. Sci. 2009, 87, 3418–3426. [Google Scholar] [CrossRef] [PubMed]
- Aarestrup, F.M.; Carstensen, B. Effect of Tylosin Used as a Growth Promoter on the Occurrence of Macrolide-Resistant Enterococci and Staphylococci in Pigs. Microb. Drug Resist. 1998, 4, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Jacob, M.E.; Fox, J.T.; Narayanan, S.K.; Drouillard, J.S.; Renter, D.G.; Nagaraja, T.G. Effects of feeding wet corn distillers grains with solubles with or without monensin and tylosin on the prevalence and antimicrobial susceptibilities of fecal foodborne pathogenic and commensal bacteria in feedlot cattle1. J. Anim. Sci. 2008, 86, 1182–1190. [Google Scholar] [CrossRef] [Green Version]
- Zaheer, R.; Cook, S.R.; Klima, C.L.; Stanford, K.; Alexander, T.; Topp, E.; Read, R.R.; McAllister, T.A. Effect of subtherapeutic vs. therapeutic administration of macrolides on antimicrobial resistance in Mannheimia haemolytica and enterococci isolated from beef cattle. Front. Microbiol. 2013, 4, 133. [Google Scholar] [CrossRef] [Green Version]
- Cazer, C.L.; Eldermire, E.R.; Lhermie, G.; Murray, S.A.; Scott, H.M.; Gröhn, Y.T. The effect of tylosin on antimicrobial resistance in beef cattle enteric bacteria: A systematic review and meta-analysis. Prev. Veter. Med. 2020, 176, 104934. [Google Scholar] [CrossRef]
- Zaheer, R.; Cook, S.R.; Barbieri, R.; Goji, N.; Cameron, A.; Petkau, A.; Polo, R.O.; Tymensen, L.; Stamm, C.; Song, J.; et al. Surveillance of Enterococcus spp. reveals distinct species and antimicrobial resistance diversity across a One-Health continuum. Sci. Rep. 2020, 10, 3937. [Google Scholar] [CrossRef] [PubMed]
- Emori, T.G.; Gaynes, R.P. An overview of nosocomial infections, including the role of the microbiology laboratory. Clin. Microbiol. Rev. 1993, 6, 428–442. [Google Scholar] [CrossRef] [PubMed]
- Arias, A.C.; Murray, E.B. Emergence and management of drug-resistant enterococcal infections. Expert Rev. Anti Infect. Ther. 2008, 6, 637–655. [Google Scholar] [CrossRef]
- Arias, C.A.; Murray, B.E. The rise of the Enterococcus: Beyond vancomycin resistance. Nat. Rev. Genet. 2012, 10, 266–278. [Google Scholar] [CrossRef] [Green Version]
- CDC. Antibiotic Resistance Threats in the United States. Am. Fam. Physician 2014, 12, 938–941. [Google Scholar]
- Huycke, M.M. Multiple-Drug Resistant Enterococci: The Nature of the Problem and an Agenda for the Future. Emerg. Infect. Dis. 1998, 4, 239–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Fluharty, F.L.; St-Pierre, N.; Morrison, M.; Yu, Z. Technical note: Occurrence in fecal microbiota of genes conferring resistance to both macrolide-lincosamide-streptogramin B and tetracyclines concomitant with feeding of beef cattle with tylosin1. J. Anim. Sci. 2008, 86, 2385–2391. [Google Scholar] [CrossRef]
- WHO. Critically Important Antimicrobials for Human Medicine, 6th ed.; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- Beukers, A.G.; Zaheer, R.; Cook, S.R.; Stanford, K.; Chaves, A.V.; Ward, M.; McAllister, T.A. Effect of in-feed administration and withdrawal of tylosin phosphate on antibiotic resistance in enterococci isolated from feedlot steers. Front. Microbiol. 2015, 6, 483. [Google Scholar] [CrossRef] [Green Version]
- Müller, H.C.; Van Bibber-Krueger, C.L.; Ogunrinu, O.J.; Amachawadi, R.; Scott, H.M.; Drouillard, J.S. Effects of intermittent feeding of tylosin phosphate during the finishing period on feedlot performance, carcass characteristics, antimicrobial resistance, and incidence and severity of liver abscesses in steers1. J. Anim. Sci. 2018, 96, 2877–2885. [Google Scholar] [CrossRef]
- Huebner, K.L.; Martin, J.N.; Weissend, C.J.; Holzer, K.L.; Parker, J.K.; Lakin, S.M.; Doster, E.; Weinroth, M.; Abdo, Z.; Woerner, D.R.; et al. Effects of a Saccharomyces cerevisiae fermentation product on liver abscesses, fecal microbiome, and resistome in feedlot cattle raised without antibiotics. Sci. Rep. 2019, 9, 2559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Izquierdo, E.; Marchioni, E.; Aoude-Werner, D.; Hasselmann, C.; Ennahar, S. Smearing of soft cheese with Enterococcus faecium WHE 81, a multi-bacteriocin producer, against Listeria monocytogenes. Food Microbiol. 2009, 26, 16–20. [Google Scholar] [CrossRef]
- Murray, S.; Holbert, A.; Norman, K.; Lawhon, S.; Sawyer, J.; Scott, H. Macrolide-susceptible probiotic Enterococcus faecium ST296 exhibits faecal-environmental-oral microbial community cycling among beef cattle in feedlots. Lett. Appl. Microbiol. 2020, 70, 274–281. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Murray, S.; Amachawadi, R.; Norman, K.; Lawhon, S.; Nagaraja, T.; Drouillard, J.; Scott, H. Effects of Zinc and Menthol-Based Diets on Co-Selection of Antibiotic Resistance among E. coli and Enterococcus spp. in Beef Cattle. Animals 2021, 11, 259. [Google Scholar] [CrossRef] [PubMed]
- Lebreton, F.; van Schaik, W.; McGuire, A.M.; Godfrey, P.; Griggs, A.; Mazumdar, V.; Corander, J.; Cheng, L.; Saif, S.; Young, S.; et al. Emergence of Epidemic Multidrug-Resistant Enterococcus faecium from Animal and Commensal Strains. mBio 2013, 4, e00534-13. [Google Scholar] [CrossRef] [Green Version]
- Jackson, C.R.; Fedorka-Cray, P.J.; Barrett, J.B.; Ladely, S.R. Effects of Tylosin Use on Erythromycin Resistance in Enterococci Isolated from Swine. Appl. Environ. Microbiol. 2004, 70, 4205–4210. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, J.W.; Vikram, A.; Miller, E.; Jones, S.A.; Arthur, T.M. In-Feed Tylosin Phosphate Administration to Feedlot Cattle Minimally Affects Antimicrobial Resistance. J. Food Prot. 2020, 83, 350–364. [Google Scholar] [CrossRef]
- Amachawadi, R.; Giok, F.; Shi, X.; Soto, J.; Narayanan, S.K.; Tokach, M.D.; Apley, M.D.; Nagaraja, T.G. Antimicrobial resistance of Enterococcus faecium strains isolated from commercial probiotic products used in cattle and swine1,2. J. Anim. Sci. 2018, 96, 912–920. [Google Scholar] [CrossRef]
- Clewell, D.B.; An, F.Y.; White, B.A.; Gawron-Burke, C. Sex Pheromones and Plasmid Transfer in Streptococcus Faecalis: A Pheromone, cAM373, which is Additionally, Excreted by Staphylococcus Aureus. Plasmids Bact. 1985, 30, 489–503. [Google Scholar] [CrossRef]






| Antibiotic | Class | Range | Breakpoint |
|---|---|---|---|
| Gentamicin | Aminoglycoside | 128–1024 | ≥500 |
| Kanamycin | Aminoglycoside | 128–1024 | ≥1024 |
| Streptomycin | Aminoglycoside | 512–2048 | >1000 |
| Vancomycin | Glycopeptide | 0.25–32 | ≥32 |
| Tigecycline | Glycylcycline | 0.015–0.5 | ≥0.5 |
| Lincomycin | Lincosamide | 1–8 | ≥8 |
| Daptomycin | Lipopeptide | 0.25–16 | ≥8 |
| Erythromycin | Macrolide | 0.25–8 | ≥8 |
| Tylosin | Macrolide | 0.25–32 | ≥32 |
| Nitrofurantoin | Nitrofuran | 2–64 | ≥128 |
| Linezolid | Oxazolidinone | 0.5–8 | ≥8 |
| Penicillin | Penicillin | 0.25–16 | ≥16 |
| Chloramphenicol | Phenicol | 2–32 | ≥32 |
| Ciprofloxacin | Quinolone | 0.12–4 | ≥4 |
| Quinupristin/dalfopristin | Streptogramin | 0.5–32 | ≥4 |
| Tetracycline | Tetracycline | 1–32 | ≥16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murray, S.A.; Holbert, A.C.; Norman, K.N.; Lawhon, S.D.; Sawyer, J.E.; Scott, H.M. Effects of Tylosin, a Direct-Fed Microbial and Feedlot Pen Environment on Phenotypic Resistance among Enterococci Isolated from Beef Cattle Feces. Antibiotics 2022, 11, 106. https://doi.org/10.3390/antibiotics11010106
Murray SA, Holbert AC, Norman KN, Lawhon SD, Sawyer JE, Scott HM. Effects of Tylosin, a Direct-Fed Microbial and Feedlot Pen Environment on Phenotypic Resistance among Enterococci Isolated from Beef Cattle Feces. Antibiotics. 2022; 11(1):106. https://doi.org/10.3390/antibiotics11010106
Chicago/Turabian StyleMurray, Sarah A., Ashlyn C. Holbert, Keri N. Norman, Sara D. Lawhon, Jason E. Sawyer, and Harvey M. Scott. 2022. "Effects of Tylosin, a Direct-Fed Microbial and Feedlot Pen Environment on Phenotypic Resistance among Enterococci Isolated from Beef Cattle Feces" Antibiotics 11, no. 1: 106. https://doi.org/10.3390/antibiotics11010106
APA StyleMurray, S. A., Holbert, A. C., Norman, K. N., Lawhon, S. D., Sawyer, J. E., & Scott, H. M. (2022). Effects of Tylosin, a Direct-Fed Microbial and Feedlot Pen Environment on Phenotypic Resistance among Enterococci Isolated from Beef Cattle Feces. Antibiotics, 11(1), 106. https://doi.org/10.3390/antibiotics11010106






