Distinction between Antimicrobial Resistance and Putative Virulence Genes Characterization in Plesiomonas shigelloides Isolated from Different Sources
Abstract
:1. Introduction
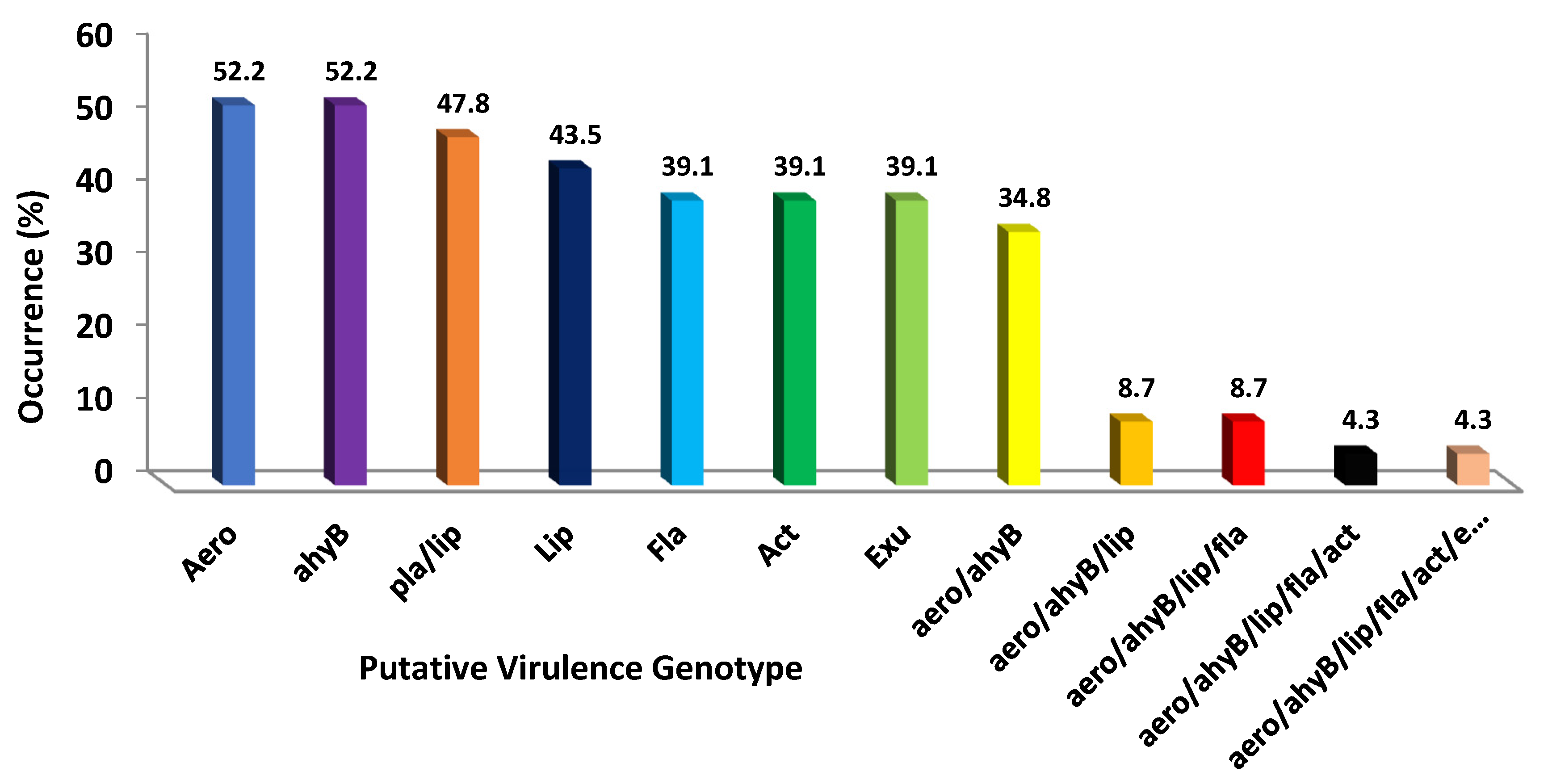
2. Results
3. Discussion
4. Materials and Methods
4.1. Conditions for the Isolation of P. shigelloides from Shellfish and Water Samples
4.2. Phenotypic Identification
4.3. Molecular Identification
4.4. Antimicrobial Susceptibility Testing
4.5. Genetic Detection of Virulence Genes
4.5.1. Nucleic Acid Isolation
4.5.2. Detection of Virulence Genes
4.6. Infectivity Test
4.7. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dubert, J.; Barja, J.L.; Romalde, J.L. New Insights into Pathogenic Vibrios Affecting Bivalves in Hatcheries: Present and Future Prospects. Front. Microbiol. 2017, 8, 762. [Google Scholar] [CrossRef] [Green Version]
- Pereira, C.; Costa, P.; Duarte, J.; Balcão, V.M.; Almeida, A. Phage Therapy as a Potential Approach in the Biocontrol of Pathogenic Bacteria Associated with Shellfish Consumption. Int. J. Food Microbiol. 2021, 338, 108995. [Google Scholar] [CrossRef] [PubMed]
- Robertson, L.J. The Potential for Marine Bivalve Shellfish to Act as Transmission Vehicles for Outbreaks of Protozoan Infections in Humans: A Review. Int. J. Food Microbiol. 2007, 120, 201–216. [Google Scholar] [CrossRef] [PubMed]
- Bosch, A.; Pintó, R.M.; Le Guyader, F.S. Viral Contaminants of Molluscan Shellfish: Detection and Characterisation. In Shellfish Safety and Quality; Elsevier: Amsterdam, The Netherlands, 2009; pp. 83–107. ISBN 978-1-84569-152-3. [Google Scholar]
- Martinez-Albores, A.; Lopez-Santamarina, A.; Rodriguez, J.A.; Ibarra, I.S.; del Mondragón, A.C.; Miranda, J.M.; Lamas, A.; Cepeda, A. Complementary Methods to Improve the Depuration of Bivalves: A Review. Foods 2020, 9, 129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mateus, L.; Costa, L.; Silva, Y.J.; Pereira, C.; Cunha, A.; Almeida, A. Efficiency of Phage Cocktails in the Inactivation of Vibrio in Aquaculture. Aquaculture 2014, 424–425, 167–173. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L. The Enterobacteria; Lippincott-Raven: Philadelphia, PA, USA, 1998. [Google Scholar]
- Wong, T.; Tsui, H.; So, M.; Lai, J.; Lai, S.; Tse, C.; Ng, T. Plesiomonas Shigelloides Infection in Hong Kong: Retrospective Study of 167 Laboratory-Confirmed Cases. Hong Kong Med. J. 2000, 6, 375–380. [Google Scholar] [PubMed]
- Doyle, M.P.; Beuchat, L.R.; Montville, T.J. Food Microbiology: Fundamentals and Frontiers, 2nd ed.; ASM Press: Washington, DC, USA, 2001; ISBN 978-1-55581-208-9. [Google Scholar]
- Gashgari, R.M.; Selim, S.A. Detection and Characterization of Antimicrobial Resistance and Putative Virulence Genes in Aeromonas Veronii Biovar Sobria Isolated from Gilthead Sea Bream (Sparus Aurata, L.). Foodborne Pathog. Dis. 2015, 12, 806–811. [Google Scholar] [CrossRef]
- Murray, P.R. American Society for Microbiology. In Manual of Clinical Microbiology, 7th ed.; ASM Press: Washington, DC, USA, 1999; ISBN 978-1-55581-126-6. [Google Scholar]
- Balebona, M.C.; Krovacek, K.; Moriñigo, M.A.; Mansson, I.; Faris, A.; Borrego, J.J. Neurotoxic Effect on Two Fish Species and a PC12 Cell Line of the Supernate of Vibrio Alginolyticus and Vibrio Anguillarum. Vet. Microbiol. 1998, 63, 61–69. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L.; McIver, C.J. Plesiomonas Shigelloides Revisited. Clin. Microbiol. Rev. 2016, 29, 349–374. [Google Scholar] [CrossRef] [Green Version]
- Kaszowska, M.; Stojkovic, K.; Niedziela, T.; Lugowski, C. The O-Antigen of Plesiomonas Shigelloides Serotype O36 Containing Pseudaminic Acid. Carbohydr. Res. 2016, 434, 1–5. [Google Scholar] [CrossRef]
- Nawaz, M.; Khan, S.A.; Khan, A.A.; Sung, K.; Tran, Q.; Kerdahi, K.; Steele, R. Detection and Characterization of Virulence Genes and Integrons in Aeromonas Veronii Isolated from Catfish. Food Microbiol. 2010, 27, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Sen, K. Development of a Rapid Identification Method for Aeromonas Species by Multiplex-PCR. Can. J. Microbiol. 2005, 51, 957–966. [Google Scholar] [CrossRef]
- Austin, B.; Austin, D.A. Bacterial Fish Pathogens: Disease of Farmed and Wild Fish, 4th ed.; Springer-Praxis books in aquaculture and fisheries; Praxis Pub: Dordrecht, The Netherland; Springer: Chichester, UK, 2007; ISBN 978-1-4020-6068-7. [Google Scholar]
- Gerhardt, P. American Society for Microbiology. In Manual of Methods for General Bacteriology; American Society for Microbiology: Washington, DC, USA, 1981; ISBN 978-0-8357-7509-0. [Google Scholar]
- EUCAST. Plesiomonas Shigelloides Calibration of Zone Diameter Breakpoints to MIC Values Plesiomonas Shigelloides MIC and Zone Diameter Correlates; European Committee on Antimicrobial Susceptibility Testing (EUCAST): Basel, Switzerland, 2018. [Google Scholar]
- Salerno, A.; Čižnár, I.; Krovacek, K.; Conte, M.; Dumontet, S.; González-Rey, C.; Pasquale, V. Phenotypic Characterization and Putative Virulence Factors of Human, Animal and Environmental Isolates of Plesiomonas Shigelloides. Folia Microbiol. 2010, 55, 641–647. [Google Scholar] [CrossRef] [PubMed]
- Nikbin, V.S.; Aslani, M.M.; Sharafi, Z.; Hashemipour, M.; Shahcheraghi, F.; Ebrahimipour, G.H. Molecular Identification and Detection of Virulence Genes among Pseudomonas Aeruginosa Isolated from Different Infectious Origins. Iran J. Microbiol. 2012, 4, 118–123. [Google Scholar]
- Deng, J.; Fu, L.; Wang, R.; Yu, N.; Ding, X.; Jiang, L.; Fang, Y.; Jiang, C.; Lin, L.; Wang, Y.; et al. Comparison of MALDI-TOF MS, Gene Sequencing and the Vitek 2 for Identification of Seventy-Three Clinical Isolates of Enteropathogens. J. Thorac. Dis. 2014, 6, 539–544. [Google Scholar]
- Kubelová, M.; Koláčková, I.; Gelbíčová, T.; Florianová, M.; Kalová, A.; Karpíšková, R. Virulence Properties of Mcr-1-Positive Escherichia Coli Isolated from Retail Poultry Meat. Microorganisms 2021, 9, 308. [Google Scholar] [CrossRef]
- Zorrilla, I.; Arijo, S.; Chabrillon, M.; Diaz, P.; Martinez-Manzanares, E.; Balebona, M.C.; Morinigo, M.A. Vibrio Species Isolated from Diseased Farmed Sole, Solea Senegalensis (Kaup), and Evaluation of the Potential Virulence Role of Their Extracellular Products. J. Fish Dis. 2003, 26, 103–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, H.A.; Hussein, M.A.; El-Ashram, A.M. Seafood a Potential Source of Some Zoonotic Bacteria in Zagazig, Egypt, with the Molecular Detection of Listeria Monocytogenes Virulence Genes. Vet. Ital. 2013, 49, 299–308. [Google Scholar] [CrossRef]
- Cabral, J.P.S. Water Microbiology. Bacterial Pathogens and Water. Int. J. Environ. Res. Public Health 2010, 7, 3657–3703. [Google Scholar] [CrossRef]
- Weltgesundheitsorganisation; FAO. OIE–World Organisation for Animal Health. In Brucellosis in Humans and Animals; World Health Organization: Geneva, Switzerland, 2006; ISBN 978-92-4-154713-0. [Google Scholar]
- Jansen, K.U.; Knirsch, C.; Anderson, A.S. The Role of Vaccines in Preventing Bacterial Antimicrobial Resistance. Nat. Med. 2018, 24, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Forsythe, S.J. Emerging Foodborne Enteric Bacterial Pathogens. In Encyclopedia of Food and Health; Elsevier: Amsterdam, The Netherlands, 2016; pp. 487–497. ISBN 978-0-12-384953-3. [Google Scholar]
- Krog, J.S.; Larsen, L.E.; Schultz, A.C. Enteric Porcine Viruses in Farmed Shellfish in Denmark. Int. J. Food Microbiol. 2014, 186, 105–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramadan, H.; Ibrahim, N.; Samir, M.; Abd El-Moaty, A.; Gad, T. Aeromonas Hydrophila from Marketed Mullet (Mugil Cephalus) in Egypt: PCR Characterization of β -Lactam Resistance and Virulence Genes. J. Appl. Microbiol. 2018, 124, 1629–1637. [Google Scholar] [CrossRef] [PubMed]
- Uçar, A.; Yilmaz, M.V.; Çakiroglu, F.P. Food Safety–Problems and Solutions. In Significance, Prevention and Control of Food Related Diseases; Makun, H.A., Ed.; InTech: London, UK, 2016; ISBN 978-953-51-2277-7. [Google Scholar]
- Reed, L.J.; Muench, H. A Simple method of estimating fifty per cent endpoints. Am. J. Epidem. 1938, 27, 493–497. [Google Scholar] [CrossRef]
| Antimicrobial Agent | Susceptible * | Resistance |
|---|---|---|
| Nalidixic acid | 6 (26.1%) ± 0.015 | 17 (73.9%) ± 1.18 |
| Cephalothin | 12 (52.3%) ± 0.23 | 11(47.8%) ± 0.96 |
| Ciprofloxacin | 17 (73.9%) ± 0.007 | 6 (26.1%) ± 0.79 |
| Carbenicillin | 11 (47.8%) ± 1.069 | 12 (52.3%) ± 0.009 |
| Erythromycin | 12 (52.3%) ± 1.30 | 11(47.8%) ± 0.12 |
| Kanamycin | 20 (87%) ± 0.99 | 3 (13%) ± 0.19 |
| Tetracycline | 20 (87%) ± 1.22 | 3 (13%) ± 2.36 |
| Genotypes | LD50 (cfu/shellfish) # |
|---|---|
| Control * | 1 × 106 ± 0.22 |
| Aerolysin/Elastase | 12 × 108 ± 1.34 |
| Aerolysin/Elastase/Hidrolipase | 12 × 109 ± 0.19 |
| Aerolysin/Elastase/Hidrolipase/Flagellin | 1 × 1010 ± 3.02 |
| Aerolysin/Elastase/Hidrolipase/Flagellin/Enterotoxin | 2 × 1012 ± 2.52 |
| Aerolysin/Elastase/Hidrolipase/Flagellin/Enterotoxin/DNases | 3 × 1012 ± 0.77 |
| Target Gene | Primer | Sequence (3ʹ-5ʹ) | Size (bp) | Reference |
|---|---|---|---|---|
| Elastase gene | ahyB-F | ACACGGTCAAGGAGATCAAC | 540 | [4] |
| ahyB-R | CGCTGGTGTTGGCCAGCAGG | |||
| Lipase gene | pla/lip-F | ATCTTCTCCGACTGGTTCGG | 383–389 | [4] |
| pla/lip-R | CCGTGCCAGGACTGGGTCTT | |||
| Hidrolipase gene | lip-F | AACCTGGTTCCGCTCAAGCCGTTG | 65 | [4] |
| lip-R | TTGCTCGCCTCGGCCCAGCAGCT | |||
| Flagellin gene | fla-F | TCCAACCGTYTGACCTC | 608 | [12] |
| fla-R | GMYTGGTTGCGRATGGT | |||
| Enterotoxin gene | act-F | AGAAGGTGACCACCACCAAGAACA | 232 | [12] |
| act-R | AACTGACATCGGCCTTGAACTC | |||
| DNases gene | exu-F | AGACATGCACAACCTCTTCC | 323 | [12] |
| exu-R | GATTGGTATTGCCCTGCAAC | |||
| Aerolysin gene | aeroF | TGGTAGCTAATAACTGCCAG | 1170 | [15] |
| aeroR | GGCTTCTCTCGTTTGGCGT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Selim, S.; Almuhayawi, M.S.; Zakai, S.A.; Salama, A.A.; Warrad, M. Distinction between Antimicrobial Resistance and Putative Virulence Genes Characterization in Plesiomonas shigelloides Isolated from Different Sources. Antibiotics 2022, 11, 85. https://doi.org/10.3390/antibiotics11010085
Selim S, Almuhayawi MS, Zakai SA, Salama AA, Warrad M. Distinction between Antimicrobial Resistance and Putative Virulence Genes Characterization in Plesiomonas shigelloides Isolated from Different Sources. Antibiotics. 2022; 11(1):85. https://doi.org/10.3390/antibiotics11010085
Chicago/Turabian StyleSelim, Samy, Mohammed S. Almuhayawi, Shadi Ahmed Zakai, Ahmed Attia Salama, and Mona Warrad. 2022. "Distinction between Antimicrobial Resistance and Putative Virulence Genes Characterization in Plesiomonas shigelloides Isolated from Different Sources" Antibiotics 11, no. 1: 85. https://doi.org/10.3390/antibiotics11010085
APA StyleSelim, S., Almuhayawi, M. S., Zakai, S. A., Salama, A. A., & Warrad, M. (2022). Distinction between Antimicrobial Resistance and Putative Virulence Genes Characterization in Plesiomonas shigelloides Isolated from Different Sources. Antibiotics, 11(1), 85. https://doi.org/10.3390/antibiotics11010085







