Multidrug-Resistant High-Risk Escherichia coli and Klebsiella pneumoniae Clonal Lineages Occur in Black-Headed Gulls from Two Conservation Islands in Germany
Abstract
:1. Introduction
2. Results
2.1. The Presence of ESBL-Producing E. coli and K. pneumoniae in Two Black-Headed Gull Colonies
2.2. Resistance Profiles of the Isolated Strains
2.3. Virulence Profiles of the Isolated Strains
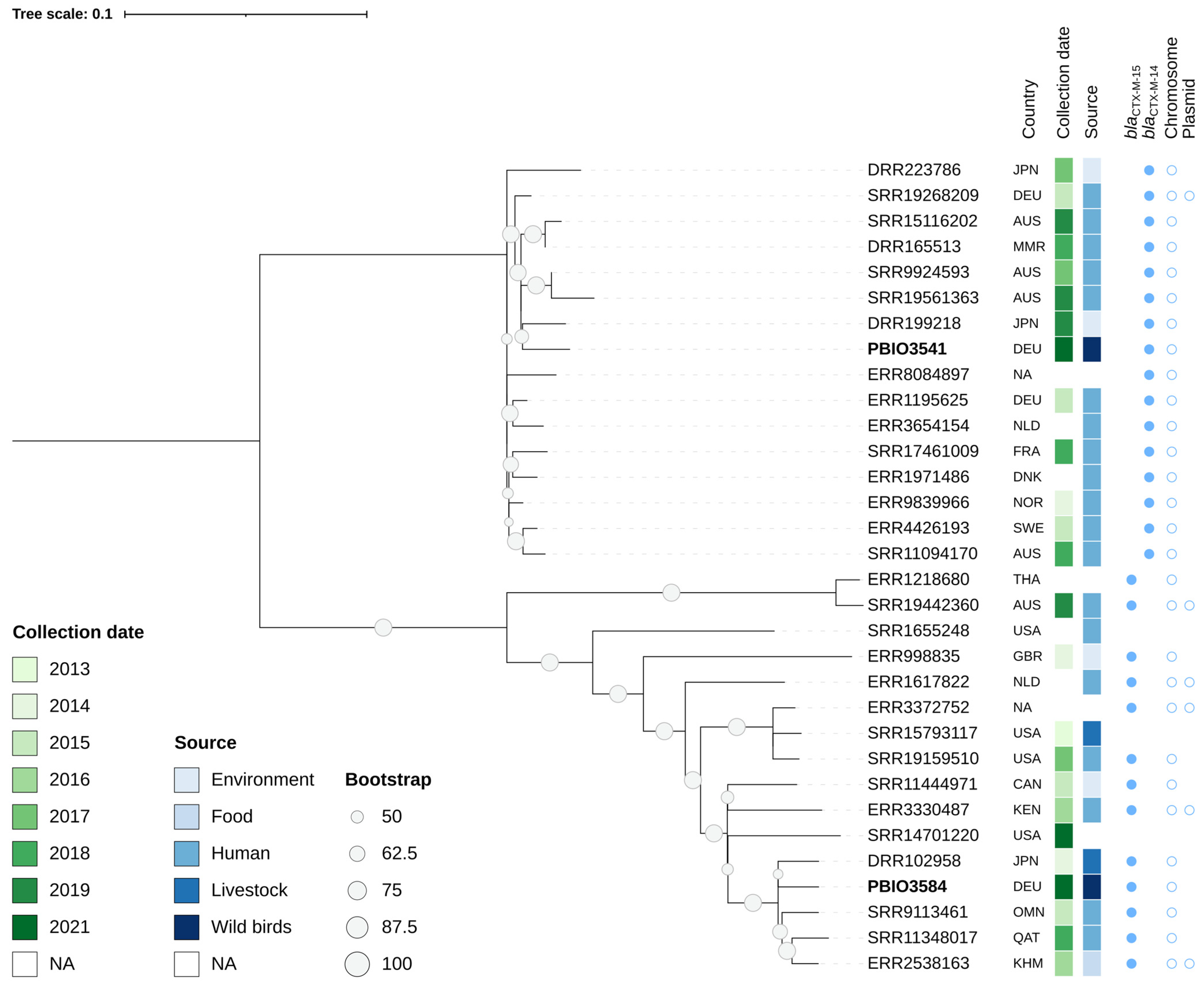
2.4. Phylogenetic Relationship
3. Discussion
4. Materials and Methods
4.1. Sampling Strategy and Bacterial Isolation
4.2. Phenotypic Antimicrobial Susceptibility Testing
4.3. Whole-Genome Sequencing
4.4. Sequence Assembly and Genomic Analyses
4.5. Phylogeny
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Sherry, N.L.; Gorrie, C.L.; Kwong, J.C.; Higgs, C.; Stuart, R.L.; Marshall, C.; Ballard, S.A.; Sait, M.; Korman, T.M.; Slavin, M.A.; et al. Controlling Superbugs Study Group. Multi-site implementation of whole genome sequencing for hospital infection control: A prospective genomic epidemiological analysis. Lancet Reg. Health-West. Pac. 2022, 23, 100446. [Google Scholar] [CrossRef] [PubMed]
- Tewari, R.; Ganaie, F.; Venugopal, N.; Mitra, S.; Shome, R.; Shome, B.R. Occurrence and characterization of genetic determinants of beta-lactam-resistance in Escherichia coli clinical isolates. Infect. Genet. Evol. 2022, 100, 105257. [Google Scholar] [CrossRef] [PubMed]
- Klees, S.; Effelsberg, N.; Stuhrenberg, B.; Mellmann, A.; Schwarz, S.; Kock, R. Prevalence and epidemiology of multidrug-resistant pathogens in the food chain and the urban environment in Northwestern Germany. Antibiotics 2020, 9, 708. [Google Scholar] [CrossRef] [PubMed]
- Wareth, G.; Neubauer, H. The Animal-foods-environment interface of Klebsiella pneumoniae in Germany: An observational study on pathogenicity, resistance development and the current situation. Vet. Res. 2021, 52, 16. [Google Scholar] [CrossRef]
- Bennani, H.; Mateus, A.; Mays, N.; Eastmure, E.; Stark, K.D.C.; Hasler, B. Overview of evidence of antimicrobial use and antimicrobial resistance in the food chain. Antibiotics 2020, 9, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, J.S.; Song, W.; Park, H.M.; Oh, J.Y.; Chae, J.C.; Jeong, S.; Jeong, S.H. Molecular characterization of fecal extended-spectrum beta-lactamase- and AmpC beta-lactamase-producing Escherichia coli from healthy companion animals and cohabiting humans in South Korea. Front. Microbiol. 2020, 11, 674. [Google Scholar] [CrossRef] [Green Version]
- Homeier-Bachmann, T.; Heiden, S.E.; Lubcke, P.K.; Bachmann, L.; Bohnert, J.A.; Zimmermann, D.; Schaufler, K. Antibiotic-resistant Enterobacteriaceae in wastewater of abattoirs. Antibiotics 2021, 10, 568. [Google Scholar] [CrossRef] [PubMed]
- Homeier-Bachmann, T.; Schutz, A.K.; Dreyer, S.; Glanz, J.; Schaufler, K.; Conraths, F.J. Genomic analysis of ESBL-producing E. coli in wildlife from North-Eastern Germany. Antibiotics 2022, 11, 123. [Google Scholar] [CrossRef] [PubMed]
- Schierack, P.; Heiden, S.E.; Khan, M.M.; Nikolaus, L.; Kolenda, R.; Stubbe, M.; Lkhagvasuren, D.; Rodiger, S.; Guenther, S.; Schaufler, K. Genomic and phenotypic analysis of an ESBL-producing E. coli ST1159 clonal lineage from wild birds in Mongolia. Front. Microbiol. 2020, 11, 1699. [Google Scholar] [CrossRef]
- Bonnedahl, J.; Drobni, P.; Johansson, A.; Hernandez, J.; Melhus, A.; Stedt, J.; Olsen, B.; Drobni, M. Characterization, and comparison, of human clinical and black-headed gull (Larus ridibundus) extended-spectrum beta-lactamase-producing bacterial isolates from Kalmar, on the southeast coast of Sweden. J. Antimicrob. Chemother. 2010, 65, 1939–1944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velhner, M.; Todorovic, D.; Novovic, K.; Jovcic, B.; Lazic, G.; Kojic, M.; Kehrenberg, C. Characterization of antibiotic resistance in Escherichia coli isolates from black-headed gulls (Larus ridibundus) present in the city of Novi Sad, Serbia. Vet. Res. Commun. 2021, 45, 199–209. [Google Scholar] [CrossRef]
- Dolejska, M.; Cizek, A.; Literak, I. High prevalence of antimicrobial-resistant genes and integrons in Escherichia coli isolates from black-headed gulls in the Czech Republic. J. Appl. Microbiol. 2007, 103, 11–19. [Google Scholar] [CrossRef]
- Nicolas-Chanoine, M.H.; Blanco, J.; Leflon-Guibout, V.; Demarty, R.; Alonso, M.P.; Canica, M.M.; Park, Y.J.; Lavigne, J.P.; Pitout, J.; Johnson, J.R. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 2008, 61, 273–281. [Google Scholar] [CrossRef] [Green Version]
- Ewers, C.; Grobbel, M.; Stamm, I.; Kopp, P.A.; Diehl, I.; Semmler, T.; Fruth, A.; Beutlich, J.; Guerra, B.; Wieler, L.H.; et al. Emergence of human pandemic O25:H4-ST131 CTX-M-15 extended-spectrum-beta-lactamase-producing Escherichia coli among companion animals. J. Antimicrob. Chemother. 2010, 65, 651–660. [Google Scholar] [CrossRef] [Green Version]
- Guenther, S.; Aschenbrenner, K.; Stamm, I.; Bethe, A.; Semmler, T.; Stubbe, A.; Stubbe, M.; Batsajkhan, N.; Glupczynski, Y.; Wieler, L.H.; et al. Comparable high rates of extended-spectrum-beta-lactamase-producing Escherichia coli in birds of prey from Germany and Mongolia. PLoS ONE 2012, 7, e53039. [Google Scholar] [CrossRef] [Green Version]
- Skurnik, D.; Clermont, O.; Guillard, T.; Launay, A.; Danilchanka, O.; Pons, S.; Diancourt, L.; Lebreton, F.; Kadlec, K.; Roux, D.; et al. Emergence of antimicrobial-resistant Escherichia coli of animal origin spreading in humans. Mol. Biol. Evol. 2016, 33, 898–914. [Google Scholar] [CrossRef] [Green Version]
- Schaufler, K.; Semmler, T.; Wieler, L.H.; Wohrmann, M.; Baddam, R.; Ahmed, N.; Muller, K.; Kola, A.; Fruth, A.; Ewers, C.; et al. Clonal spread and interspecies transmission of clinically relevant ESBL-producing Escherichia coli of ST410-another successful pandemic clone? FEMS Microbiol. Ecol. 2016, 92, fiv155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guenther, S.; Semmler, T.; Stubbe, A.; Stubbe, M.; Wieler, L.H.; Schaufler, K. Chromosomally encoded ESBL genes in Escherichia coli of ST38 from Mongolian wild birds. J. Antimicrob. Chemother. 2017, 72, 1310–1313. [Google Scholar] [CrossRef]
- Schaufler, K.; Nowak, K.; Dux, A.; Semmler, T.; Villa, L.; Kourouma, L.; Bangoura, K.; Wieler, L.H.; Leendertz, F.H.; Guenther, S. Clinically relevant ESBL-producing K. pneumoniae ST307 and E. coli ST38 in an urban West African rat population. Front. Microbiol. 2018, 9, 150. [Google Scholar] [CrossRef]
- Eger, E.; Domke, M.; Heiden, S.E.; Paditz, M.; Balau, V.; Huxdorff, C.; Zimmermann, D.; Homeier-Bachmann, T.; Schaufler, K. Highly virulent and multidrug-resistant Escherichia coli sequence type 58 from a sausage in Germany. Antibiotics 2022, 11, 1006. [Google Scholar] [CrossRef]
- Fuga, B.; Sellera, F.P.; Cerdeira, L.; Esposito, F.; Cardoso, B.; Fontana, H.; Moura, Q.; Cardenas-Arias, A.; Sano, E.; Ribas, R.M.; et al. WHO critical priority Escherichia coli as One Health challenge for a post-pandemic scenario: Genomic surveillance and analysis of current trends in Brazil. Microbiol. Spectr. 2022, 10, e0125621. [Google Scholar] [CrossRef]
- Franklin-Alming, F.V.; Kaspersen, H.; Hetland, M.A.K.; Bakksjo, R.J.; Nesse, L.L.; Leangapichart, T.; Lohr, I.H.; Telke, A.A.; Sunde, M. Exploring Klebsiella pneumoniae in healthy poultry reveals high genetic diversity, good biofilm-forming abilities and higher prevalence in turkeys than broilers. Front. Microbiol. 2021, 12, 725414. [Google Scholar] [CrossRef]
- Heiden, S.E.; Hübner, N.-O.; Bohnert, J.A.; Heidecke, C.-D.; Kramer, A.; Balau, V.; Gierer, W.; Schaefer, S.; Eckmanns, T.; Gatermann, S.; et al. A Klebsiella pneumoniae ST307 outbreak clone from Germany demonstrates features of extensive drug resistance, hypermucoviscosity, and enhanced iron acquisition. Genome Med. 2020, 12, 1–15. [Google Scholar] [CrossRef]
- Melzer, M.; Petersen, I. Mortality following bacteraemic infection caused by extended spectrum beta-lactamase (ESBL) producing E. coli compared to non-ESBL producing E. coli. J. Infect. 2007, 55, 254–259. [Google Scholar] [CrossRef]
- Schwaber, M.J.; Carmeli, Y. Mortality and delay in effective therapy associated with extended-spectrum beta-lactamase production in Enterobacteriaceae bacteraemia: A systematic review and meta-analysis. J. Antimicrob. Chemother. 2007, 60, 913–920. [Google Scholar] [CrossRef] [Green Version]
- Piro, S.; Schmitz Ornés, A. Nest site tenacity and mate fidelity in the black-headed gull (Chroicocephalus ridibundus). Avian Res. 2021, 12, 63. [Google Scholar] [CrossRef]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef] [Green Version]
- Touchon, M.; Hoede, C.; Tenaillon, O.; Barbe, V.; Baeriswyl, S.; Bidet, P.; Bingen, E.; Bonacorsi, S.; Bouchier, C.; Bouvet, O.; et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009, 5, e1000344. [Google Scholar] [CrossRef] [Green Version]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef]
- Mathers, A.J.; Peirano, G.; Pitout, J.D. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 2015, 28, 565–591. [Google Scholar] [CrossRef] [Green Version]
- Yoon, E.J.; Choi, Y.J.; Kim, D.; Won, D.; Choi, J.R.; Jeong, S.H. Amplification of the chromosomal blaCTX-M-14 gene in Escherichia coli expanding the spectrum of resistance under antimicrobial pressure. Microbiol. Spectr. 2022, 10, e0031922. [Google Scholar] [CrossRef]
- Eger, E.; Heiden, S.E.; Korolew, K.; Bayingana, C.; Ndoli, J.M.; Sendegeya, A.; Gahutu, J.B.; Kurz, M.S.E.; Mockenhaupt, F.P.; Müller, J.; et al. Circulation of extended-spectrum beta-lactamase-producing Escherichia coli of pandemic sequence types 131, 648, and 410 among hospitalized patients, caregivers, and the community in Rwanda. Front. Microbiol. 2021, 12, 662575. [Google Scholar] [CrossRef]
- Canton, R.; Coque, T.M. The CTX-M beta-lactamase pandemic. Curr. Opin. Microbiol. 2006, 9, 466–475. [Google Scholar] [CrossRef]
- Hidron, A.I.; Edwards, J.R.; Patel, J.; Horan, T.C.; Sievert, D.M.; Pollock, D.A.; Fridkin, S.K.; National Healthcare Safety Network Team; Participating National Healthcare Safety Network Facilities. NHSN annual update: Antimicrobial-resistant pathogens associated with healthcare-associated infections: Annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect. Control Hosp. Epidemiol. 2008, 29, 996–1011. [Google Scholar] [CrossRef] [Green Version]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Olaitan, A.O.; Morand, S.; Rolain, J.M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.; Lee, J.Y.; Lee, H.; Park, M.; Kang, K.; Lim, S.K.; Shin, D.; Ko, K.S. Comparison of fitness cost and virulence in chromosome- and plasmid-mediated colistin-resistant Escherichia coli. Front. Microbiol. 2020, 11, 798. [Google Scholar] [CrossRef]
- Pitout, J.D. Extraintestinal pathogenic Escherichia coli: A combination of virulence with antibiotic resistance. Front. Microbiol. 2012, 3, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beceiro, A.; Tomas, M.; Bou, G. Antimicrobial resistance and virulence: A successful or deleterious association in the bacterial world? Clin. Microbiol. Rev. 2013, 26, 185–230. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef] [PubMed]
- Mulvey, M.A. Adhesion and entry of uropathogenic Escherichia coli. Cell. Microbiol. 2002, 4, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Lam, M.M.C.; Wick, R.R.; Watts, S.C.; Cerdeira, L.T.; Wyres, K.L.; Holt, K.E. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 2021, 12, 4188. [Google Scholar] [CrossRef]
- Arimizu, Y.; Kirino, Y.; Sato, M.P.; Uno, K.; Sato, T.; Gotoh, Y.; Auvray, F.; Brugere, H.; Oswald, E.; Mainil, J.G.; et al. Large-scale genome analysis of bovine commensal Escherichia coli reveals that bovine-adapted E. coli lineages are serving as evolutionary sources of the emergence of human intestinal pathogenic strains. Genome Res. 2019, 29, 1495–1505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Lopez, A.; Sundararaju, S.; Al-Mana, H.; Tsui, K.M.; Hasan, M.R.; Suleiman, M.; Janahi, M.; Al Maslamani, E.; Tang, P. Molecular characterization of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae among the pediatric population in Qatar. Front. Microbiol. 2020, 11, 581711. [Google Scholar] [CrossRef] [PubMed]
- Atterby, C.; Borjesson, S.; Ny, S.; Jarhult, J.D.; Byfors, S.; Bonnedahl, J. ESBL-producing Escherichia coli in Swedish gulls-A case of environmental pollution from humans? PLoS ONE 2017, 12, e0190380. [Google Scholar] [CrossRef]
- Athanasakopoulou, Z.; Diezel, C.; Braun, S.D.; Sofia, M.; Giannakopoulos, A.; Monecke, S.; Gary, D.; Krahmer, D.; Chatzopoulos, D.C.; Touloudi, A.; et al. Occurrence and characteristics of ESBL- and carbapenemase- producing Escherichia coli from wild and feral birds in Greece. Microorganisms 2022, 10, 1217. [Google Scholar] [CrossRef] [PubMed]
- Ewbank, A.C.; Fuentes-Castillo, D.; Sacristan, C.; Cardoso, B.; Esposito, F.; Fuga, B.; de Macedo, E.C.; Lincopan, N.; Catao-Dias, J.L. Extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli survey in wild seabirds at a pristine atoll in the southern Atlantic Ocean, Brazil: First report of the O25b-ST131 clone harboring blaCTX-M-8. Sci. Total Environ. 2022, 806, 150539. [Google Scholar] [CrossRef] [PubMed]
- Fashae, K.; Engelmann, I.; Monecke, S.; Braun, S.D.; Ehricht, R. Molecular characterisation of extended-spectrum β-lactamase producing Escherichia coli in wild birds and cattle, Ibadan, Nigeria. BMC Vet. Res. 2021, 17, 33. [Google Scholar] [CrossRef] [PubMed]
- Zendri, F.; Maciuca, I.E.; Moon, S.; Jones, P.H.; Wattret, A.; Jenkins, R.; Baxter, A.; Timofte, D. Occurrence of ESBL-producing Escherichia coli ST131, including the H30-Rx and C1-M27 subclones, among urban seagulls from the United Kingdom. Microb. Drug Resist. 2020, 26, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Atterby, C.; Ramey, A.M.; Hall, G.G.; Jarhult, J.; Borjesson, S.; Bonnedahl, J. Increased prevalence of antibiotic-resistant E. coli in gulls sampled in Southcentral Alaska is associated with urban environments. Infect. Ecol. Epidemiol. 2016, 6, 32334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alonso, C.A.; Gonzalez-Barrio, D.; Ruiz-Fons, F.; Ruiz-Ripa, L.; Torres, C. High frequency of B2 phylogroup among non-clonally related fecal Escherichia coli isolates from wild boars, including the lineage ST131. FEMS Microbiol. Ecol. 2017, 93, fix016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDougall, F.K.; Boardman, W.S.J.; Power, M.L. Characterization of beta-lactam-resistant Escherichia coli from Australian fruit bats indicates anthropogenic origins. Microb. Genom. 2021, 7, 000571. [Google Scholar] [CrossRef]
- Awawdeh, L.; Turni, C.; Mollinger, J.L.; Henning, J.; Cobbold, R.N.; Trott, D.J.; Wakeham, D.L.; Gibson, J.S. Antimicrobial susceptibility, plasmid replicon typing, phylogenetic grouping, and virulence potential of avian pathogenic and faecal Escherichia coli isolated from meat chickens in Australia. Avian Pathol. 2022, 51, 349–360. [Google Scholar] [CrossRef]
- Loucif, L.; Chelaghma, W.; Cherak, Z.; Bendjama, E.; Beroual, F.; Rolain, J.M. Detection of NDM-5 and MCR-1 antibiotic resistance encoding genes in Enterobacterales in long-distance migratory bird species Ciconia ciconia, Algeria. Sci. Total Environ. 2022, 814, 152861. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.W.; Van Kessel, J.A.S.; Haley, B.J. Draft genome sequence of Escherichia coli ARS-CC7049, a sequence type 38 Strain isolated from dry cow feces on a commercial dairy operation. Microbiol. Resour. Announc. 2022, 11, e0094321. [Google Scholar] [CrossRef]
- Jamborova, I.; Dolejska, M.; Vojtech, J.; Guenther, S.; Uricariu, R.; Drozdowska, J.; Papousek, I.; Pasekova, K.; Meissner, W.; Hordowski, J.; et al. Plasmid-mediated resistance to cephalosporins and fluoroquinolones in various Escherichia coli sequence types isolated from rooks wintering in Europe. Appl. Environ. Microbiol. 2015, 81, 648–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, X.; Yan, X.; Li, Y.; Zhang, D.; Li, L.; Geng, Y.; Su, F.; Yue, C.; Hou, R.; Liu, S. Identification of extended-spectrum beta-lactamase (CTX-M)-producing Klebsiella pneumoniae belonging to ST37, ST290, and ST2640 in captive giant pandas. BMC Vet. Res. 2022, 18, 186. [Google Scholar] [CrossRef]
- Wang, Z.; Li, M.; Shen, X.; Wang, L.; Liu, L.; Hao, Z.; Duan, J.; Yu, F. Outbreak of blaNDM-5-harboring Klebsiella pneumoniae ST290 in a Tertiary Hospital in China. Microb. Drug Resist. 2019, 25, 1443–1448. [Google Scholar] [CrossRef] [PubMed]
- Cienfuegos-Gallet, A.V.; Zhou, Y.; Ai, W.; Kreiswirth, B.N.; Yu, F.; Chen, L. Multicenter genomic analysis of carbapenem-resistant Klebsiella pneumoniae from bacteremia in China. Microbiol. Spectr. 2022, 10, e0229021. [Google Scholar] [CrossRef]
- Janecko, N.; Halova, D.; Jamborova, I.; Papousek, I.; Masarikova, M.; Dolejska, M.; Literak, I. Occurrence of plasmid-mediated quinolone resistance genes in Escherichia coli and Klebsiella spp. recovered from Corvus brachyrhynchos and Corvus corax roosting in Canada. Lett. Appl. Microbiol. 2018, 67, 130–135. [Google Scholar] [CrossRef]
- Dolejska, M.; Masarikova, M.; Dobiasova, H.; Jamborova, I.; Karpiskova, R.; Havlicek, M.; Carlile, N.; Priddel, D.; Cizek, A.; Literak, I. High prevalence of Salmonella and IMP-4-producing Enterobacteriaceae in the silver gull on Five Islands, Australia. J. Antimicrob. Chemother. 2016, 71, 63–70. [Google Scholar] [CrossRef] [Green Version]
- Savin, M.; Bierbaum, G.; Schmithausen, R.M.; Heinemann, C.; Kreyenschmidt, J.; Schmoger, S.; Akbaba, I.; Kasbohrer, A.; Hammerl, J.A. Slaughterhouse wastewater as a reservoir for extended-spectrum beta-lactamase (ESBL)-producing, and colistin-resistant Klebsiella spp. and their impact in a “One Health” perspective. Sci. Total Environ. 2022, 804, 150000. [Google Scholar] [CrossRef] [PubMed]
- Siller, P.; Daehre, K.; Rosen, K.; Munch, S.; Bartel, A.; Funk, R.; Nubel, U.; Amon, T.; Roesler, U. Low airborne tenacity and spread of ESBL-/AmpC-producing Escherichia coli from fertilized soil by wind erosion. Environ. Microbiol. 2021, 23, 7497–7511. [Google Scholar] [CrossRef]
- Poulin-Laprade, D.; Brouard, J.S.; Gagnon, N.; Turcotte, A.; Langlois, A.; Matte, J.J.; Carrillo, C.D.; Zaheer, R.; McAllister, T.; Topp, E.; et al. Resistance determinants and their genetic context in enterobacteria from a longitudinal study of pigs reared under various husbandry conditions. Appl. Environ. Microbiol. 2021, 87, e02612–e02620. [Google Scholar] [CrossRef] [PubMed]
- Akhil Prakash, E.; Hromadkova, T.; Jabir, T.; Vipindas, P.V.; Krishnan, K.P.; Mohamed Hatha, A.A.; Briedis, M. Dissemination of multidrug resistant bacteria to the polar environment-Role of the longest migratory bird Arctic tern (Sterna paradisaea). Sci. Total Environ. 2022, 815, 152727. [Google Scholar] [CrossRef] [PubMed]
- Velazquez-Meza, M.E.; Galarde-Lopez, M.; Carrillo-Quiroz, B.; Alpuche-Aranda, C.M. Antimicrobial resistance: One Health approach. Vet. World 2022, 15, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, I.; Thomas, K.; Van Essen, A.; Schink, A.K.; Day, M.; Chattaway, M.; Wu, G.; Mevius, D.; Helmuth, R.; Guerra, B.; et al. Chromosomal location of blaCTX-M genes in clinical isolates of Escherichia coli from Germany, The Netherlands and the UK. Int. J. Antimicrob. Agents 2014, 43, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Ciesinski, L.; Guenther, S.; Pieper, R.; Kalisch, M.; Bednorz, C.; Wieler, L.H. High dietary zinc feeding promotes persistence of multi-resistant E. coli in the swine gut. PLoS ONE 2018, 13, e0191660. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Wright, M.S.; Stepanauskas, R.; McArthur, J.V. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006, 14, 176–182. [Google Scholar] [CrossRef]
- Moser, C.; Pedersen, H.T.; Lerche, C.J.; Kolpen, M.; Line, L.; Thomsen, K.; Hoiby, N.; Jensen, P.O. Biofilms and host response-Helpful or harmful. J. Pathol. Microbiol. Immunol. 2017, 125, 320–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 32nd ed.; CLSI Guideline M100: Wayne, PA, USA, 2022. [Google Scholar]
- Illumina. bcl-Convert: A Proprietary Illumina Software for the Conversion of bcl Files to Basecalls. Available online: https://support-docs.illumina.com/SW/BCL_Convert/Content/SW/FrontPages/BCL_Convert.htm (accessed on 7 March 2022).
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes de novo assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.F.; Mohamed, K.; Fan, Y.; Agama Study, G.; Achtman, M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef] [Green Version]
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Moller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Beghain, J.; Bridier-Nahmias, A.; Le Nagard, H.; Denamur, E.; Clermont, O. ClermonTyping: An easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genom. 2018, 4, e000192. [Google Scholar] [CrossRef] [PubMed]
- Wyres, K.L.; Wick, R.R.; Gorrie, C.; Jenney, A.; Follador, R.; Thomson, N.R.; Holt, K.E. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb. Genom. 2016, 2, e000102. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Croucher, N.J.; Page, A.J.; Connor, T.R.; Delaney, A.J.; Keane, J.A.; Bentley, S.D.; Parkhill, J.; Harris, S.R. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015, 43, e15. [Google Scholar] [CrossRef] [Green Version]
- Page, A.J.; Taylor, B.; Delaney, A.J.; Soares, J.; Seemann, T.; Keane, J.A.; Harris, S.R. SNP-sites: Rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genom. 2016, 2, e000056. [Google Scholar] [CrossRef] [Green Version]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]



| Strain | Species | ST a | Phylogroup | Island c | Source | Replicon Type | ESBL Gene |
|---|---|---|---|---|---|---|---|
| PBIO3518 | E. coli | 34 | A | RW | Collective feces | FII, I1, X1/4, Y | blaCTX-M-14 |
| PBIO3538 | E. coli | 58 | B1 | B | Collective feces | Q2 | blaCTX-M-15 |
| PBIO3541 | E. coli | 38 | D | B | Collective feces | blaCTX-M-14 | |
| PBIO3583 | E. coli | 453 | B1 | B | ICS d nestling | X4/8, Y | blaCTX-M-15 |
| PBIO3584 | E. coli | 38 | D | B | ICS nestling | blaCTX-M-15 | |
| PBIO3592 | E. coli | 453 | B1 | B | ICS nestling | Y | blaCTX-M-15 |
| PBIO3678 | E. coli | 744 | A | B | ICS nestling | FIA/B/C, FII, Q1, X1 | blaCTX-M-55 |
| PBIO3687 | E. coli | 131 | B2 | B | Collective feces | FIA/B, FII | blaCTX-M-27 |
| PBIO3688 | E. coli | 617 | A | B | Collective feces | FIB, FII, I1, Q1 | blaCTX-M-15 |
| PBIO3691 | K. pneumoniae | 290 | NA b | B | Collective feces | FIB, FII | blaCTX-M-15 |
| PBIO3707 | E. coli | 1598 | A | B | ICS adult | FII, R | blaCTX-M-15 |
| PBIO3708 | E. coli | 58 | B1 | B | ICS adult | FIB, FII, I1, Q1 | blaCTX-M-32 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brendecke, J.; Homeier-Bachmann, T.; Schmitz Ornés, A.; Guenther, S.; Heiden, S.E.; Schwabe, M.; Eger, E.; Schaufler, K. Multidrug-Resistant High-Risk Escherichia coli and Klebsiella pneumoniae Clonal Lineages Occur in Black-Headed Gulls from Two Conservation Islands in Germany. Antibiotics 2022, 11, 1357. https://doi.org/10.3390/antibiotics11101357
Brendecke J, Homeier-Bachmann T, Schmitz Ornés A, Guenther S, Heiden SE, Schwabe M, Eger E, Schaufler K. Multidrug-Resistant High-Risk Escherichia coli and Klebsiella pneumoniae Clonal Lineages Occur in Black-Headed Gulls from Two Conservation Islands in Germany. Antibiotics. 2022; 11(10):1357. https://doi.org/10.3390/antibiotics11101357
Chicago/Turabian StyleBrendecke, Jana, Timo Homeier-Bachmann, Angela Schmitz Ornés, Sebastian Guenther, Stefan E. Heiden, Michael Schwabe, Elias Eger, and Katharina Schaufler. 2022. "Multidrug-Resistant High-Risk Escherichia coli and Klebsiella pneumoniae Clonal Lineages Occur in Black-Headed Gulls from Two Conservation Islands in Germany" Antibiotics 11, no. 10: 1357. https://doi.org/10.3390/antibiotics11101357







