Azole-Based Compounds That Are Active against Candida Biofilm: In Vitro, In Vivo and In Silico Studies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemistry
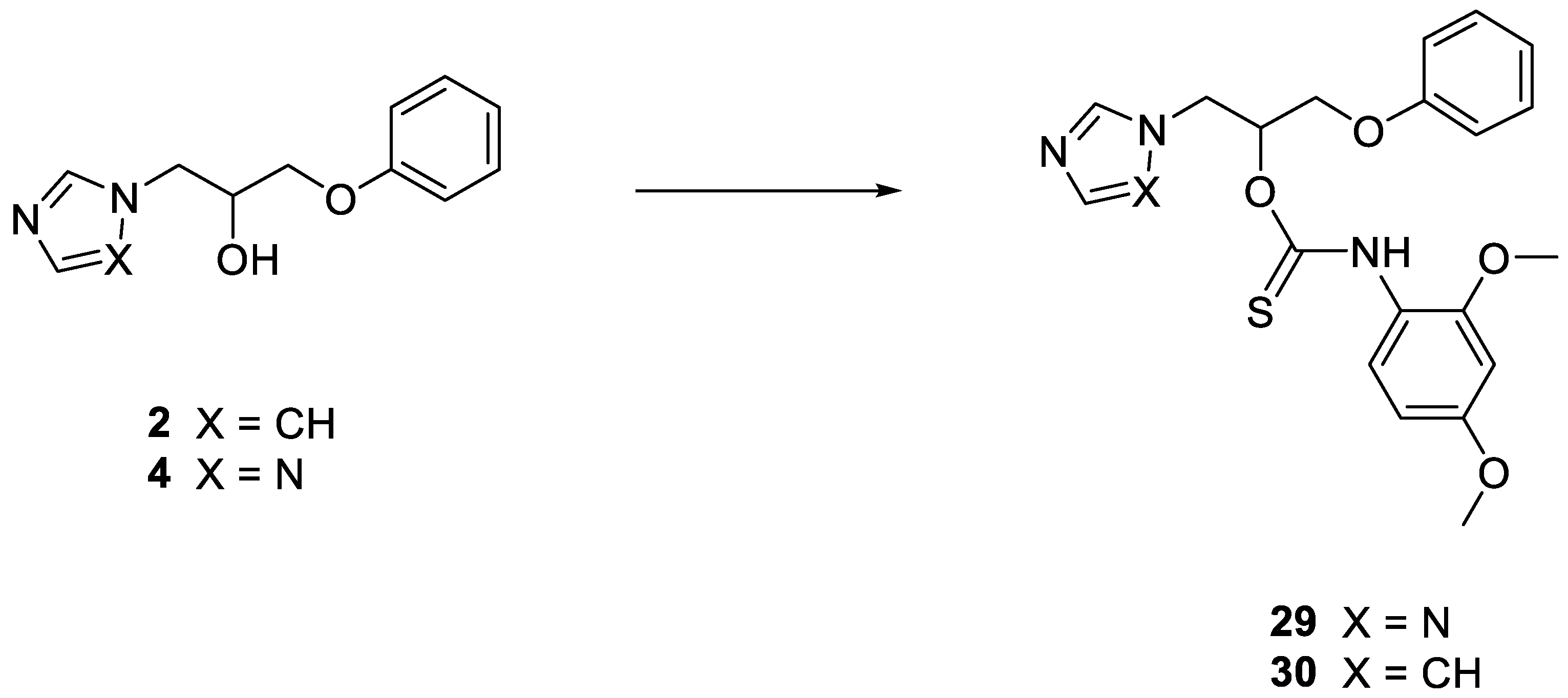
2.1.1. General Procedure for the Synthesis of Thiocarbamates 29 and 30
1-Phenoxy-3-(1H-1,2,4-triazol-1-yl) propan-2-yl (2,4-dimethoxyphenyl)thiocarbamate 29
1-(1H-imidazol-1-yl)-3-phenoxypropan-2-yl (2,4-dimethoxyphenyl)thiocarbamate 30
2.2. Antifungal Susceptibility
2.2.1. Organisms
2.2.2. Antifungal Susceptibility Testing
2.2.3. In Vitro Activity of the Compounds against C. albicans, C. glabrata and C. krusei Biofilms
2.2.4. G. mellonella Survival Assay
2.2.5. In Vivo Toxicity Studies
2.3. Statistical Analysis
2.4. Molecular Modelling Studies
2.4.1. Docking Studies
2.4.2. Molecular Dynamic Simulations
3. Results
3.1. In Vitro Antifungal Activity Evaluation
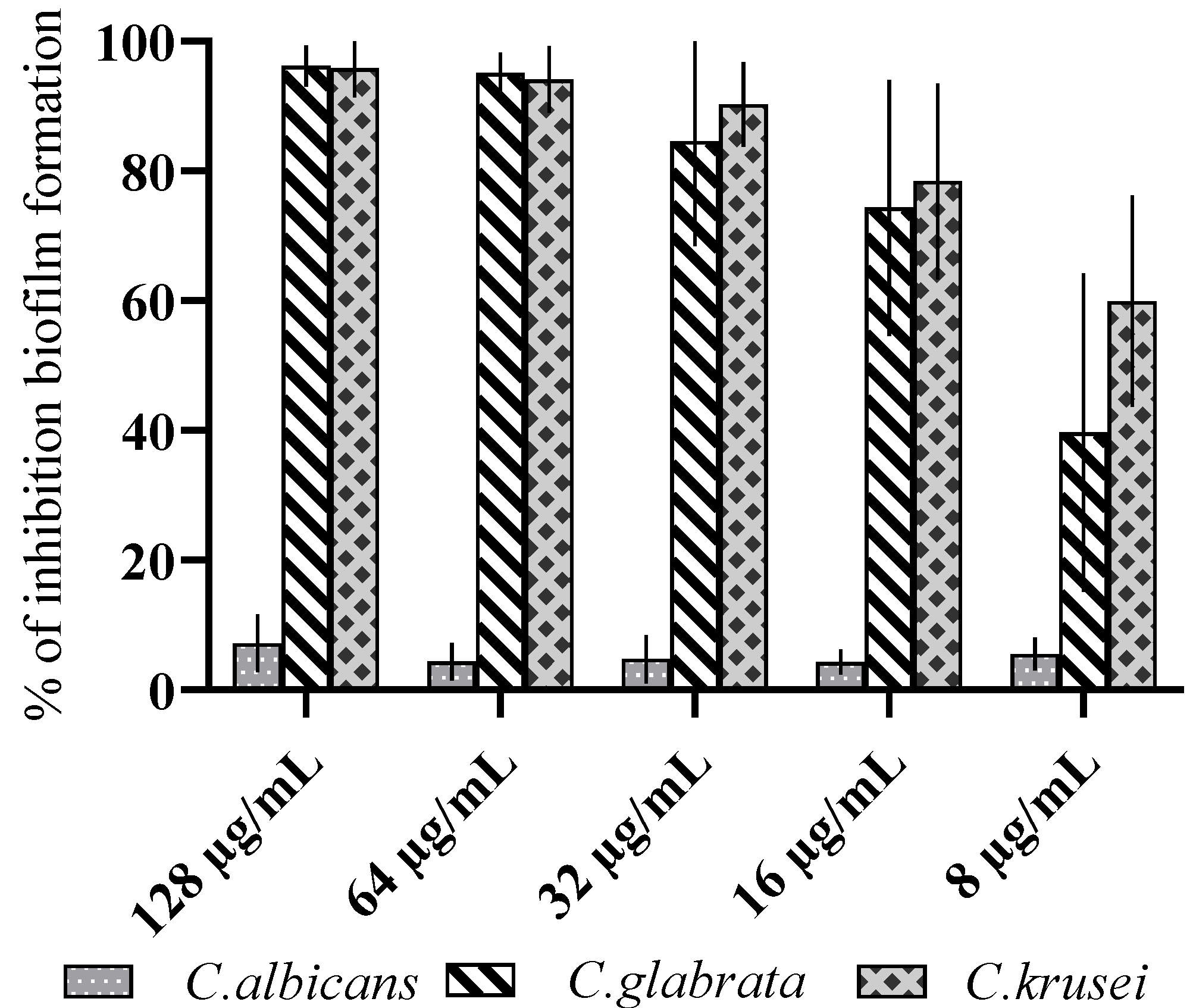
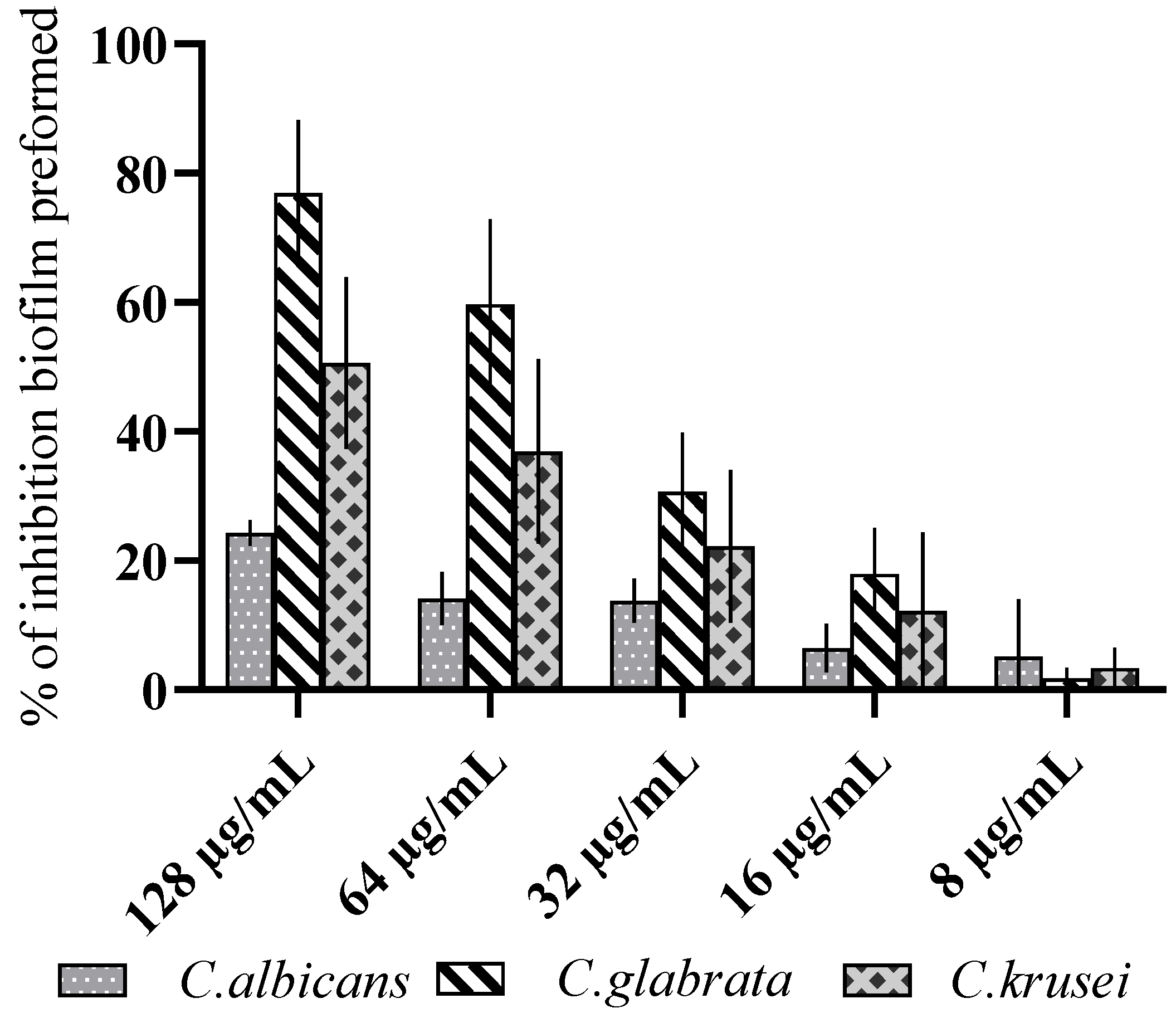
3.2. Antibiofilm Activity
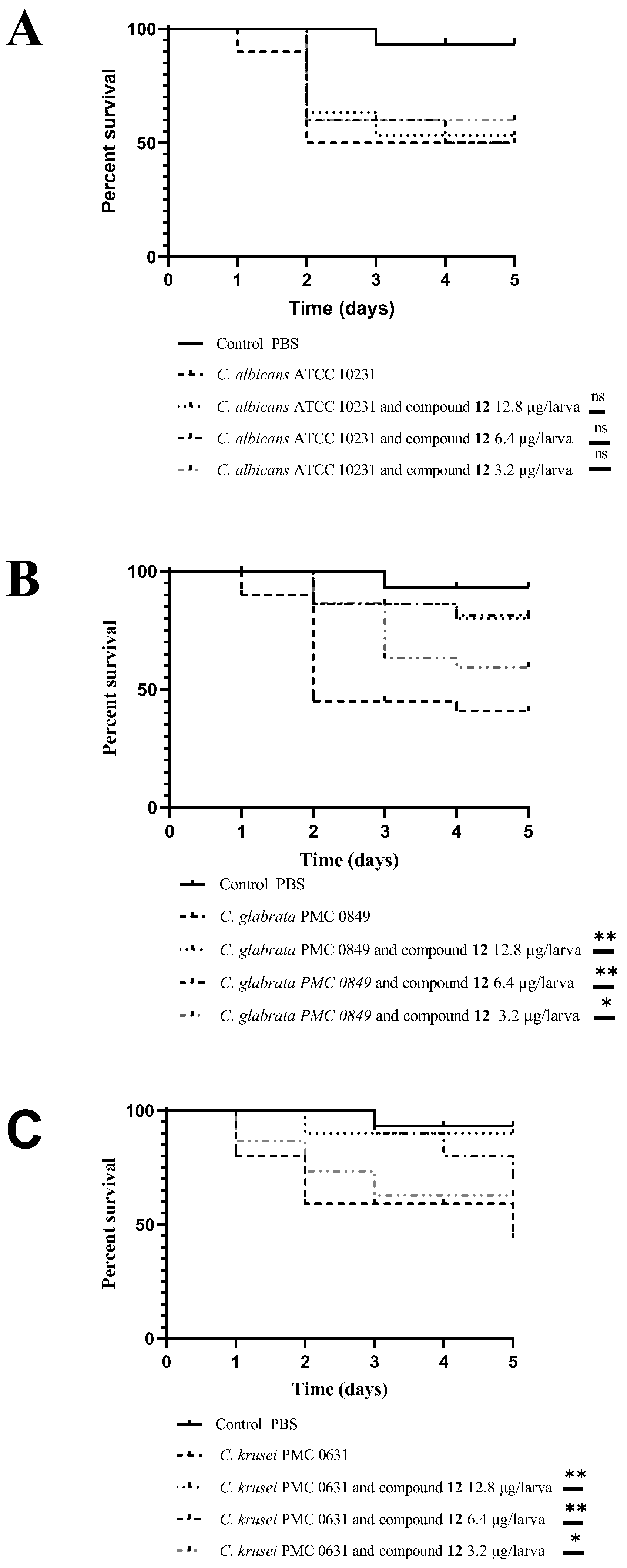
3.3. In Vivo Antifungal Activity Evaluation
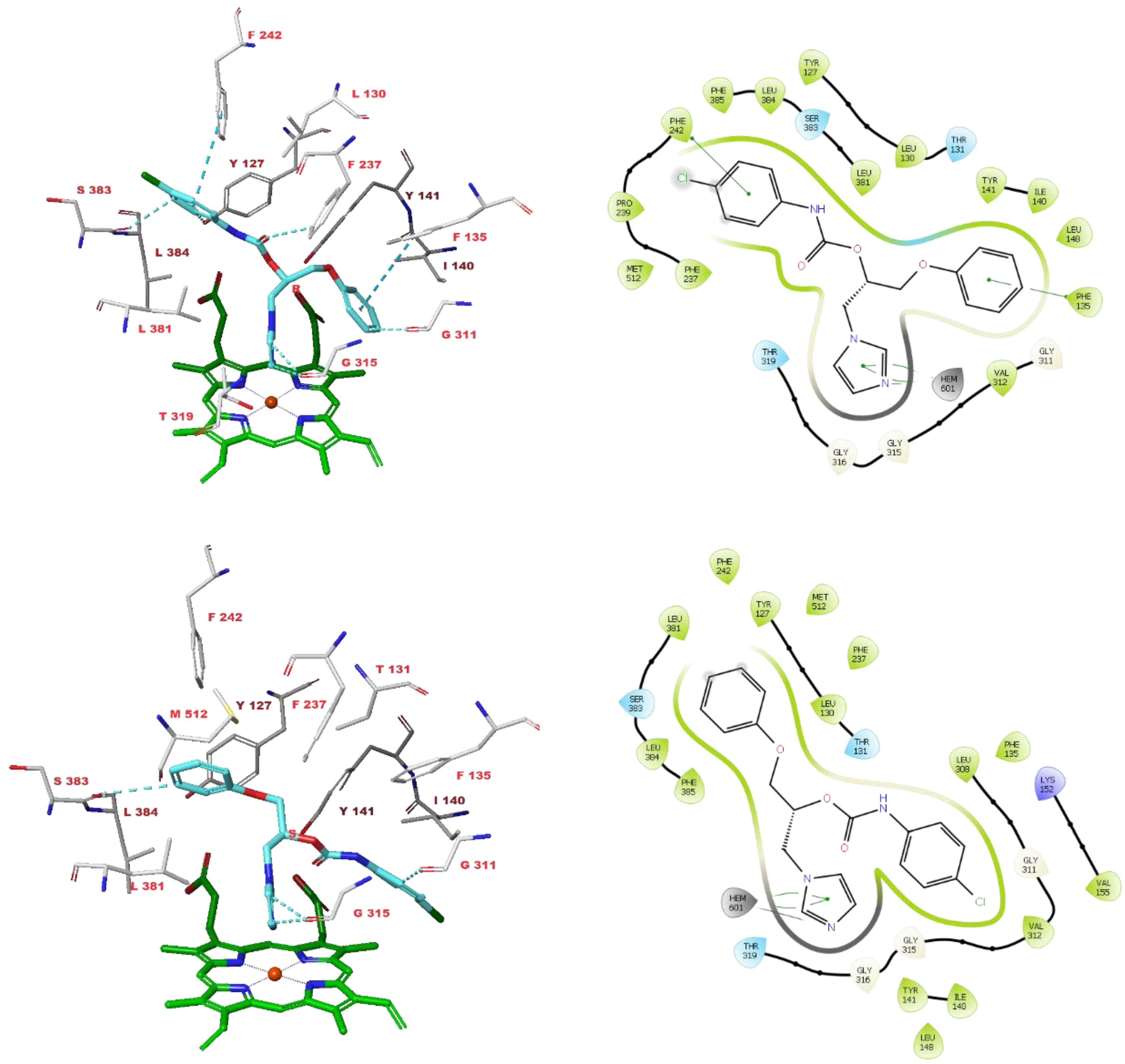
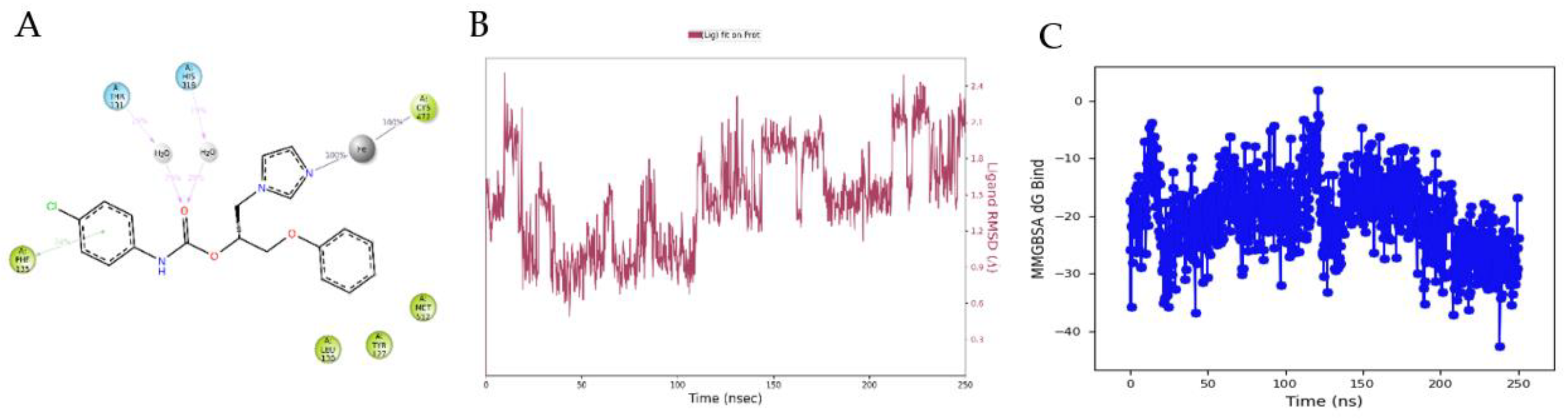
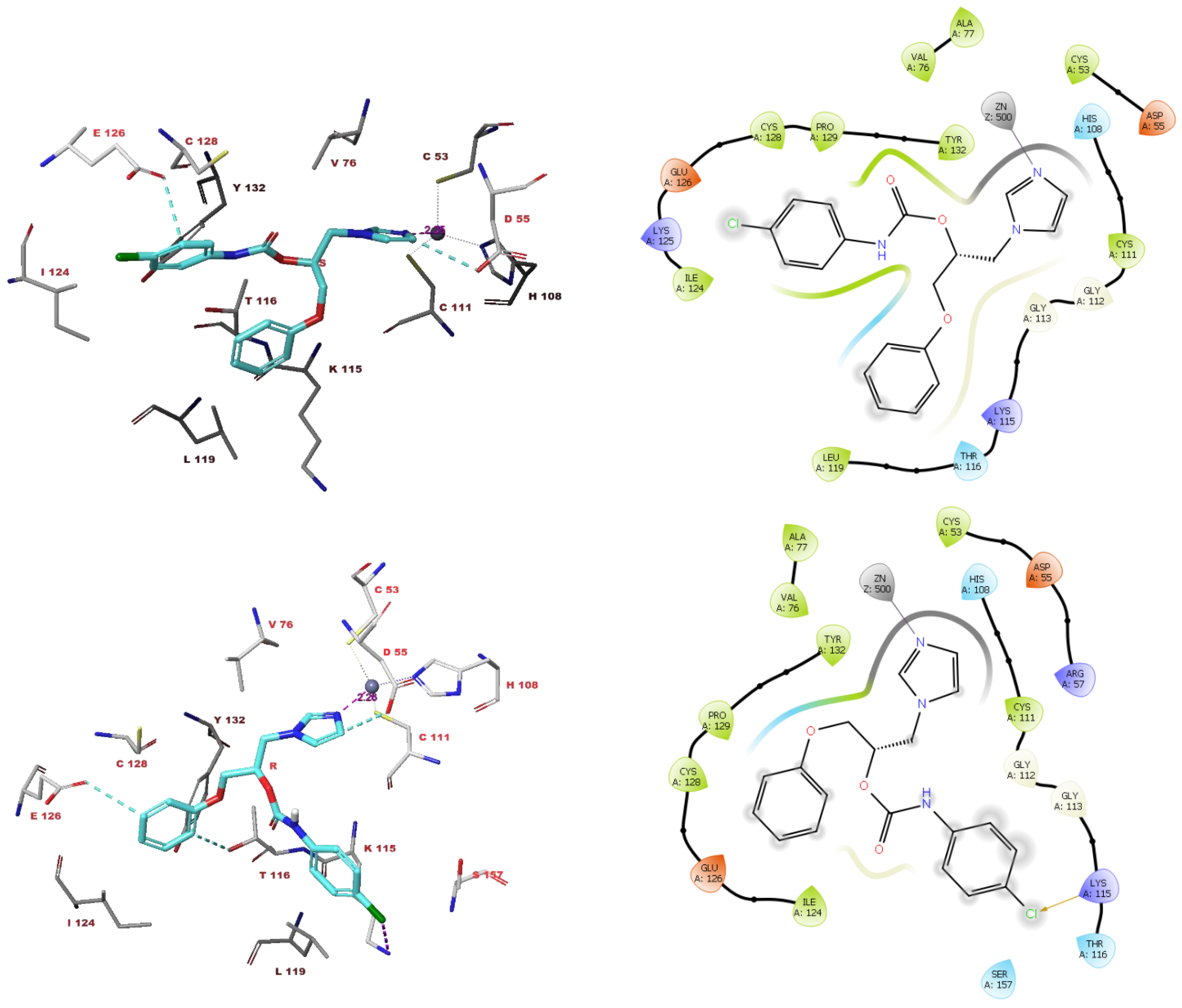
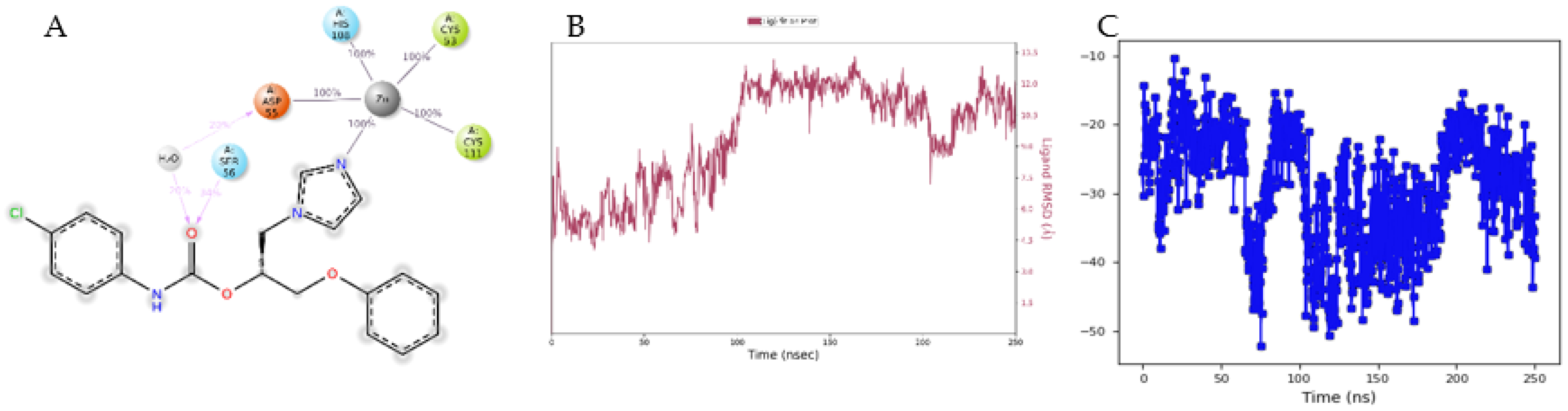
3.4. Molecular Modelling Studies
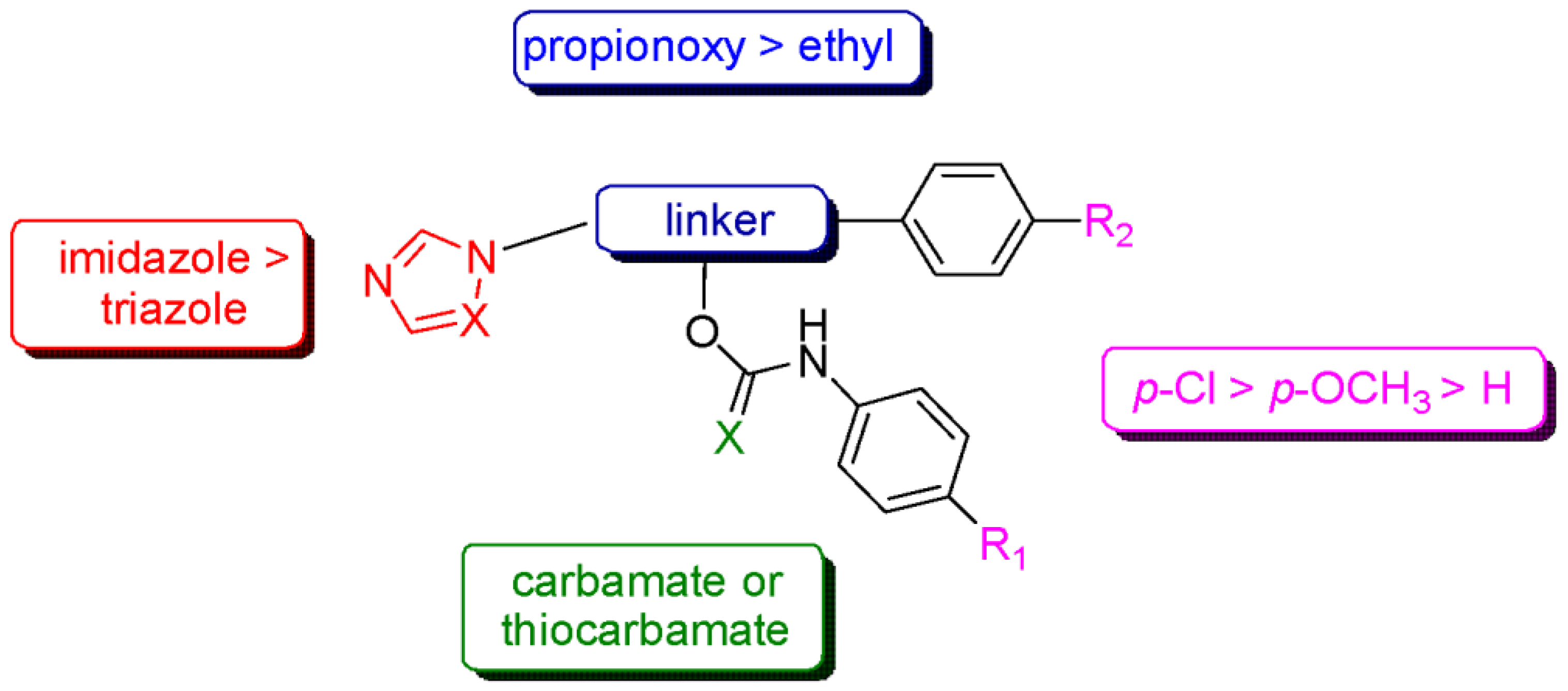
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Almeida, F.; Rodrigues, M.L.; Coelho, C. The Still Underestimated Problem of Fungal Diseases Worldwide. Front. Microbiol. 2019, 10, 214. [Google Scholar] [CrossRef] [Green Version]
- Vallabhaneni, S.; Mody, R.K.; Walker, T.; Chiller, T. The Global Burden of Fungal Diseases. Infect. Dis. Clin. North Am. 2016, 30, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and Multi-National Prevalence of Fungal Diseases—Estimate Precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef]
- Hay, R. Therapy of Skin, Hair and Nail Fungal Infections. J. Fungi 2018, 4, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Revie, N.M.; Iyer, K.R.; Robbins, N.; Cowen, L.E. Antifungal drug resistance: Evolution, mechanisms and impact. Curr. Opin. Microbiol. 2018, 45, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Prasad, R.; Banerjee, A.; Shah, A.H. Resistance to antifungal therapies. Essays Biochem. 2017, 61, 157–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perlin, D.S.; Rautemaa-Richardson, R.; Alastruey-Izquierdo, A. The global problem of antifungal resistance: Prevalence, mechanisms, and management. Lancet Infect. Dis. 2017, 17, e383–e392. [Google Scholar] [CrossRef]
- Monk, B.C.; Sagatova, A.A.; Hosseini, P.; Ruma, Y.N.; Wilson, R.K.; Keniya, M.V. Fungal Lanosterol 14α-demethylase: A target for next-generation antifungal design. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2020, 1868, 140206. [Google Scholar] [CrossRef]
- Allen, D.; Wilson, D.; Drew, R.; Perfect, J. Azole antifungals: 35 years of invasive fungal infection management. Expert Rev. Anti-infective Ther. 2015, 13, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Peyton, L.R.; Gallagher, S.; Hashemzadeh, M. Triazole antifungals: A review. Drugs Today 2015, 51, 705–718. [Google Scholar] [CrossRef]
- Ramage, G.; Rajendran, R.; Sherry, L.; Williams, C. Fungal Biofilm Resistance. Int. J. Microbiol. 2012, 2012, 528521. [Google Scholar] [CrossRef] [PubMed]
- Whaley, S.G.; Berkow, E.L.; Rybak, J.M.; Nishimoto, A.T.; Barker, K.S.; Rogers, P.D. Azole Antifungal Resistance in Candida albicans and Emerging Non-albicans Candida Species. Front. Microbiol. 2017, 7, 2173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gulati, M.; Nobile, C.J. Candida albicans biofilms: Development, regulation, and molecular mechanisms. Microbes Infect. 2016, 18, 310–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polke, M.; Hube, B.; Jacobsen, I.D. Candida Survival Strategies. Adv. Appl. Microbiol. 2015, 91, 139–235. [Google Scholar] [CrossRef]
- Marichal, P.; Bossche, H.V. Mechanisms of resistance to azole antifungals. Acta Biochim. Pol. 1995, 42, 509–516. [Google Scholar] [CrossRef] [Green Version]
- Vanden Bossche, H.; Dromer, F.; Improvisi, I.; Lozano-Chiu, M.; Rex, J.H.; Sanglard, D. Antifungal drug resistance in pathogenic fungi. Med. Mycol. 1998, 36 (Suppl. S1), 119–128. [Google Scholar] [PubMed]
- Pristov, K.E.; Ghannoum, M.A. Resistance of Candida to azoles and echinocandins worldwide. Clin. Microbiol. Infect. 2019, 25, 792–798. [Google Scholar] [CrossRef] [PubMed]
- Nishimoto, A.T.; Sharma, C.; Rogers, P.D. Molecular and genetic basis of azole antifungal resistance in the opportunistic pathogenic fungus Candida albicans. J. Antimicrob. Chemother. 2020, 75, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Ammazzalorso, A.; Gallorini, M.; Fantacuzzi, M.; Gambacorta, N.; De Filippis, B.; Giampietro, L.; Maccallini, C.; Nicolotti, O.; Cataldi, A.; Amoroso, R. Design, synthesis and biological evaluation of imidazole and triazole-based carbamates as novel aromatase inhibitors. Eur. J. Med. Chem. 2021, 211, 113115. [Google Scholar] [CrossRef] [PubMed]
- Maccallini, C.; Gallorini, M.; Sisto, F.; Akdemir, A.; Ammazzalorso, A.; De Filippis, B.; Fantacuzzi, M.; Giampietro, L.; Carradori, S.; Cataldi, A.; et al. New azolyl-derivatives as multitargeting agents against breast cancer and fungal infections: Synthesis, biological evaluation and docking study. J. Enzym. Inhib. Med. Chem. 2021, 36, 1631–1644. [Google Scholar] [CrossRef] [PubMed]
- Brennan, M.; Thomas, D.Y.; Whiteway, M.; Kavanagh, K. Correlation between virulence of Candida albicans mutants in mice and Galleria mellonellalarvae. FEMS Immunol. Med. Microbiol. 2002, 34, 153–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moghimi, S.; Shafiei, M.; Foroumadi, A. Drug design strategies for the treatment azole-resistant candidiasis. Expert Opin. Drug Discov. 2022, 17, 879–895. [Google Scholar] [CrossRef]
- Supuran, C.; Capasso, C. A Highlight on the Inhibition of Fungal Carbonic Anhydrases as Drug Targets for the Antifungal Armamentarium. Int. J. Mol. Sci. 2021, 22, 4324. [Google Scholar] [CrossRef] [PubMed]
- Güzel-Akdemir, Ö.; Carradori, S.; Grande, R.; Demir-Yazıcı, K.; Angeli, A.; Supuran, C.T.; Akdemir, A. Development of Thiazolidinones as Fungal Carbonic Anhydrase Inhibitors. Int. J. Mol. Sci. 2020, 21, 2960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, B.-W.; Shi, Y.; Li, W.-C.; Wang, Q.; Pan, M.; Wu, Q.; Fu, H.-Z. Synthesis and biological evaluation of Vinpocetine derivatives. Bioorganic Med. Chem. Lett. 2020, 30, 126472. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts, 3rd ed.; Approved Standard. CLSI M27-A3(28); Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008. [Google Scholar]
- Clinical and Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi, 2nd ed.; Approved Standard. CLSI Document M38-A2; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts—Fourth Informational Supplement, Document M27-A3; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Ourabah, A.; Atmani-Kilani, D.; Debbache-Benaida, N.; Kolesova, O.; Azib, L.; Yous, F.; Benloukil, M.; Botta, B.; Atmani, D.; Simonetti, G. Anti-Candida albicans biofilm activity of extracts from two selected indigenous Algerian plants: Clematis flammula and Fraxinus angustifolia. J. Herb. Med. 2020, 20, 100319. [Google Scholar] [CrossRef]
- Simonetti, G.; Palocci, C.; Valletta, A.; Kolesova, O.; Chronopoulou, L.; Donati, L.; Di Nitto, A.; Brasili, E.; Tomai, P.; Gentili, A.; et al. Anti-Candida Biofilm Activity of Pterostilbene or Crude Extract from Non-Fermented Grape Pomace Entrapped in Biopolymeric Nanoparticles. Molecules 2019, 24, 2070. [Google Scholar] [CrossRef] [Green Version]
- Cairone, F.; Simonetti, G.; Orekhova, A.; Casadei, M.; Zengin, G.; Cesa, S. Health Potential of Clery Strawberries: Enzymatic Inhibition and Anti-Candida Activity Evaluation. Molecules 2021, 26, 1731. [Google Scholar] [CrossRef]
- Ohshima, T.; Ikawa, S.; Kitano, K.; Maeda, N. A Proposal of Remedies for Oral Diseases Caused by Candida: A Mini Review. Front. Microbiol. 2018, 9, 1522. [Google Scholar] [CrossRef] [Green Version]
- Wall, G.; Montelongo-Jauregui, D.; Bonifacio, B.V.; Lopez-Ribot, J.L.; Uppuluri, P. Candida albicans biofilm growth and dispersal: Contributions to pathogenesis. Curr. Opin. Microbiol. 2019, 52, 1–6. [Google Scholar] [CrossRef]
- De Vita, D.; Friggeri, L.; D’Auria, F.D.; Pandolfi, F.; Piccoli, F.; Panella, S.; Palamara, A.T.; Simonetti, G.; Scipione, L.; Di Santo, R.; et al. Activity of caffeic acid derivatives against Candida albicans biofilm. Bioorganic Med. Chem. Lett. 2014, 24, 1502–1505. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Gupta, H.; Poluri, K.M. Geraniol eradicates Candida glabrata biofilm by targeting multiple cellular pathways. Appl. Microbiol. Biotechnol. 2021, 105, 5589–5605. [Google Scholar] [CrossRef] [PubMed]
- Jamiu, A.T.; Albertyn, J.; Sebolai, O.; Gcilitshana, O.; Pohl, C.H. Inhibitory effect of polyunsaturated fatty acids alone or in combination with fluconazole on Candida krusei biofilms in vitro and in Caenorhabditis elegans. Med. Mycol. 2021, 59, 1225–1237. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.N.; de Mello, T.P.; de Souza, R.L.; Branquinha, M.H.; dos Santos, A.L.S. New and Promising Chemotherapeutics for Emerging Infections Involving Drug-resistant Non-albicans Candida Species. Curr. Top. Med. Chem. 2019, 19, 2527–2553. [Google Scholar] [CrossRef] [PubMed]
- Černáková, L.; Light, C.; Salehi, B.; Rogel-Castillo, C.; Victoriano, M.; Martorell, M.; Sharifi-Rad, J.; Martins, N.; Rodrigues, C.F. Novel Therapies for Biofilm-Based Candida spp. Infections. Adv. Exp. Med. Biol. 2019, 1214, 93–123. [Google Scholar] [CrossRef]
- Smith, D.F.Q.; Casadevall, A. Fungal immunity and pathogenesis in mammals versus the invertebrate model organism Galleria mellonella. Pathog. Dis. 2021, 79, ftab013. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, I.D. Galleria mellonellaas a model host to study virulence of Candida. Virulence 2014, 5, 237–239. [Google Scholar] [CrossRef] [Green Version]
- Kaskatepe, B.; Erdem, S.A.; Ozturk, S.; Oz, Z.S.; Subasi, E.; Koyuncu, M.; Vlainić, J.; Kosalec, I. Antifungal and Anti-Virulent Activity of Origanum majorana L. Essential Oil on Candida albicans and In Vivo Toxicity in the Galleria mellonella Larval Model. Molecules 2022, 27, 663. [Google Scholar] [CrossRef]
- Piatek, M.; Sheehan, G.; Kavanagh, K. Galleria mellonella: The Versatile Host for Drug Discovery, In Vivo Toxicity Testing and Characterising Host-Pathogen Interactions. Antibiotics 2021, 10, 1545. [Google Scholar] [CrossRef]
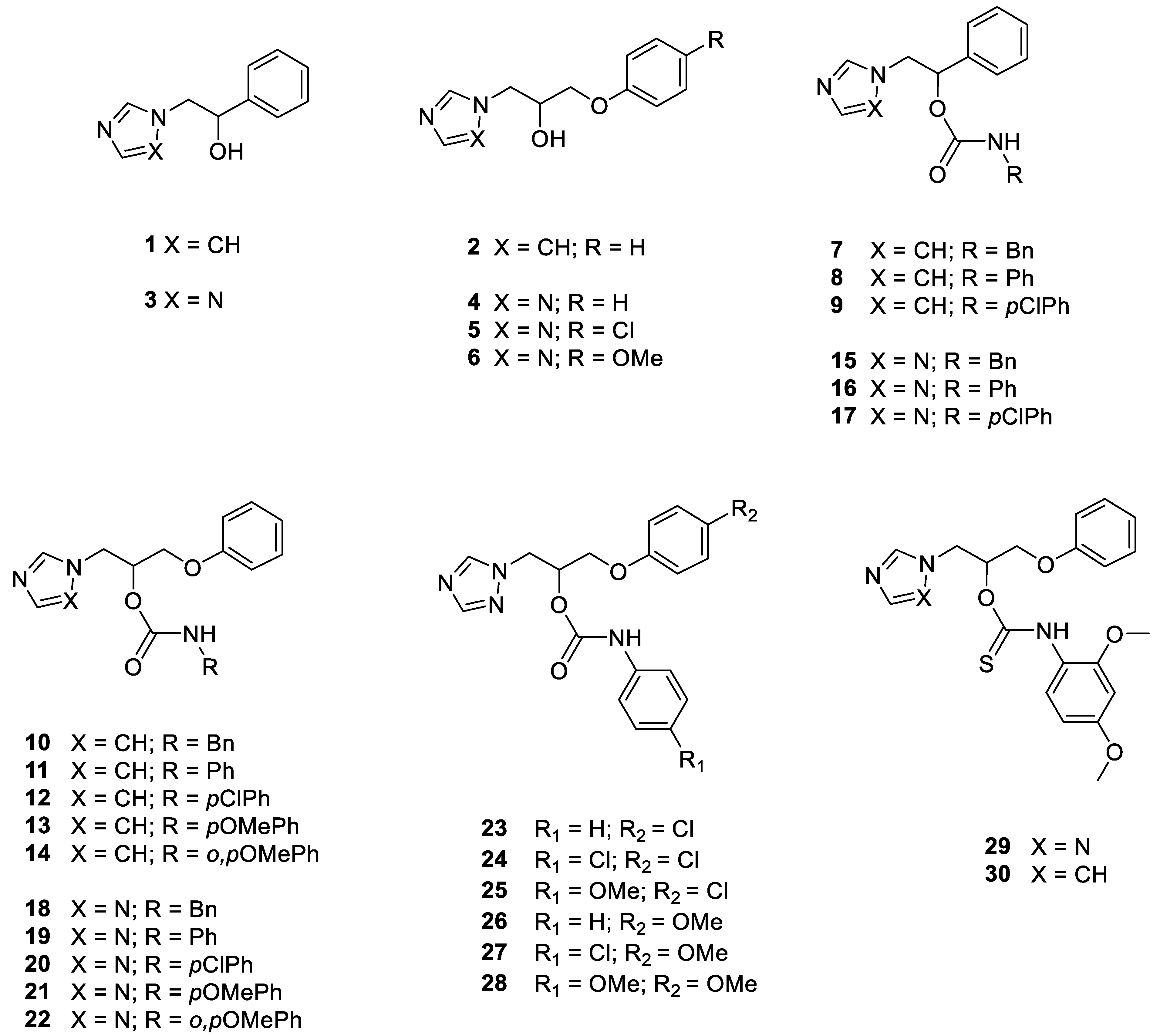
| Cpd | Structure | Candida albicans | Candida glabrata | Candida krusei | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MIC50 μg/mL Median Value | MIC50 μg/mL Median Value | MIC50 μg/mL Median Value | ||||||||
| ATCC 10231 | 3153A | ATCCC 24433 | PMC 0849 | PMC 0822 | PMC 0805 | PMC 0603 | PMC 0624 | PMC 0613 | ||
| 1 | 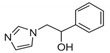 | >128 | >128 | >128 | 48 | >128 | >128 | >128 | >128 | >128 |
| 2 | 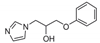 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 128 |
| 3 | 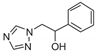 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 |
| 4 |  | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 |
| 5 | 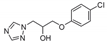 | >128 | >128 | >128 | 64 | >128 | >128 | >128 | >128 | >128 |
| 6 | 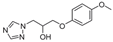 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 128 |
| 7 | 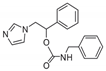 | >128 | 128 | >128 | 12 | 128 | >128 | 64 | 128 | 32 |
| 8 | 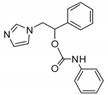 | >128 | >128 | >128 | 20 | 128 | 128 | 32 | >128 | >128 |
| 9 |  | >128 | 128 | >128 | 6 | 128 | 128 | 128 | >128 | 64 |
| 10 | 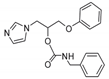 | 128 | >128 | >128 | 8 | 128 | 128 | 96 | 96 | 16 |
| 11 | 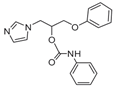 | >128 | >128 | >128 | 3 | >128 | >128 | 32 | 96 | >128 |
| 12 | 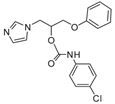 | 32 | 64 | 32 | 1 | 8 | 16 | 4 | 24 | 12 |
| 13 |  | 128 | 128 | 128 | 4 | 64 | 64 | 16 | 64 | >128 |
| 14 | 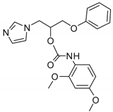 | 128 | >128 | >128 | 8 | 128 | 128 | 128 | >128 | Nd |
| 15 | 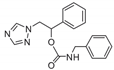 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 |
| 16 |  | >128 | >128 | >128 | 16 | >128 | >128 | >128 | >128 | >128 |
| 17 | 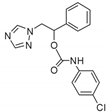 | 64 | 80 | >128 | 12 | 96 | >128 | 128 | >128 | >128 |
| 18 | 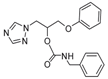 | >128 | >128 | >128 | >128 | >128 | 128 | >128 | >128 | >128 |
| 19 | 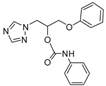 | >128 | >128 | >128 | 40 | >128 | >128 | >128 | >128 | >128 |
| 20 | 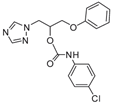 | >128 | >128 | >128 | 8 | 128 | 128 | >128 | >128 | >128 |
| 21 | 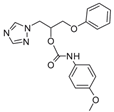 | >128 | >128 | >128 | 16 | >128 | >128 | >128 | >128 | >128 |
| 22 | 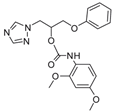 | >128 | >128 | >128 | >128 | >128 | 128 | >128 | >128 | >128 |
| 23 | 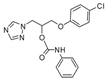 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 128 | >128 |
| 24 | 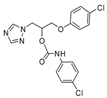 | >128 | >128 | >128 | 4 | 32 | 64 | 48 | 128 | 16 |
| 25 | 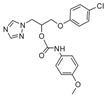 | >128 | >128 | >128 | 4 | >128 | >128 | >128 | >128 | 96 |
| 26 | 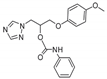 | >128 | >128 | >128 | 24 | 128 | >128 | >128 | >128 | >128 |
| 27 | 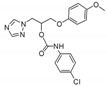 | >128 | >128 | >128 | 8 | 64 | 128 | >128 | >128 | >128 |
| 28 | 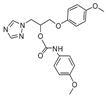 | >128 | >128 | >128 | 32 | >128 | >128 | >128 | >128 | >128 |
| 29 |  | >128 | >128 | >128 | >128 | 64 | 64 | >128 | >128 | 64 |
| 30 | 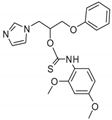 | >128 | >128 | >128 | 4 | 128 | 128 | 32 | >128 | 48 |
| FLC | 2 | 4 | 2 | 0.5 | 0.5 | 1 | 2 | 4 | 2 | |
| Compound | LD50 (mg kg−1) * | Solvent |
|---|---|---|
| 12 | >36.5 mg/kg | 100 H2O: 1 DMSO |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carradori, S.; Ammazzalorso, A.; De Filippis, B.; Şahin, A.F.; Akdemir, A.; Orekhova, A.; Bonincontro, G.; Simonetti, G. Azole-Based Compounds That Are Active against Candida Biofilm: In Vitro, In Vivo and In Silico Studies. Antibiotics 2022, 11, 1375. https://doi.org/10.3390/antibiotics11101375
Carradori S, Ammazzalorso A, De Filippis B, Şahin AF, Akdemir A, Orekhova A, Bonincontro G, Simonetti G. Azole-Based Compounds That Are Active against Candida Biofilm: In Vitro, In Vivo and In Silico Studies. Antibiotics. 2022; 11(10):1375. https://doi.org/10.3390/antibiotics11101375
Chicago/Turabian StyleCarradori, Simone, Alessandra Ammazzalorso, Barbara De Filippis, Ahmet Fatih Şahin, Atilla Akdemir, Anastasia Orekhova, Graziana Bonincontro, and Giovanna Simonetti. 2022. "Azole-Based Compounds That Are Active against Candida Biofilm: In Vitro, In Vivo and In Silico Studies" Antibiotics 11, no. 10: 1375. https://doi.org/10.3390/antibiotics11101375
APA StyleCarradori, S., Ammazzalorso, A., De Filippis, B., Şahin, A. F., Akdemir, A., Orekhova, A., Bonincontro, G., & Simonetti, G. (2022). Azole-Based Compounds That Are Active against Candida Biofilm: In Vitro, In Vivo and In Silico Studies. Antibiotics, 11(10), 1375. https://doi.org/10.3390/antibiotics11101375










