Genome Mining Discovery of a New Benzazepine Alkaloid Pseudofisnin A from the Marine Fungus Neosartorya pseudofischeri F27-1
Abstract
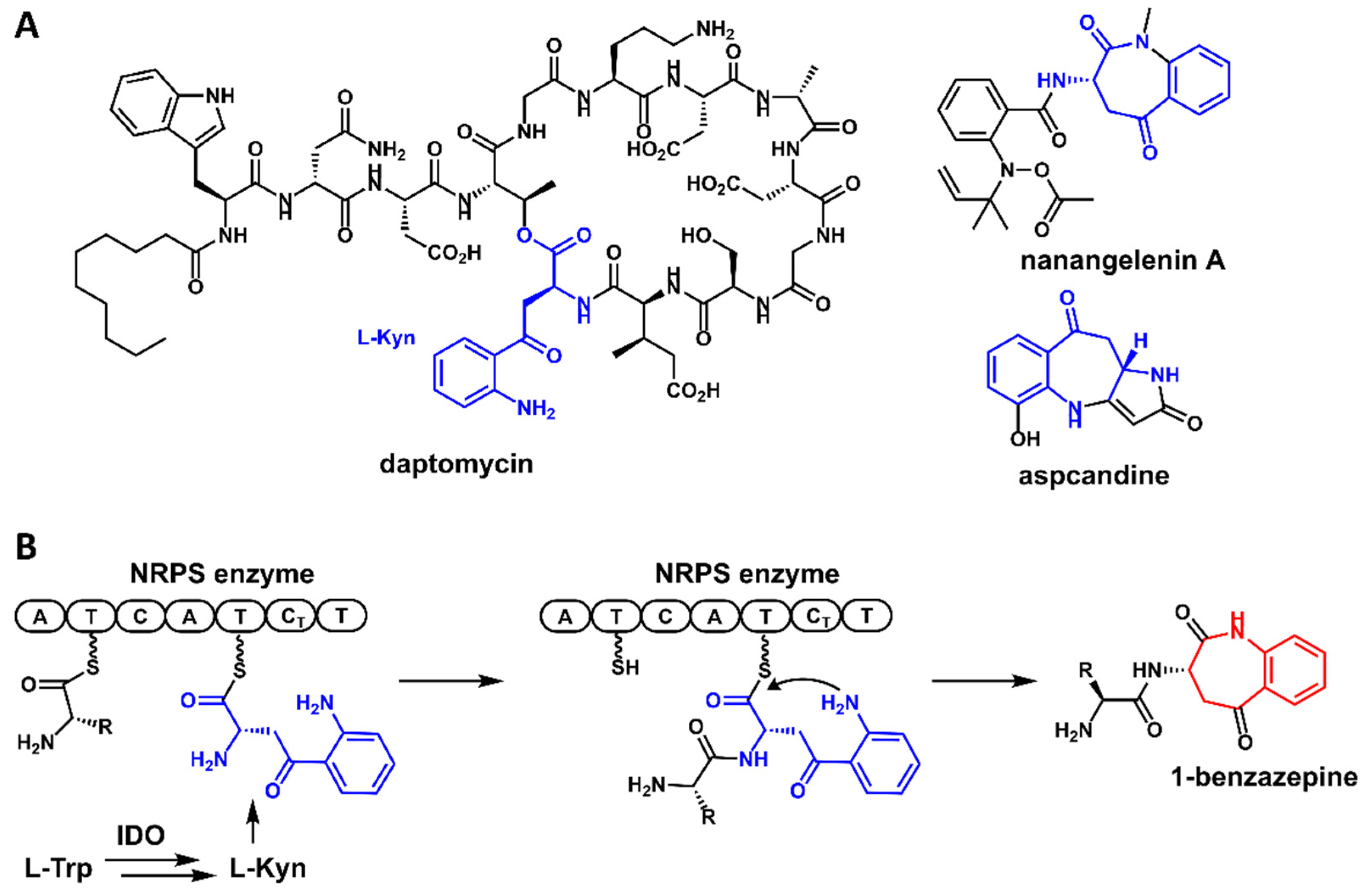
:1. Introduction
2. Results and Discussion
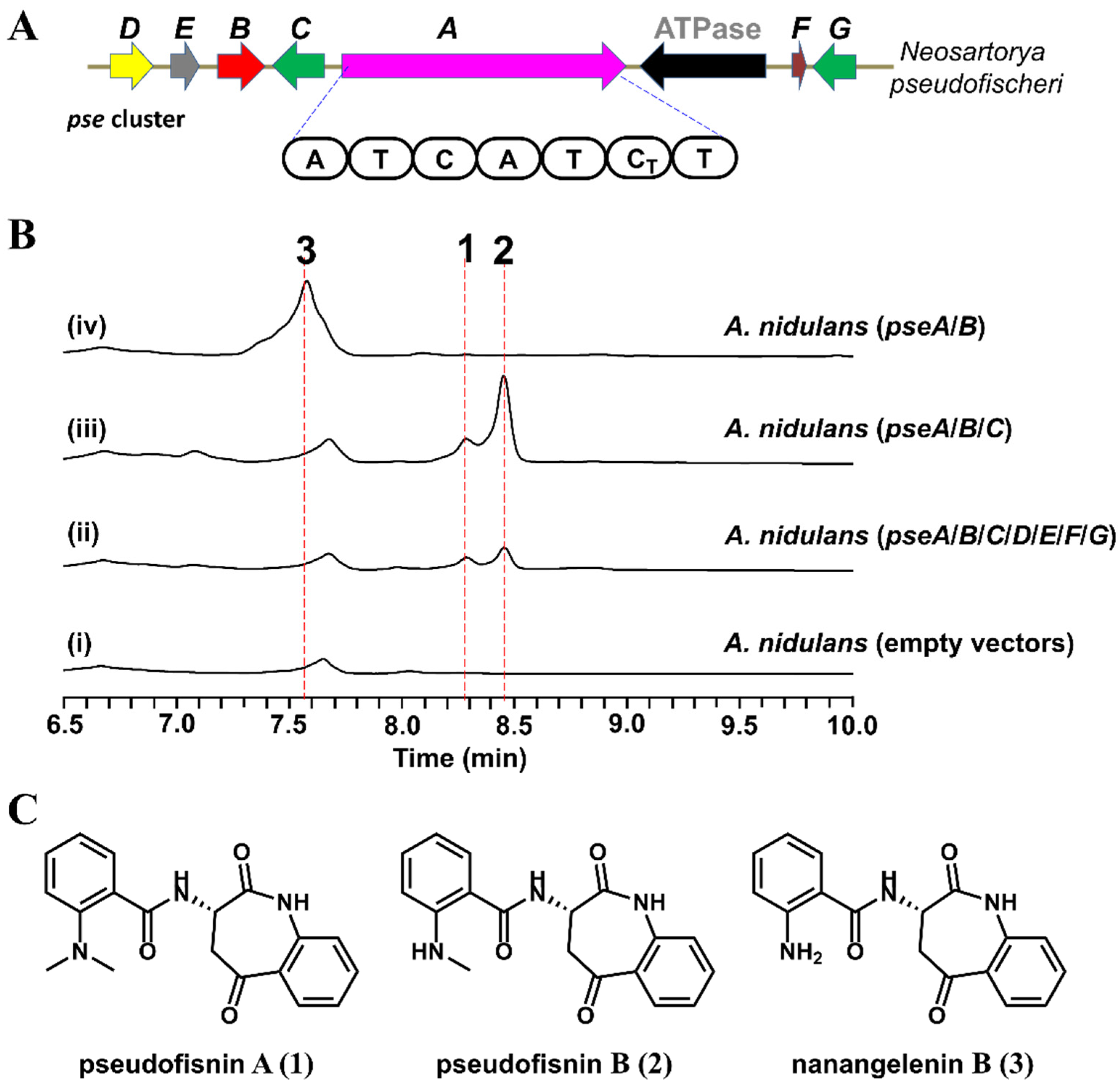
2.1. Identification of the Pse Cluster through Core Biosynthetic Enzyme-Guided Genome Mining
2.2. Heterologous Expression of the Pse Cluster and Characterization of the Products
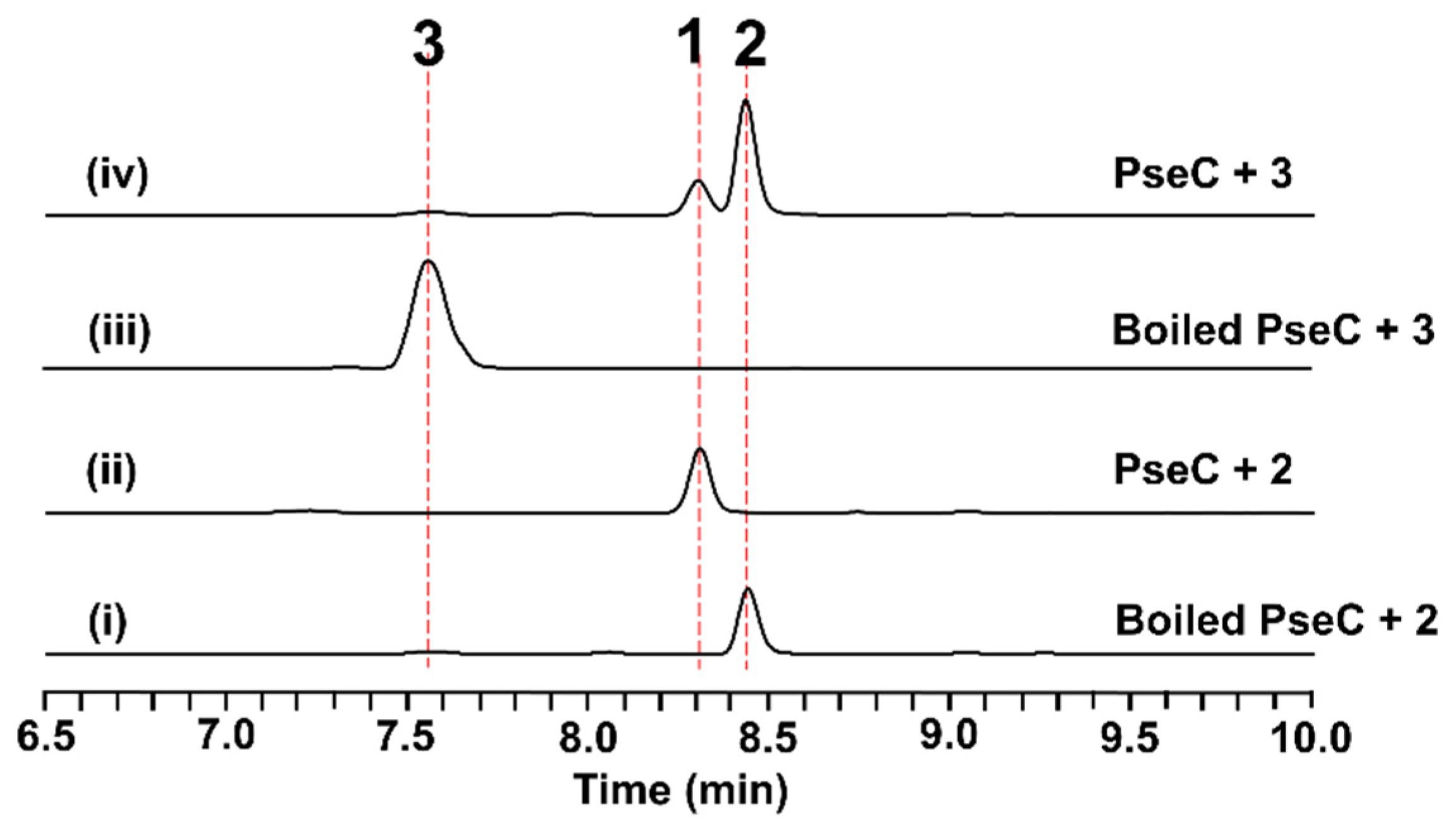
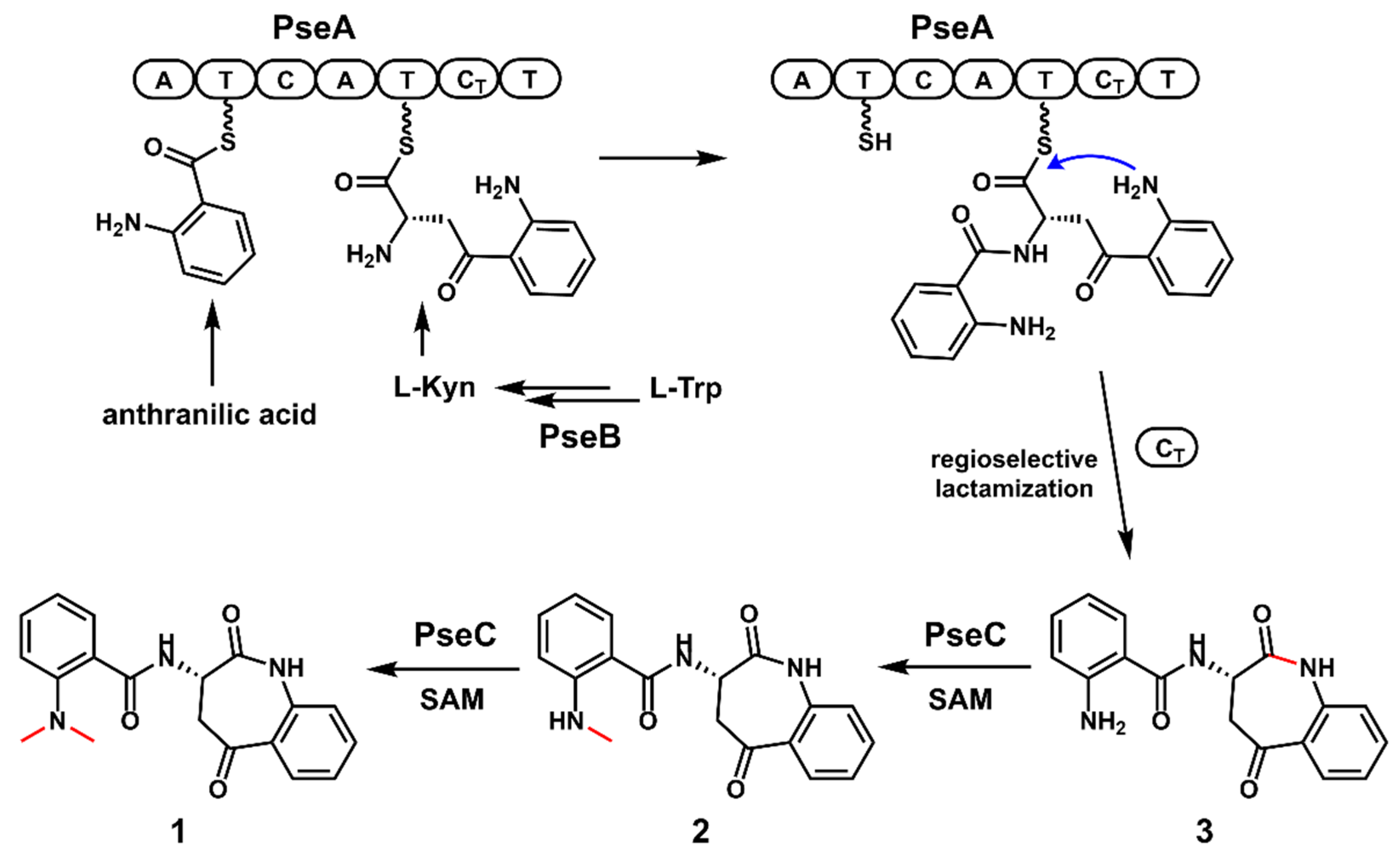
2.3. Functional Identification of the Genes Involved in the Biosynthesis of Pseudofisnins
2.4. In Vitro Characterization of PseC as an Iterative Methyltransferase
2.5. The Proposed Biosynthetic Pathway of 1
3. Conclusions
4. Materials and Methods
4.1. Strains and Culture Conditions
4.2. General DNA Manipulation Techniques
4.3. Plasmids Construction
4.4. Protoplast Preparation and Transformation of A. nidulans
4.5. Chemical Analysis and Compound Isolation and Characterization
4.6. Protein Expression and Purification of PseC
4.7. Enzymatic Assays of PseC In Vitro
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cervenka, I.; Agudelo, L.Z.; Ruas, J.L. Kynurenines: Tryptophan’s metabolites in exercise, inflammation, and mental health. Science 2017, 357, eaaf9794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heyes, M.P.; Achim, C.L.; Wiley, C.A.; Major, E.O.; Saito, K.; Markey, S.P. Human microglia convert l-tryptophan into the neurotoxin quinolinic acid. Biochem. J. 1996, 320, 595–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batabyal, D.; Yeh, S.R. Human tryptophan dioxygenase: A comparison to indoleamine 2,3-dioxygenase. J. Am. Chem. Soc. 2007, 129, 15690–15701. [Google Scholar] [CrossRef] [PubMed]
- Stone, T.W.; Darlington, L.G. Endogenous kynurenines as targets for drug discovery and development. Nat. Rev. Drug Disc. 2002, 1, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Malpass, K. Neurodegenerative disease: The kynurenine pathway--promising new targets and therapies for neurodegenerative disease. Nat. Rev. Neurol. 2011, 7, 417. [Google Scholar] [CrossRef] [PubMed]
- Karas, J.A.; Carter, G.P.; Howden, B.P.; Turner, A.M.; Paulin, O.K.A.; Swarbrick, J.D.; Baker, M.A.; Li, J.; Velkov, T. Structure-Activity Relationships of Daptomycin Lipopeptides. J. Med. Chem. 2020, 63, 13266–13290. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Gilchrist, C.L.M.; Phan, C.S.; Lacey, H.J.; Vuong, D.; Moggach, S.A.; Lacey, E.; Piggott, A.M.; Chooi, Y.H. Biosynthesis of a New Benzazepine Alkaloid Nanangelenin A from Aspergillus nanangensis Involves an Unusual l-Kynurenine-Incorporating NRPS Catalyzing Regioselective Lactamization. J. Am. Chem. Soc. 2020, 142, 7145–7152. [Google Scholar] [CrossRef] [PubMed]
- Caesar, L.K.; Robey, M.T.; Swyers, M.; Islam, M.N.; Ye, R.; Vagadia, P.P.; Schiltz, G.E.; Thomas, P.M.; Wu, C.C.; Kelleher, N.L.; et al. Heterologous Expression of the Unusual Terreazepine Biosynthetic Gene Cluster Reveals a Promising Approach for Identifying New Chemical Scaffolds. mBio 2020, 11, e01691-20. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Tang, J.W.; Liu, Y.Y.; Matsuda, Y. Aspcandine: A Pyrrolobenzazepine Alkaloid Synthesized by a Fungal Nonribosomal Peptide Synthetase-Polyketide Synthase Hybrid. Org. Lett. 2022, 24, 4816–4819. [Google Scholar] [CrossRef] [PubMed]
- Jigar, H.S.; Rama, M.H.; Hari, N.P. Pharmacological and biological activities of benzazepines: An overview. Curr. Bioact. Compd. 2015, 11, 170–188. [Google Scholar]
- Webb, R.L.; Miller, D.; Traina, V.; Gomez, H.J. Benazepril. Cardiovasc. Drug Rev. 1990, 8, 89–104. [Google Scholar] [CrossRef]
- Lin, H.-C.; Chiou, G.; Chooi, Y.-H.; McMahon, T.C.; Xu, W.; Garg, N.K.; Tang, Y. Elucidation of the concise biosynthetic pathway of the communesin indole alkaloids. Angew. Chem. Int. Ed. 2015, 54, 3004–3007. [Google Scholar] [CrossRef] [PubMed]
- Rönsch, H. Chapter 1 Rhoeadine alkaloids. In The Alkaloids: Chemistry and Pharmacology; Brossi, A., Ed.; Academic Press: Orlando, FL, USA, 1986; Volume 28, pp. 1–93. [Google Scholar]
- Li, L.; Yu, P.; Tang, M.C.; Zou, Y.; Gao, S.S.; Hung, Y.S.; Zhao, M.; Watanabe, K.; Houk, K.N.; Tang, Y. Biochemical Characterization of a Eukaryotic Decalin-Forming Diels-Alderase. J. Am. Chem. Soc. 2016, 138, 15837–15840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, N.; Hung, Y.S.; Gao, S.S.; Hang, L.; Zou, Y.; Chooi, Y.H.; Tang, Y. Identification and Heterologous Production of a Benzoyl-Primed Tricarboxylic Acid Polyketide Intermediate from the Zaragozic Acid A Biosynthetic Pathway. Org. Lett. 2017, 19, 3560–3563. [Google Scholar] [CrossRef] [PubMed]
- Cubero, O.F.; Crespo, A.; Fatehi, J.; Bridge, P.D. DNA extraction and PCR amplification method suitable for fresh, herbarium-stored, lichenized, and other fungi. Plant Syst. Evol. 1999, 216, 243–249. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, X.-X.; Chen, L.; Tang, M.-C. Genome Mining Discovery of a New Benzazepine Alkaloid Pseudofisnin A from the Marine Fungus Neosartorya pseudofischeri F27-1. Antibiotics 2022, 11, 1444. https://doi.org/10.3390/antibiotics11101444
Xue X-X, Chen L, Tang M-C. Genome Mining Discovery of a New Benzazepine Alkaloid Pseudofisnin A from the Marine Fungus Neosartorya pseudofischeri F27-1. Antibiotics. 2022; 11(10):1444. https://doi.org/10.3390/antibiotics11101444
Chicago/Turabian StyleXue, Xiao-Xin, Lin Chen, and Man-Cheng Tang. 2022. "Genome Mining Discovery of a New Benzazepine Alkaloid Pseudofisnin A from the Marine Fungus Neosartorya pseudofischeri F27-1" Antibiotics 11, no. 10: 1444. https://doi.org/10.3390/antibiotics11101444
APA StyleXue, X. -X., Chen, L., & Tang, M. -C. (2022). Genome Mining Discovery of a New Benzazepine Alkaloid Pseudofisnin A from the Marine Fungus Neosartorya pseudofischeri F27-1. Antibiotics, 11(10), 1444. https://doi.org/10.3390/antibiotics11101444




