Histidine 19 Residue Is Essential for Cell Internalization of Antifungal Peptide SmAPα1-21 Derived from the α-Core of the Silybum marianum Defensin DefSm2-D in Fusarium graminearum
Abstract
:1. Introduction
2. Results
2.1. Antifungal Peptide Design, Synthesis, and Characterization
2.2. Peptide SmAP2H19A Derived from SmAPα1-21 Is Less Active than Parent Peptide
2.3. Peptides Are Inserted onto Lipid Monolayers and Permeabilize Fungal Membrane
2.4. SmAPα1-21 Induces Endogenous ROS and Peroxisome Biogenesis
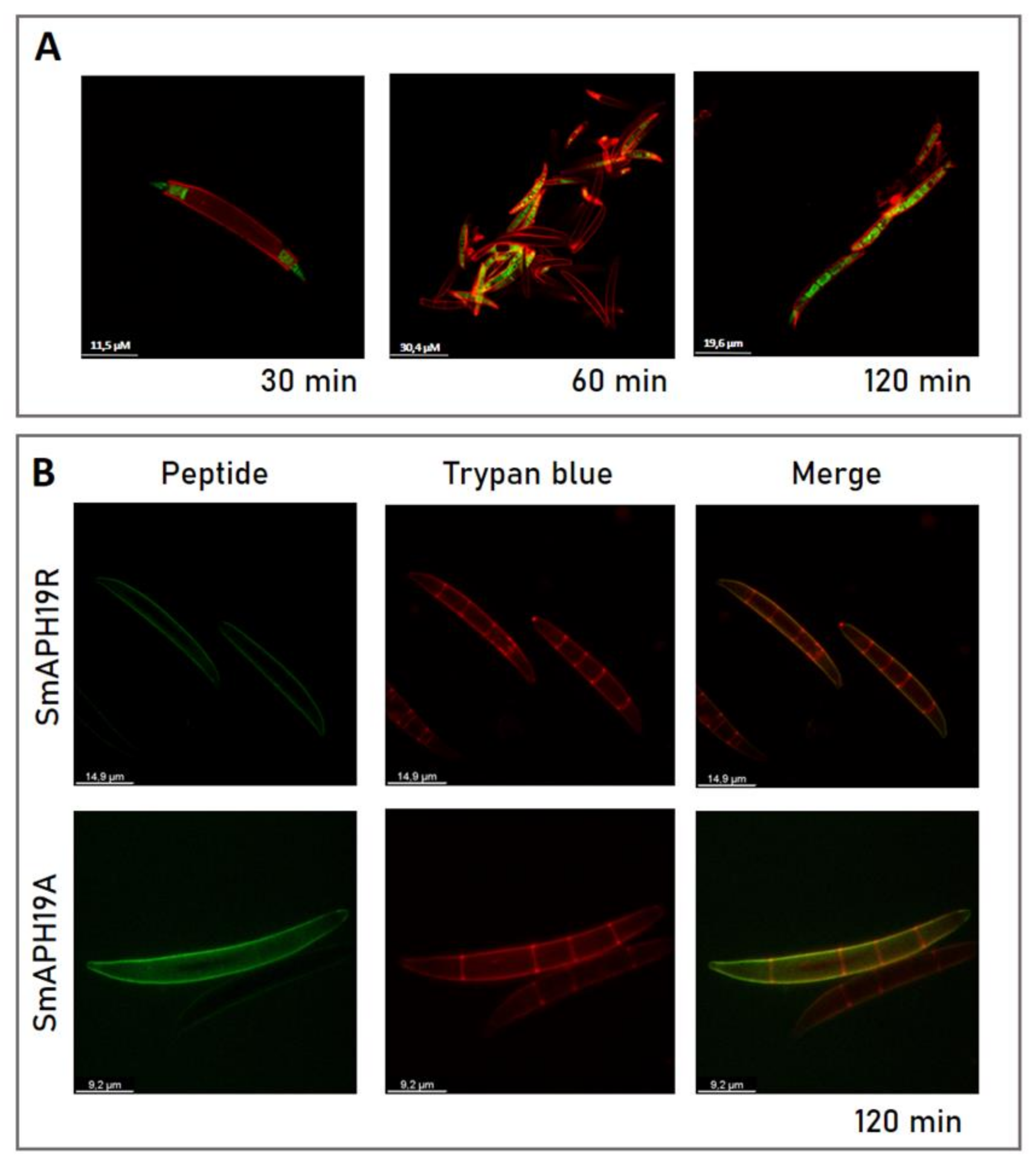
2.5. His19 Is Essential for SmAPα1-21 Internalization
3. Discussion
4. Materials and Methods
4.1. Biological Material
4.2. Peptide Design and Synthesis
4.3. Peptide Purification and Characterization
4.4. In vitro Antifungal Assays
4.5. Surface Pressure Measurement
4.6. Evaluation of Membrane Integrity
4.7. Reactive Oxygen Species (ROS) Detection
4.8. TEM Imaging
4.9. Peptide Derivatization and Subcellular Localization of Derivatized Peptides
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brauer, V.S.; Rezende, C.P.; Pessoni, A.M.; De Paula, R.G.; Rangappa, K.S.; Nayaka, S.C.; Gupta, V.K.; Almeida, F. Antifungal Agents in Agriculture: Friends and Foes of Public Health. Biomolecules 2019, 9, 521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jampilek, J. Potential of Agricultural Fungicides for Antifungal Drug Discovery. Expert Opin. Drug Discov. 2016, 11, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perfect, J.R. Is There an Emerging Need for New Antifungals? Expert Opin. Emerg. Drugs 2016, 21, 129–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wegulo, S.N.; Baenziger, P.S.; Hernandez Nopsa, J.; Bockus, W.W.; Hallen-Adams, H. Management of Fusarium Head Blight of Wheat and Barley. Crop Prot. 2015, 73, 100–107. [Google Scholar] [CrossRef]
- Omotayo, O.P.; Omotayo, A.O.; Mwanza, M.; Babalola, O.O. Prevalence of Mycotoxins and Their Consequences on Human Health. Toxicol. Res. 2019, 35, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Struyfs, C.; Cammue, B.P.A.; Thevissen, K. Membrane-Interacting Antifungal Peptides. Front. Cell Dev. Biol. 2021, 9, 649875. [Google Scholar] [CrossRef]
- Buda De Cesare, G.; Cristy, S.A.; Garsin, D.A.; Lorenz, M.C. Antimicrobial Peptides: A New Frontier in Antifungal Therapy. mBio 2020, 11, e02123-20. [Google Scholar] [CrossRef]
- Teixeira, V.; Feio, M.J.; Bastos, M. Role of Lipids in the Interaction of Antimicrobial Peptides with Membranes. Prog. Lipid Res. 2012, 51, 149–177. [Google Scholar] [CrossRef]
- Rogozhin, E.A.; Slezina, M.P.; Slavokhotova, A.A.; Istomina, E.A.; Korostyleva, T.V.; Smirnov, A.N.; Grishin, E.V.; Egorov, T.A.; Odintsova, T.I. A Novel Antifungal Peptide from Leaves of the Weed Stellaria media L. Biochimie 2015, 116, 125–132. [Google Scholar] [CrossRef]
- Slezina, M.P.; Istomina, E.A.; Kulakovskaya, E.V.; Abashina, T.N.; Odintsova, T.I. Synthetic Oligopeptides Mimicking γ-Core Regions of Cysteine-Rich Peptides of Solanum Lycopersicum Possess Antimicrobial Activity against Human and Plant Pathogens. Issues Mol. Biol. 2021, 43, 1226–1242. [Google Scholar] [CrossRef]
- Silverstein, K.A.T.; Moskal, W.A.; Wu, H.C.; Underwood, B.A.; Graham, M.A.; Town, C.D.; VandenBosch, K.A. Small Cysteine-Rich Peptides Resembling Antimicrobial Peptides Have Been under-Predicted in Plants. Plant J. 2007, 51, 262–280. [Google Scholar] [CrossRef]
- Toledo, E.B.; Lucas, D.R.; Simão, T.L.B.V.; Calixto, S.D.; Lassounskaia, E.; Muzitano, M.F.; Damica, F.Z.; Gomes, V.M.; de Oliveira Carvalho, A. Design of Improved Synthetic Antifungal Peptides with Targeted Variations in Charge, Hydrophobicity and Chirality Based on a Correlation Study between Biological Activity and Primary Structure of Plant Defensin γ-Cores. Amino Acids 2021, 53, 219–237. [Google Scholar] [CrossRef]
- Poon, I.K.H.; Baxter, A.A.; Lay, F.T.; Mills, G.D.; Adda, C.G.; Payne, J.A.E.; Phan, T.K.; Ryan, G.F.; White, J.A.; Veneer, P.K.; et al. Phosphoinositide-Mediated Oligomerization of a Defensin Induces Cell Lysis. eLife 2014, 3, e01808. [Google Scholar] [CrossRef]
- Kvansakul, M.; Lay, F.T.; Adda, C.G.; Veneer, P.K.; Baxter, A.A.; Phan, T.K.; Poon, I.K.H.; Hulett, M.D. Binding of Phosphatidic Acid by NsD7 Mediates the Formation of Helical Defensin-Lipid Oligomeric Assemblies and Membrane Permeabilization. Proc. Natl. Acad. Sci. USA 2016, 113, 11202–11207. [Google Scholar] [CrossRef] [Green Version]
- De Medeiros, L.N.; Angeli, R.; Sarzedas, C.G.; Barreto-Bergter, E.; Valente, A.P.; Kurtenbach, E.; Almeida, F.C.L. Backbone Dynamics of the Antifungal Psd1 Pea Defensin and Its Correlation with Membrane Interaction by NMR Spectroscopy. Biochim. Biophys. Acta Biomembr. 2010, 1798, 105–113. [Google Scholar] [CrossRef] [Green Version]
- De Medeiros, L.N.; Domitrovic, T.; de Andrade, P.C.; Faria, J.; Bergter, E.B.; Weissmuller, G.; Kurtenbach, E. Psd1 Binding Affinity toward Fungal Membrane Components as Assessed by SPR: The Role of Glucosylceramide in Fungal Recognition and Entry. Biopolym.-Pept. Sci. Sect. 2014, 102, 456–464. [Google Scholar] [CrossRef]
- Yount, N.Y.; Yeaman, M.R. Multidimensional Signatures in Antimicrobial Peptides. Proc. Natl. Acad. Sci. USA 2004, 101, 7363–7368. [Google Scholar] [CrossRef] [Green Version]
- Sagaram, U.S.; Pandurangi, R.; Kaur, J.; Smith, T.J.; Shah, D.M. Structure-Activity Determinants in Antifungal Plant Defensins Msdef1 and Mtdef4 with Different Modes of Action against Fusarium Graminearum. PLoS ONE 2011, 6, e18550. [Google Scholar] [CrossRef] [Green Version]
- Kaewklom, S.; Euanorasetr, J.; Intra, B.; Panbangred, W.; Aunpad, R. Antimicrobial Activities of Novel Peptides Derived from Defensin Genes of Brassica Hybrid Cv Pule. Int. J. Pept. Res. Ther. 2016, 22, 93–100. [Google Scholar] [CrossRef]
- Garrigues, S.; Gandía, M.; Borics, A.; Marx, F.; Manzanares, P.; Marcos, J.F. Mapping and Identification of Antifungal Peptides in the Putative Antifungal Protein AfpB from the Filamentous Fungus Penicillium Digitatum. Front. Microbiol. 2017, 8, 592. [Google Scholar] [CrossRef]
- Fernández, A.; Colombo, M.L.; Curto, L.M.; Gómez, G.E.; Delfino, J.M.; Guzmán, F.; Bakás, L.; Malbrán, I.; Vairo Cavalli, S. Peptides Derived From the α -Core and γ -Core Regions of a Putative Silybum Marianum Flower Defensin Show Antifungal Activity Against Fusarium graminearum. Front. Microbiol. 2021, 12, 632008. [Google Scholar] [CrossRef] [PubMed]
- Killian, J.A.; Von Heijne, G. How Proteins Adapt to a Membrane-Water Interface. Trends Biochem. Sci. 2000, 25, 429–434. [Google Scholar] [CrossRef]
- Marsh, D. Lateral Pressure in Membranes. Biochim. Et Biophys. Acta (BBA)-Rev. Biomembr. 1996, 1286, 183–223. [Google Scholar] [CrossRef]
- Calvez, P.; Bussières, S.; Demers, É.; Salesse, C. Parameters Modulating the Maximum Insertion Pressure of Proteins and Peptides in Lipid Monolayers. Biochimie 2009, 91, 718–733. [Google Scholar] [CrossRef] [PubMed]
- Astafieva, A.A.; Rogozhin, E.A.; Odintsova, T.I.; Khadeeva, N.V.; Grishin, E.V.; Egorov, T.A. Discovery of Novel Antimicrobial Peptides with Unusual Cysteine Motifs in Dandelion Taraxacum officinale Wigg. Flowers. Peptides 2012, 36, 266–271. [Google Scholar] [CrossRef]
- Parisi, K.; Shafee, T.M.A.; Quimbar, P.; van der Weerden, N.L.; Bleackley, M.R.; Anderson, M.A. The Evolution, Function and Mechanisms of Action for Plant Defensins. Semin Cell Dev Biol 2019, 88, 107–118. [Google Scholar] [CrossRef]
- Farkas, A.; Maróti, G.; Kereszt, A.; Kondorosi, É. Comparative Analysis of the Bacterial Membrane Disruption Effect of Two Natural Plant Antimicrobial Peptides. Front. Microbiol. 2017, 8, 51. [Google Scholar] [CrossRef] [Green Version]
- Hitchner, M.A.; Santiago-Ortiz, L.E.; Necelis, M.R.; Shirley, D.J.; Palmer, T.J.; Tarnawsky, K.E.; Vaden, T.D.; Caputo, G.A. Activity and Characterization of a PH-Sensitive Antimicrobial Peptide. Biochim. Biophys. Acta Biomembr. 2019, 1861, 182984. [Google Scholar] [CrossRef]
- Liao, S.-M.; Du, Q.-S.; Meng, J.-Z.; Pang, Z.-W.; Huang, R.-B. The Multiple Roles of Histidine in Protein Interactions. Chem. Cent. J. 2013, 7, 44. [Google Scholar] [CrossRef] [Green Version]
- Pirtskhalava, M.; Vishnepolsky, B.; Grigolava, M.; Managadze, G. Physicochemical Features and Peculiarities of Interaction of Amp with the Membrane. Pharmaceuticals 2021, 14, 471. [Google Scholar] [CrossRef]
- Heyda, J.; Mason, P.E.; Jungwirth, P. Attractive Interactions between Side Chains of Histidine-Histidine and Histidine-Arginine-Based Cationic Dipeptides in Water. J. Phys. Chem. B 2010, 114, 8744–8749. [Google Scholar] [CrossRef]
- Cools, T.L.; Struyfs, C.; Cammue, B.P.; Thevissen, K. Antifungal Plant Defensins: Increased Insight in Their Mode of Action as a Basis for Their Use to Combat Fungal Infections. Future Microbiol. 2017, 12, 441–454. [Google Scholar] [CrossRef]
- Baxter, A.A.; Richter, V.; Lay, F.T.; Poon, I.K.H.; Adda, C.G.; Veneer, P.K.; Phan, T.K.; Bleackley, M.R.; Anderson, M.A.; Kvansakul, M.; et al. The Tomato Defensin TPP3 Binds Phosphatidylinositol (4,5)-Bisphosphate via a Conserved Dimeric Cationic Grip Conformation To Mediate Cell Lysis. Mol. Cell Biol. 2015, 35, 1964–1978. [Google Scholar] [CrossRef] [Green Version]
- Vazquez, R.F.; Maté, S.M.; Bakás, L.S.; Fernández, M.M.; Malchiodi, E.L.; Herlax, V.S. Novel Evidence for the Specific Interaction between Cholesterol and α-Haemolysin of Escherichia coli. Biochem. J. 2014, 458, 481–489. [Google Scholar] [CrossRef]
- Cytryńska, M.; Zdybicka-Barabas, A. Defense Peptides: Recent Developments. Biomol. Concepts 2015, 6, 237–251. [Google Scholar] [CrossRef]
- Thevissen, K.; François, I.E.J.A.; Takemoto, J.Y.; Ferket, K.K.A.; Meert, E.M.K.; Cammue, B.P.A. DmAMP1, an Antifungal Plant Defensin from Dahlia (Dahlia Merckii), Interacts with Sphingolipids from Saccharomyces cerevisiae. FEMS Microbiol. Lett. 2003, 226, 169–173. [Google Scholar] [CrossRef] [Green Version]
- Seelig, A. Local Anesthetics and Pressure: A Comparison of Dibucaine Binding to Lipid Monolayers and Bilayers. BBA-Biomembr. 1987, 899, 196–204. [Google Scholar] [CrossRef]
- Velivelli, S.; Islam, K.T.; Hobson, E.; Shah, D.M. Modes of Action of a Bi-Domain Plant Defensin MtDef5 against a Bacterial Pathogen Xanthomonas Campestris. Front. Microbiol. 2018, 9, 934. [Google Scholar] [CrossRef]
- Miyazawa, K.; Yoshimi, A.; Abe, K. The Mechanisms of Hyphal Pellet Formation Mediated by Polysaccharides, α-1,3-Glucan and Galactosaminogalactan, in Aspergillus Species. Fungal Biol. Biotechnol. 2020, 7, 10. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, J. The Filamentous Fungal Pellet and Forces Driving Its Formation. Crit. Rev. Biotechnol. 2016, 36, 1066–1077. [Google Scholar] [CrossRef]
- Falter, C.; Reumann, S. The Essential Role of Fungal Peroxisomes in Plant Infection. Mol. Plant Pathol. 2022, 23, 781–794. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-L.; Wang, Z.; Liu, C. Roles of Peroxisomes in the Rice Blast Fungus. BioMed Res. Int. 2016, 2016, 9343417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Min, K.; Son, H.; Lee, J.; Choi, G.J.; Kim, J.C.; Lee, Y.W. Peroxisome Function Is Required for Virulence and Survival of Fusarium graminearum. Mol. Plant-Microbe Interact. 2012, 25, 1617–1627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navarro-Espíndola, R.; Suaste-Olmos, F.; Peraza-Reyes, L. Dynamic Regulation of Peroxisomes and Mitochondria during Fungal Development. J. Fungi 2020, 6, 302. [Google Scholar] [CrossRef] [PubMed]
- Bartoszewska, M.; Opaliński, Ł.; Veenhuis, M.; van der Klei, I.J. The Significance of Peroxisomes in Secondary Metabolite Biosynthesis in Filamentous Fungi. Biotechnol. Lett. 2011, 33, 1921–1931. [Google Scholar] [CrossRef] [Green Version]
- Aerts, A.M.; Bammens, L.; Govaert, G.; Carmona-Gutierrez, D.; Madeo, F.; Cammue, B.P.A.; Thevissen, K. The Antifungal Plant Defensin HsAFP1 from Heuchera Sanguinea Induces Apoptosis in Candida albicans. Front. Microbiol. 2011, 2, 47. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Velivelli, S.L.S.; Shah, D.M. Antifungal Potency and Modes of Action of a Novel Olive Tree Defensin Against Closely Related Ascomycete Fungal Pathogens. Mol. Plant-Microbe Interact. 2019, 32, 1649–1664. [Google Scholar] [CrossRef]
- Aerts, A.M.; François, I.E.J.A.; Thevissen, K.; Meert, E.M.K.; Li, Q.; Cammue, B.P.A. The Antifungal Activity of RsAFP2, a Plant Defensin from Raphanus Sativus, Involves the Induction of Reactive Oxygen Species in Candida albicans. J. Mol. Microbiol. Biotechnol. 2007, 13, 243–247. [Google Scholar] [CrossRef]
- Van der Weerden, N.L.; Lay, F.T.; Anderson, M.A. The Plant Defensin, NaD1, Enters the Cytoplasm of Fusarium Oxysporum Hyphae. J. Biol. Chem. 2008, 283, 14445–14452. [Google Scholar] [CrossRef] [Green Version]
- Cools, T.L.; Vriens, K.; Struyfs, C.; Verbandt, S.; Ramada, M.H.S.; Brand, G.D.; Bloch, C.; Koch, B.; Traven, A.; Drijfhout, J.W.; et al. The Antifungal Plant Defensin HsAFP1 Is a Phosphatidic Acid-Interacting Peptide Inducing Membrane Permeabilization. Front. Microbiol. 2017, 8, 2295. [Google Scholar] [CrossRef]
- Van der Weerden, N.L.; Hancock, R.E.W.; Anderson, M.A. Permeabilization of Fungal Hyphae by the Plant Defensin NaD1 Occurs through a Cell Wall-Dependent Process. J. Biol. Chem. 2010, 285, 37513–37520. [Google Scholar] [CrossRef] [Green Version]
- Malbrán, I.; Mourelos, C.A.; Girotti, J.R.; Aulicino, M.B.; Balatti, P.A.; Lori, G.A. Aggressiveness Variation of Fusarium Graminearum Isolates from Argentina Following Point Inoculation of Field Grown Wheat Spikes. Crop Prot. 2012, 42, 234–243. [Google Scholar] [CrossRef]
- Malbrán, I.; Mourelos, C.A.; Girotti, J.R.; Balatti, P.A.; Lori, G.A. Toxigenic Capacity and Trichothecene Production by Fusarium Graminearum Isolates from Argentina and Their Relationship with Aggressiveness and Fungal Expansion in the Wheat Spike. Phytopathology 2014, 104, 357–364. [Google Scholar] [CrossRef] [Green Version]
- Luna, O.F.; Gomez, J.; Cárdenas, C.; Albericio, F.; Marshall, S.H.; Guzmán, F. Deprotection Reagents in Fmoc Solid Phase Peptide Synthesis: Moving Away from Piperidine? Molecules 2016, 21, 1542. [Google Scholar] [CrossRef]
- Bleackley, M.R.; Dawson, C.S.; Mckenna, J.A.; Quimbar, P.; Hayes, B.M.E.; van der Weerden, N.L.; Anderson, M.A. Synergistic Activity between Two Antifungal Proteins, the Plant Defensin. mSphere 2017, 2, e00390-17. [Google Scholar] [CrossRef] [Green Version]
- Di Rienzo, J.A.; Casanoves, F.; Balzarini, M.G.; Gonzalez, L.; Tablada, M.; Robledo, C.W. InfoStat V 2018; Grupo InfoStat, FCA, Universidad Nacional de Córdoba: Córdoba, Argentina, 2018. [Google Scholar]
- Vázquez, R.F.; Daza Millone, M.A.; Pavinatto, F.J.; Herlax, V.S.; Bakás, L.S.; Oliveira, O.N.; Vela, M.E.; Maté, S.M. Interaction of Acylated and Unacylated Forms of E. Coli Alpha-Hemolysin with Lipid Monolayers: A PM-IRRAS Study. Colloids Surf B Biointerfaces 2017, 158, 76–83. [Google Scholar] [CrossRef]
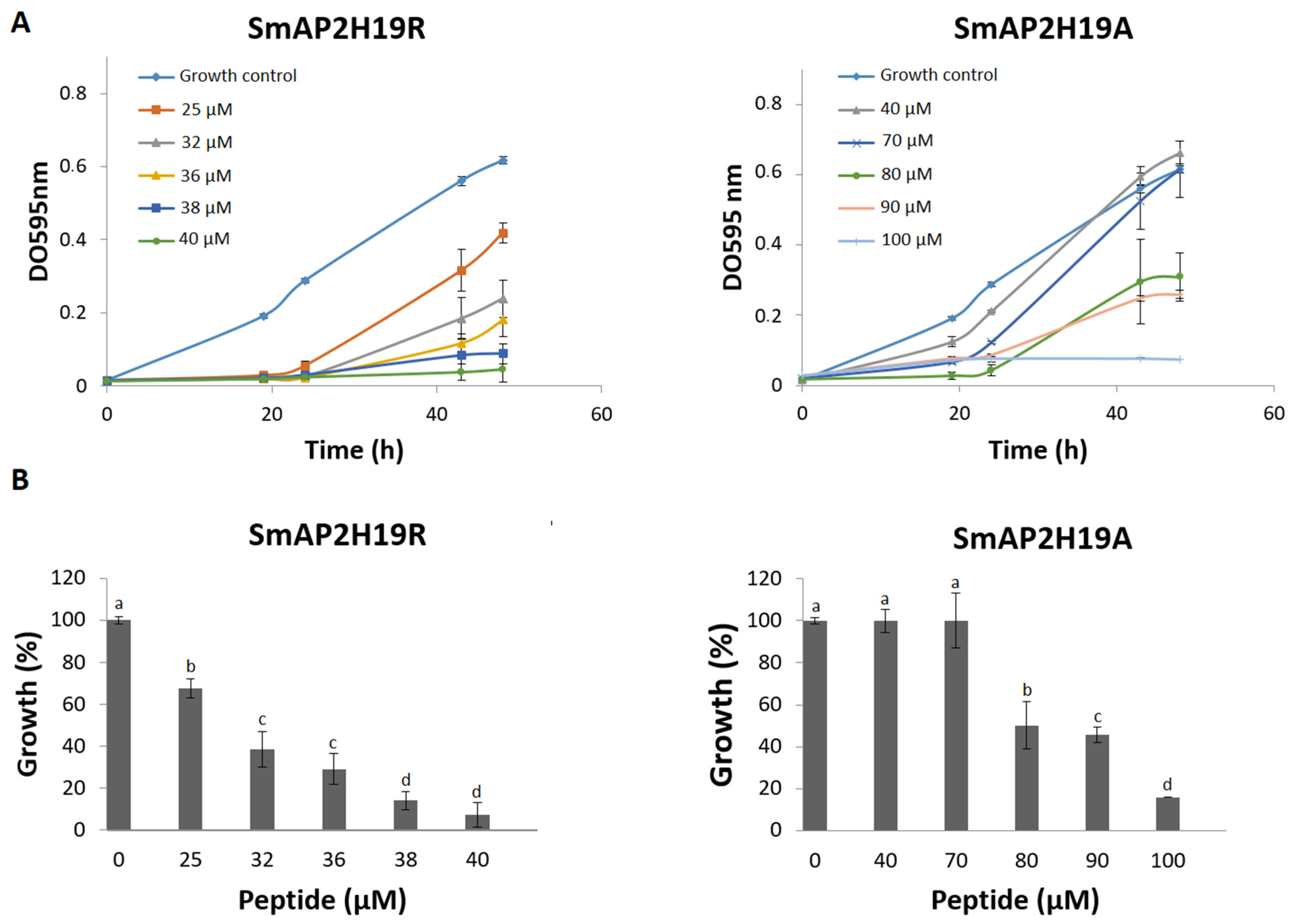

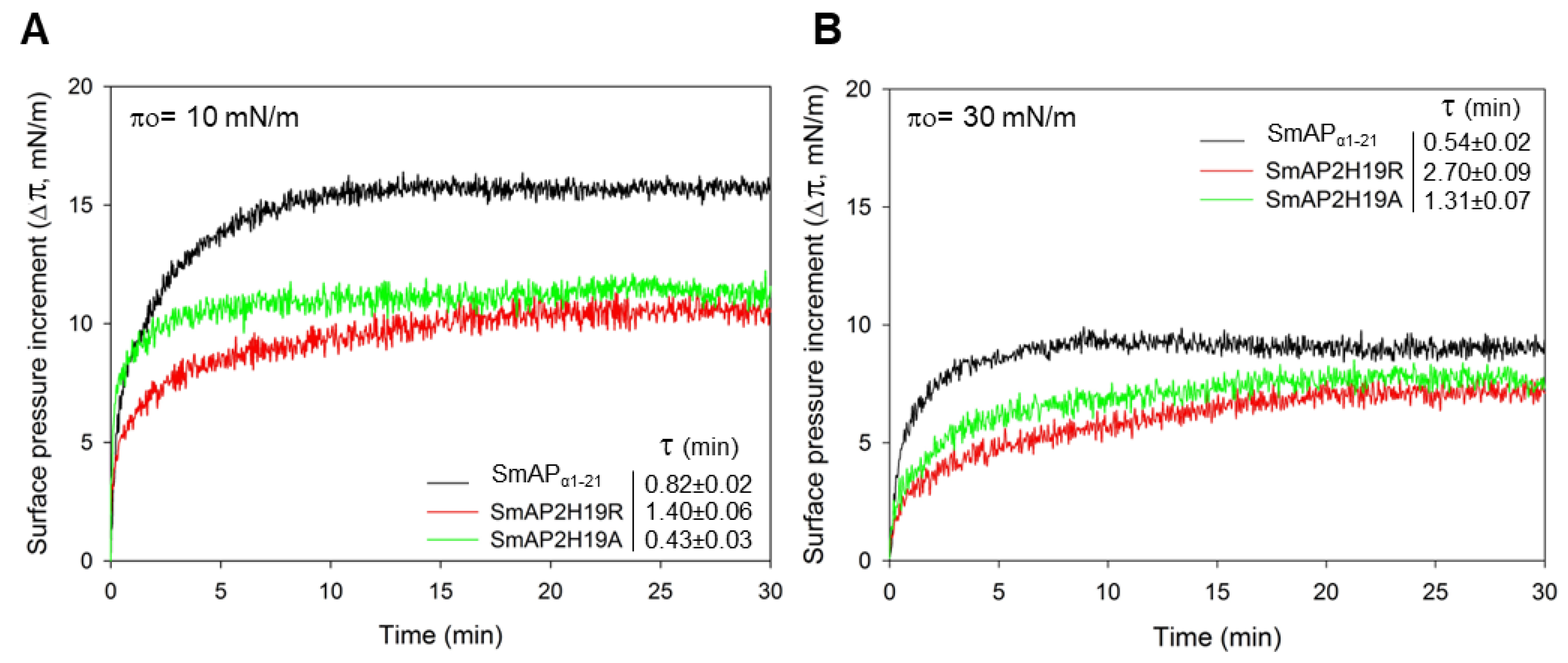


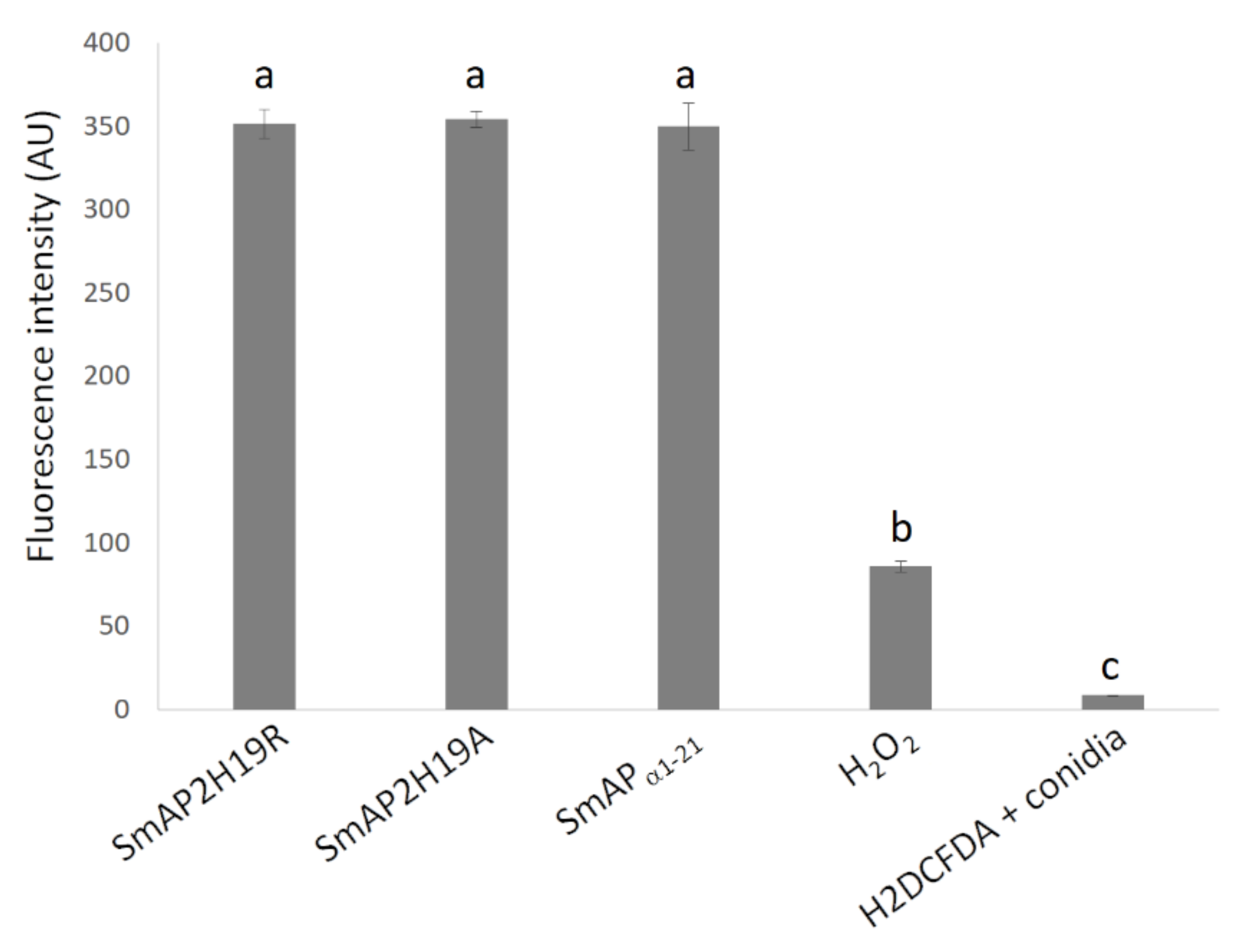
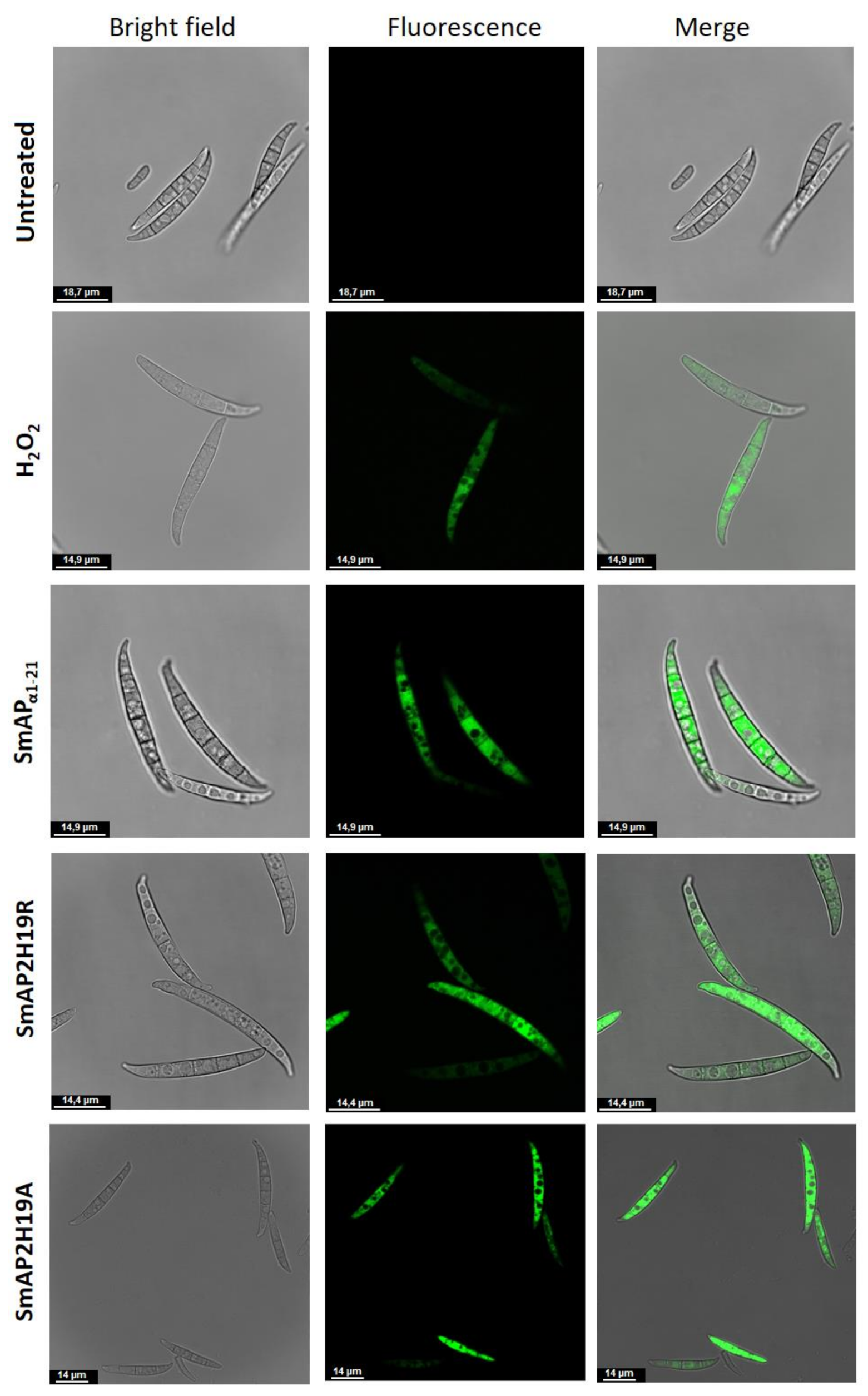

| Peptide | Sequence | Molecular Weight (Da) | pI 1 | Net Charge 2 | MIC (μM) 3 |
|---|---|---|---|---|---|
| SmAPα1-21 | KLCEKPSKTWFGNCGNPRHCG | 2361.7 | 9.1 | +4.0 | 32 |
| SmAP2H19R | KLCEKPSKTWFGNCGNPRRCG | 2383.8 | 9.5 | +4.1 | 38 |
| SmAP2H19A | KLCEKPSKTWFGNCGNPRACG | 2297.7 | 9.1 | +3.0 | 100 |
| F-SmAPα1-21 4 | 60 | ||||
| F-SmAP2H19R | 38 | ||||
| F-SmAP2H19A | 100 | ||||
| RB- SmAPα1-21 5 | 60 | ||||
| RB-SmAP2H19R | 40 | ||||
| RB-SmAP2H19A | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández, A.; González, M.; Malbrán, I.; Vázquez, R.F.; Maté, S.M.; Guzmán, F.; Bakás, L.S.; Vairo Cavalli, S. Histidine 19 Residue Is Essential for Cell Internalization of Antifungal Peptide SmAPα1-21 Derived from the α-Core of the Silybum marianum Defensin DefSm2-D in Fusarium graminearum. Antibiotics 2022, 11, 1501. https://doi.org/10.3390/antibiotics11111501
Fernández A, González M, Malbrán I, Vázquez RF, Maté SM, Guzmán F, Bakás LS, Vairo Cavalli S. Histidine 19 Residue Is Essential for Cell Internalization of Antifungal Peptide SmAPα1-21 Derived from the α-Core of the Silybum marianum Defensin DefSm2-D in Fusarium graminearum. Antibiotics. 2022; 11(11):1501. https://doi.org/10.3390/antibiotics11111501
Chicago/Turabian StyleFernández, Agustina, Mariano González, Ismael Malbrán, Romina F. Vázquez, Sabina M. Maté, Fanny Guzmán, Laura S. Bakás, and Sandra Vairo Cavalli. 2022. "Histidine 19 Residue Is Essential for Cell Internalization of Antifungal Peptide SmAPα1-21 Derived from the α-Core of the Silybum marianum Defensin DefSm2-D in Fusarium graminearum" Antibiotics 11, no. 11: 1501. https://doi.org/10.3390/antibiotics11111501
APA StyleFernández, A., González, M., Malbrán, I., Vázquez, R. F., Maté, S. M., Guzmán, F., Bakás, L. S., & Vairo Cavalli, S. (2022). Histidine 19 Residue Is Essential for Cell Internalization of Antifungal Peptide SmAPα1-21 Derived from the α-Core of the Silybum marianum Defensin DefSm2-D in Fusarium graminearum. Antibiotics, 11(11), 1501. https://doi.org/10.3390/antibiotics11111501









