Utility of Metagenomic Next-Generation Sequencing in Infective Endocarditis: A Systematic Review
Abstract
1. Introduction
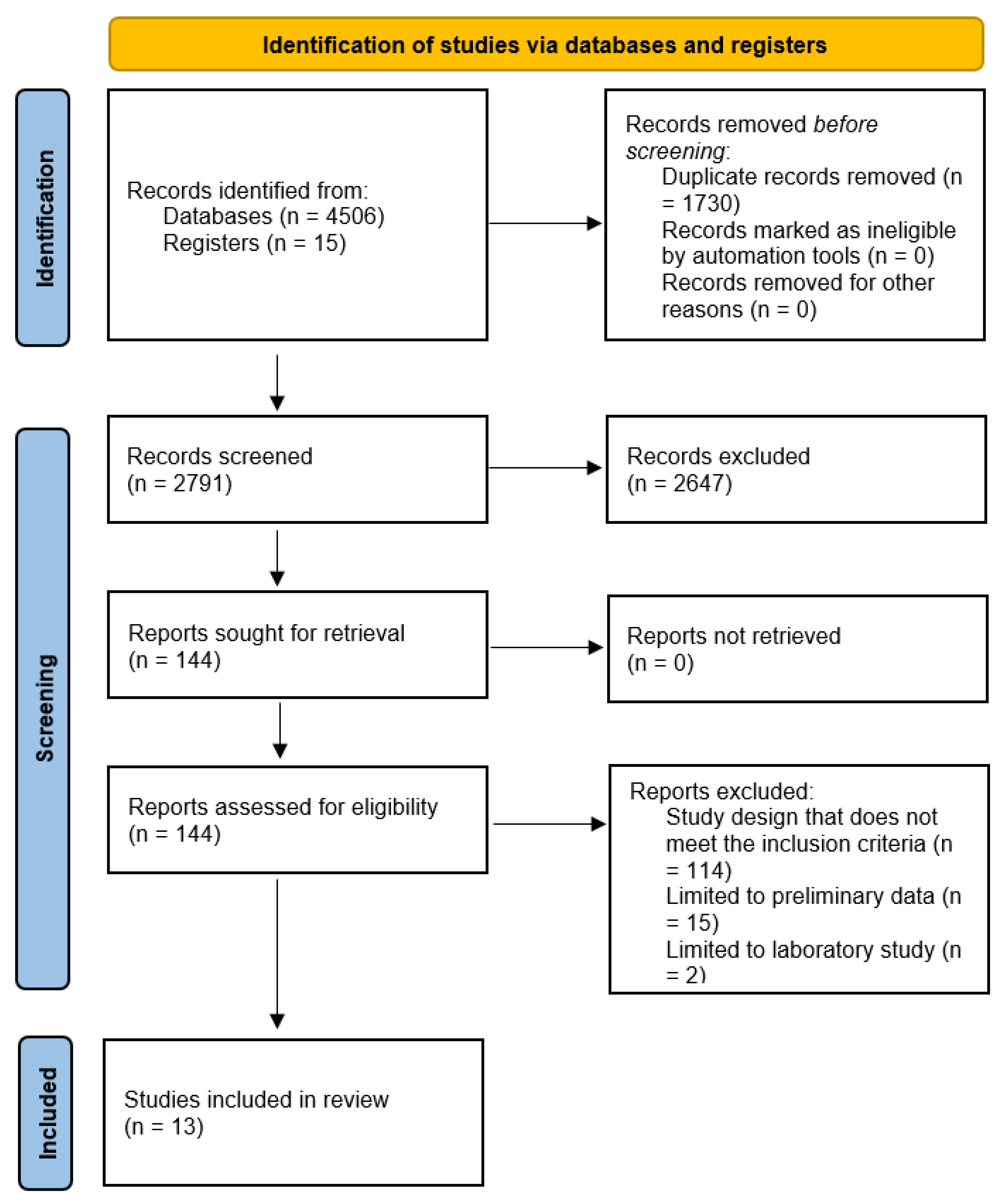
2. Methods
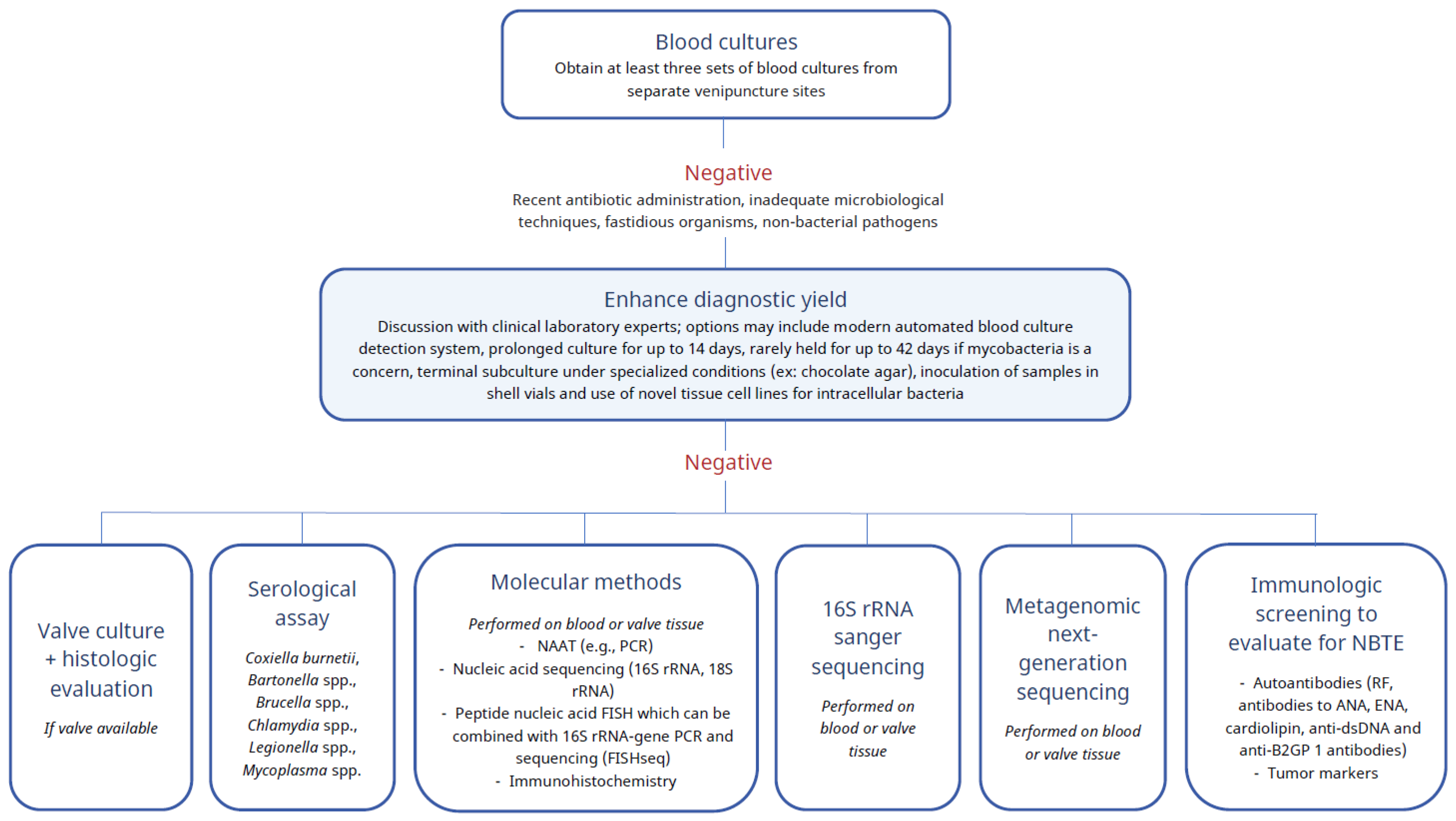
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miller, J.M.; Binnicker, M.J.; Campbell, S.; Carroll, K.C.; Chapin, K.C.; Gilligan, P.H.; Gonzalez, M.D.; Jerris, R.C.; Kehl, S.C.; Patel, R.; et al. A Guide to Utilization of the Microbiology Laboratory for Diagnosis of Infectious Diseases: 2018 Update by the Infectious Diseases Society of America and the American Society for Microbiology. Clin. Infect. Dis. 2018, 67, 813–816. [Google Scholar] [CrossRef] [PubMed]
- Baddour, L.M.; Wilson, W.R.; Bayer, A.S.; Fowler, V.G., Jr.; Tleyjeh, I.M.; Rybak, M.J.; Barsic, B.; Lockhart, P.B.; Gewitz, M.H.; Levison, M.E.; et al. Infective Endocarditis in Adults: Diagnosis, Antimicrobial Therapy, and Management of Complications: A Scientific Statement for Healthcare Professionals From the American Heart Association. Circulation 2015, 132, 1435–1486. [Google Scholar] [CrossRef] [PubMed]
- Lamas, C.C.; Fournier, P.E.; Zappa, M.; Brandão, T.J.; Januário-da-Silva, C.A.; Correia, M.G.; Barbosa, G.I.; Golebiovski, W.F.; Weksler, C.; Lepidi, H.; et al. Diagnosis of blood culture-negative endocarditis and clinical comparison between blood culture-negative and blood culture-positive cases. Infection 2016, 44, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Topan, A.; Carstina, D.; Slavcovici, A.; Rancea, R.; Capalneanu, R.; Lupse, M. Assesment of the Duke criteria for the diagnosis of infective endocarditis after twenty-years. An analysis of 241 cases. Clujul. Med. 2015, 88, 321–326. [Google Scholar] [CrossRef]
- Fournier, P.E.; Gouriet, F.; Casalta, J.P.; Lepidi, H.; Chaudet, H.; Thuny, F.; Collart, F.; Habib, G.; Raoult, D. Blood culture-negative endocarditis: Improving the diagnostic yield using new diagnostic tools. Medicine (Baltimore) 2017, 96, e8392. [Google Scholar] [CrossRef]
- Liesman, R.M.; Pritt, B.S.; Maleszewski, J.J.; Patel, R. Laboratory Diagnosis of Infective Endocarditis. J. Clin. Microbiol. 2017, 55, 2599–2608. [Google Scholar] [CrossRef]
- Godfrey, R.; Curtis, S.; Schilling, W.H.; James, P.R. Blood culture negative endocarditis in the modern era of 16S rRNA sequencing. Clin. Med. (Lond.) 2020, 20, 412–416. [Google Scholar] [CrossRef]
- Gu, W.; Miller, S.; Chiu, C.Y. Clinical Metagenomic Next-Generation Sequencing for Pathogen Detection. Annu. Rev. Pathol. 2019, 14, 319–338. [Google Scholar] [CrossRef]
- Chiu, C.Y.; Miller, S.A. Clinical metagenomics. Nat. Rev. Genet. 2019, 20, 341–355. [Google Scholar] [CrossRef]
- Barba, M.; Czosnek, H.; Hadidi, A. Historical perspective, development and applications of next-generation sequencing in plant virology. Viruses 2014, 6, 106–136. [Google Scholar] [CrossRef]
- Lamoureux, C.; Surgers, L.; Fihman, V.; Gricourt, G.; Demontant, V.; Trawinski, E.; N’Debi, M.; Gomart, C.; Royer, G.; Launay, N.; et al. Prospective Comparison Between Shotgun Metagenomics and Sanger Sequencing of the 16S rRNA Gene for the Etiological Diagnosis of Infections. Front. Microbiol. 2022, 13, 761873. [Google Scholar] [CrossRef] [PubMed]
- Oberbach, A.; Schlichting, N.; Feder, S.; Lehmann, S.; Kullnick, Y.; Buschmann, T.; Blumert, C.; Horn, F.; Neuhaus, J.; Neujahr, R.; et al. New insights into valve-related intramural and intracellular bacterial diversity in infective endocarditis. PLoS ONE 2017, 12, e0175569. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Hu, H.; Kang, Y.; Chen, W.; Fang, W.; Wang, K.; Zhang, Q.; Fu, A.; Zhou, S.; Cheng, C.; et al. Identification of pathogens in culture-negative infective endocarditis cases by metagenomic analysis. Ann. Clin. Microbiol. Antimicrob. 2018, 17, 43. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Hu, H.; Fang, W.; Shi, D.; Liang, C.; Sun, Y.; Gao, G.; Wang, H.; Zhang, Q.; Wang, L.; et al. Detection of pathogens from resected heart valves of patients with infective endocarditis by next-generation sequencing. Int. J. Infect. Dis. 2019, 83, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Yang, Y.; Pan, J.; Miao, Q.; Jin, W.; Ma, Y.; Zhou, C.; Gao, X.; Wang, C.; Hu, B. The clinical value of valve metagenomic next-generation sequencing when applied to the etiological diagnosis of infective endocarditis. Ann. Transl. Med. 2021, 9, 1490. [Google Scholar] [CrossRef]
- Zeng, X.; Wu, J.; Li, X.; Xiong, W.; Tang, L.; Li, X.; Zhuang, J.; Yu, R.; Chen, J.; Jian, X.; et al. Application of Metagenomic Next-Generation Sequencing in the Etiological Diagnosis of Infective Endocarditis During the Perioperative Period of Cardiac Surgery: A Prospective Cohort Study. Front. Cardiovasc. Med. 2022, 9, 811492. [Google Scholar] [CrossRef]
- Santibáñez, P.; García-García, C.; Portillo, A.; Santibáñez, S.; García-Álvarez, L.; de Toro, M.; Oteo, J.A. What Does 16S rRNA Gene-Targeted Next Generation Sequencing Contribute to the Study of Infective Endocarditis in Heart-Valve Tissue? Pathogens 2021, 11, 34. [Google Scholar] [CrossRef]
- Flurin, L.; Wolf, M.J.; Fisher, C.R.; Cano Cevallos, E.J.; Vaillant, J.J.; Pritt, B.S.; DeSimone, D.C.; Patel, R. Pathogen Detection in Infective Endocarditis Using Targeted Metagenomics on Whole Blood and Plasma: A Prospective Pilot Study. J. Clin. Microbiol. 2022, 60, e0062122. [Google Scholar] [CrossRef]
- To, R.K.; Ramchandar, N.; Gupta, A.; Pong, A.; Cannavino, C.; Foley, J.; Farnaes, L.; Coufal, N.G. Use of Plasma Metagenomic Next-generation Sequencing for Pathogen Identification in Pediatric Endocarditis. Pediatr. Infect. Dis. J. 2021, 40, 486–488. [Google Scholar] [CrossRef]
- Eichenberger, E.M.; Degner, N.; Scott, E.R.; Ruffin, F.; Franzone, J.; Sharma-Kuinkel, B.; Shah, P.; Hong, D.; Dalai, S.C.; Blair, L.; et al. Microbial Cell-Free DNA Identifies the Causative Pathogen in Infective Endocarditis and Remains Detectable Longer than Conventional Blood Culture in Patients with Prior Antibiotic Therapy. Clin. Infect. Dis. 2022. [Google Scholar] [CrossRef]
- Olsen, T.; Justesen, U.S.; Nielsen, J.C.; Jørgensen, O.D.; Sandgaard, N.C.F.; Ravn, C.; Gerdes, C.; Thøgersen, A.M.; Gill, S.; Fuursted, K.; et al. Microbiological diagnosis in cardiac implantable electronic device infections detected by sonication and next-generation sequencing. Heart Rhythm 2022, 19, 901–908. [Google Scholar] [CrossRef] [PubMed]
- Hogan, C.A.; Yang, S.; Garner, O.B.; Green, D.A.; Gomez, C.A.; Dien Bard, J.; Pinsky, B.A.; Banaei, N. Clinical Impact of Metagenomic Next-Generation Sequencing of Plasma Cell-Free DNA for the Diagnosis of Infectious Diseases: A Multicenter Retrospective Cohort Study. Clin. Infect. Dis. 2021, 72, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.A.; Al Dhaheri, F.; Pollock, N.R.; Sharma, T.S. Assessment of the Clinical Utility of Plasma Metagenomic Next-Generation Sequencing in a Pediatric Hospital Population. J. Clin. Microbiol. 2020, 58, e00419-20. [Google Scholar] [CrossRef] [PubMed]
- Shishido, A.A.; Noe, M.; Saharia, K.; Luethy, P. Clinical impact of a metagenomic microbial plasma cell-free DNA next-generation sequencing assay on treatment decisions: A single-center retrospective study. BMC Infect. Dis. 2022, 22, 372. [Google Scholar] [CrossRef] [PubMed]
- Million, M.; Gaudin, M.; Melenotte, C.; Chasson, L.; Edouard, S.; Verdonk, C.; Prudent, E.; Amphoux, B.; Meresse, S.; Dorent, R.; et al. Metagenomic Analysis of Microdissected Valvular Tissue for Etiological Diagnosis of Blood Culture-Negative Endocarditis. Clin. Infect. Dis. 2020, 70, 2405–2412. [Google Scholar] [CrossRef]
- Bouchiat, C.; Ginevra, C.; Benito, Y.; Gaillard, T.; Salord, H.; Dauwalder, O.; Laurent, F.; Vandenesch, F. Improving the Diagnosis of Bacterial Infections: Evaluation of 16S rRNA Nanopore Metagenomics in Culture-Negative Samples. Front. Microbiol. 2022, 13, 943441. [Google Scholar] [CrossRef]
- do Vale, A.; Cabanes, D.; Sousa, S. Bacterial Toxins as Pathogen Weapons against Phagocytes. Front. Microbiol. 2016, 7, 42. [Google Scholar] [CrossRef]
- Branger, S.; Casalta, J.P.; Habib, G.; Collard, F.; Raoult, D. Streptococcus pneumoniae endocarditis: Persistence of DNA on heart valve material 7 years after infectious episode. J. Clin. Microbiol. 2003, 41, 4435–4437. [Google Scholar] [CrossRef]
- Casalta, J.P.; Thuny, F.; Fournier, P.E.; Lepidi, H.; Habib, G.; Grisoli, D.; Raoult, D. DNA persistence and relapses questions on the treatment strategies of Enterococcus infections of prosthetic valves. PLoS ONE 2012, 7, e53335. [Google Scholar] [CrossRef]
- Lang, S.; Watkin, R.W.; Lambert, P.A.; Littler, W.A.; Elliott, T.S. Detection of bacterial DNA in cardiac vegetations by PCR after the completion of antimicrobial treatment for endocarditis. Clin. Microbiol. Infect. 2004, 10, 579–581. [Google Scholar] [CrossRef]
| Type of Specimens | Authors, Date of Publication | - Country - Study Period - Type of Study | n Value | Previous Antibiotic Exposure | Microbiology (n Values) |
|---|---|---|---|---|---|
| mNGS on valve | Cheng J et al. 2019 [14] | - China - April 2017–September 2018 - Retrospective cohort | 51: 44 IE - 41 definite IE - 3 possible IE 7 negative controls | 35 (79.5%) | Staphylococcus aureus (2) Streptococcus spp. (28) Enterococcus faecalis (1) Abiotrophia defectiva (2) Pseudomonas aeruginosa (1) Haemophilus parainfluenzae (1) Bartonella quintana (1) Coxiella burnetii (6) |
| Cai S et al. 2021 [15] | - China - June 2018–November 2020 - Retrospective cohort | 57: 49 IE - 28 culture positive - 21 culture negative 8 negative controls (43 NVE, 6 PVE) | 46 (93.9%) | Staphylococcus spp. (10) Streptococcus spp. (26) Enterococcus faecalis (1) Haemophilus parainfluenzae (1) Abiotrophia defectiva (4) Granulicatella adiacens (2) Gemella haemolysans (1) Erysipelothrix rhusiopathiae (1) Cardiobacterium hominis (1) Aggregatibacter segnis (1) Candida parapsilosis (1) | |
| Zeng X et al. 2022 [16] | - China - May 2019–December 2020 - Prospective cohort | 110: 99 IE 11 negative controls | 43 (43.4%) | Staphylococcus aureus (5) Staphylococcus spp. (5) Streptococcus spp. (51) Enterococcus faecalis (3) Granulicatella adiacens (3) Coxiella burnetii (8) Legionella drancourtii (4) Bartonella Quintana (1) Other Polymicrobial (10 patients) | |
| 16S rRNA gene targeted NGS on heart valve | Santibáñez P et al. 2021 [17] | - Spain - 2009–2017 - Retrospective cohort | 27: - 23 definite IE - 2 possible IE - 2 unavailable 4 BCNE (18 NV, 4 PV, 5 DRI) | 25 (92.6%) | Staphylococcus aureus (1) Staphylococcus spp. (3) Streptococcus spp. (7) Enterococcus faecalis (4) Haemophilus parainfluenzae (1) Coxiella burnetii (1) Polymicrobial (10 patients) |
| Whole Blood and Plasma NGS on an Illumina MiSeqTM platform | Flurin L et al. 2022 [18] | - USA - October 2020–July 2021 - Prospective cohort, pilot study | 35: - 28 definite IE - 7 possible IE 6 BCNE ( 13 NV, 22 PV) | Staphylococcus aureus (8) Staphylococcus spp. (3) Streptococcus spp. (6) Enterococcus faecalis (2) Kingella sp. (1) Cutibacterium acnes (1) Corynebacterium spp. (2) Bartonella sp. (1) | |
| Plasma mcfDNA (Karius) | To RK et al. 2021 [19] | - USA - January 2017–January 2020 - Retrospective cohort | 10 | Staphylococcus aureus (1) Staphylococcus epidermidis (1) Streptococcus spp. (2) Kingella kingae (1) Corynebacterium diphtheriae (1) Pseudomonas aeruginosa (1) Gemella bergeri (1) | |
| Eichenberger EM et al. 2022 [20] | - USA - July 2016 -January 2018 - Prospective observational cohort study | 23: - 23 definite IE (15 NV, 5 PV, 7 infected PM or cardioverter defibrillator lead) | Staphylococcus aureus (14) Staphylococcus epidermidis (2) Streptococcus agalactiae (1) Enterococcus faecalis (2) Candida albicans (2) Pantoea ananatis (1) | ||
| NGS on DRI | Olsen T et al. 2022 [21] | - Denmark - October 2016 and January 2019 - Descriptive, prospective, multicenter study | 60: Pockets, generators, leads 41 PM, 14 ICD, 3 CRT-P, 2 CRT-D | Staphylococcus aureus (25) Staphylococcus spp. (9) Streptococcus spp. (9) Enterococcus faecalis (10) Propionibacterium acnes (2) Corynebacterium sp. (1) Other Gram-negative rods (2) |
| Authors | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|
| Cheng J et al. [14] | mNGS: 97.6% BC: 46.2% VC: 17.1% GS: 51.4% | mNGS: 85.7% BC: 100% VC: 100% GS: 100% | mNGS: 97.6% BC: 100% VC: 100% GS: 100% | mNGS: 85.7% BC: 12.5% VC: 17.1% GS: 26.1% |
| Cai S et al. [15] | valve mNGS in culture positive and negative IE: 100% | mNGS is culture-positive IE: 100% | ||
| Zeng X et al. [16] | mNGS: 85.9% BC: 29.3% VC: 16.2% Combined: 89.9% | mNGS: 72.7% BC: 100% VC: 100% Combined: 72.7% | mNGS: 96.6% BC: 100% VC: 100% Combined: 96.7% | mNGS: 36.4% BC: 13.6% VC: 11.7% Combined: 44.4% |
| Santibáñez P et al. [17] | mNGS: 88.9% | mNGS: 91.7% | ||
| Flurin L et al. [18] | tMGS positive in - WB: 59% (20/34) - Plasma: 47% (16/34) - Combined: 66% (23/35) In BCPE: tMGS positive in - WB: 61% (17/28) - Plasma: 45% (13/29) - combined: 62% (18/29) In BCNE: tMGS positive in - WB: 50% (3/6) - Plasma: 45% (3/5) - Combined: 83% (5/6) BC: 83% (29) VC: 50% (6/12) 16S rRNA gene PCR on valve tissue: 60% (3/5) | Of the positive tMGS cases: 55.6% (10/18) concordant results from plasma and WB | ||
| To RK et al. [19] | mNGS: 80% BC, VC and 16S rRNA: 50% | In BCPE: 100% In BCNE: 71.4% | ||
| Eichenberger EM et al. [20] | mcfDNA: 87% BC: 87% | |||
| Olsen T et al. [21] | NGS analysis of generators: 18% (10/57) and leads: 48% (27/56) | NGS analysis of generators (~50%) and leads (~90%) |
| Microbiologic Diagnostic Tools | Advantages | Disadvantages |
|---|---|---|
| Blood culture |
|
|
| Valve Culture |
|
|
| Serology |
|
|
| Polymerase chain reaction (PCR) |
|
|
| Fluorescence in situ hybridization (FISH) |
|
|
| Immunohistochemistry |
|
|
| 16S rRNA sanger sequencing |
|
|
| Metagenomic next-generation sequencing |
|
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haddad, S.F.; DeSimone, D.C.; Chesdachai, S.; Gerberi, D.J.; Baddour, L.M. Utility of Metagenomic Next-Generation Sequencing in Infective Endocarditis: A Systematic Review. Antibiotics 2022, 11, 1798. https://doi.org/10.3390/antibiotics11121798
Haddad SF, DeSimone DC, Chesdachai S, Gerberi DJ, Baddour LM. Utility of Metagenomic Next-Generation Sequencing in Infective Endocarditis: A Systematic Review. Antibiotics. 2022; 11(12):1798. https://doi.org/10.3390/antibiotics11121798
Chicago/Turabian StyleHaddad, Sara F., Daniel C. DeSimone, Supavit Chesdachai, Danielle J. Gerberi, and Larry M. Baddour. 2022. "Utility of Metagenomic Next-Generation Sequencing in Infective Endocarditis: A Systematic Review" Antibiotics 11, no. 12: 1798. https://doi.org/10.3390/antibiotics11121798
APA StyleHaddad, S. F., DeSimone, D. C., Chesdachai, S., Gerberi, D. J., & Baddour, L. M. (2022). Utility of Metagenomic Next-Generation Sequencing in Infective Endocarditis: A Systematic Review. Antibiotics, 11(12), 1798. https://doi.org/10.3390/antibiotics11121798






