Repurposing α-Adrenoreceptor Blockers as Promising Anti-Virulence Agents in Gram-Negative Bacteria
Abstract
:1. Introduction
2. Results
2.1. Double-Staged Multiple Biological Target Docking Investigation
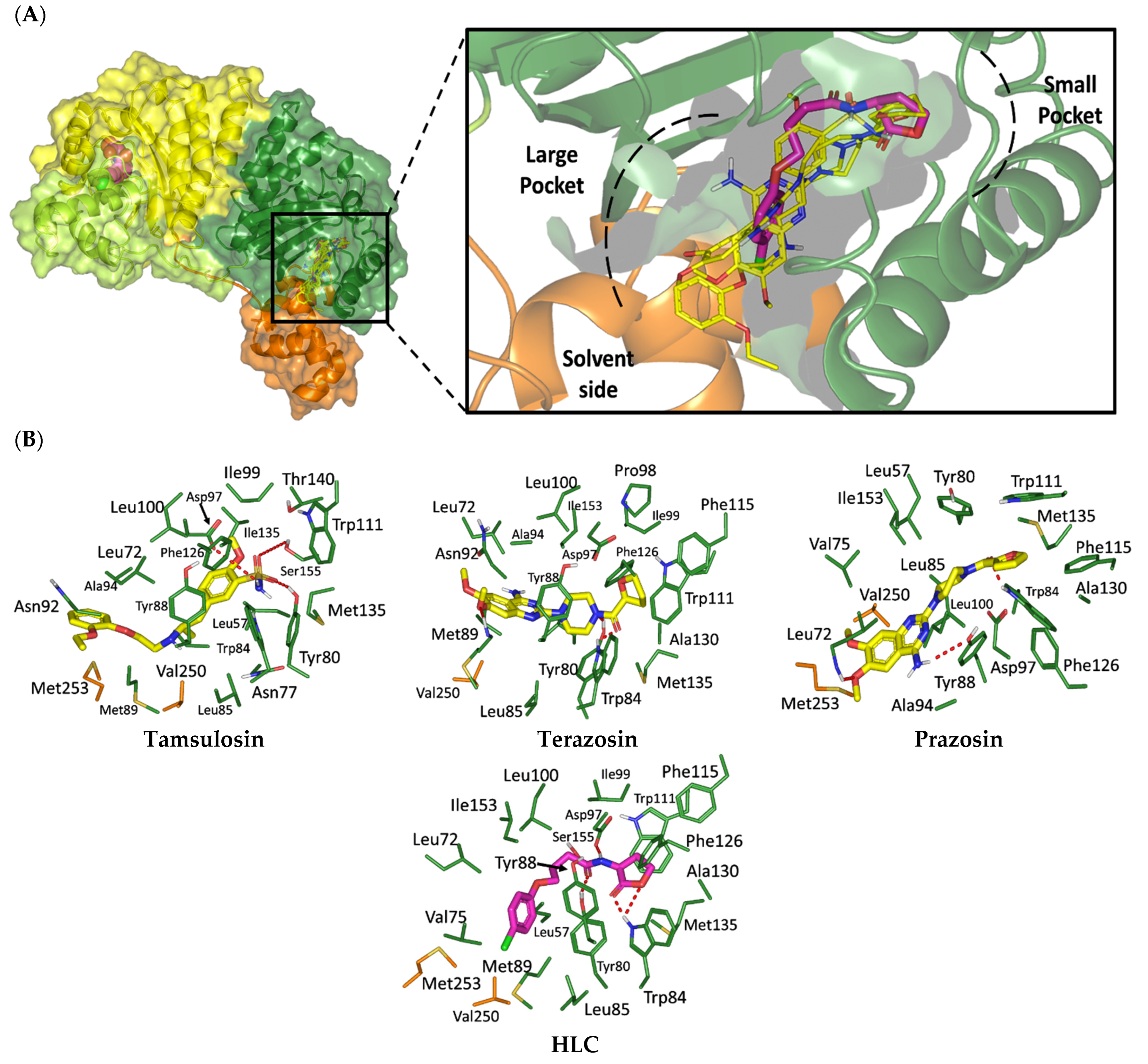
2.1.1. Analysis of Ligand–QscR P. aeruginosa Binding Interactions
2.1.2. Analysis of Ligand–CviR C. violaceum Binding Interactions
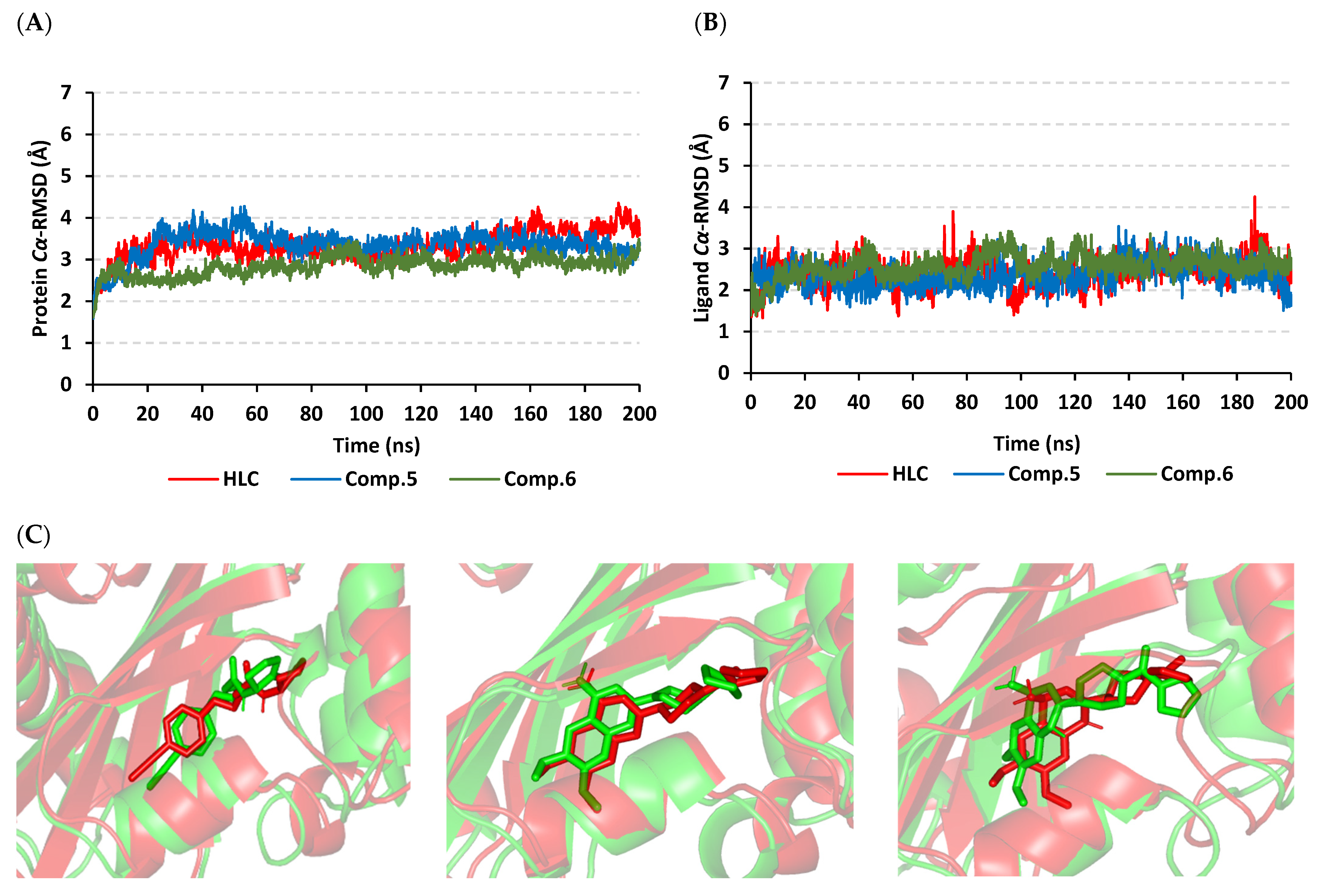
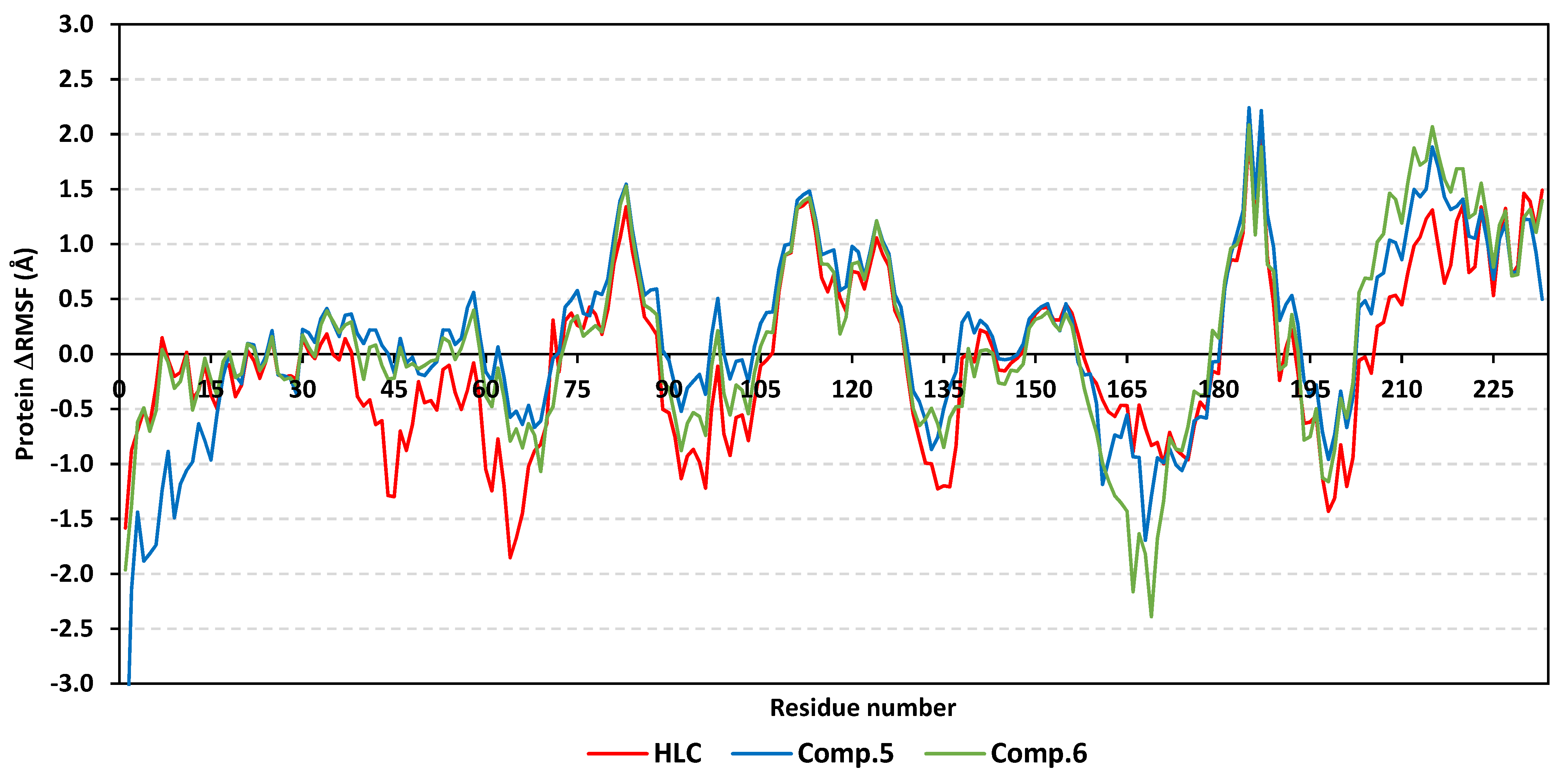
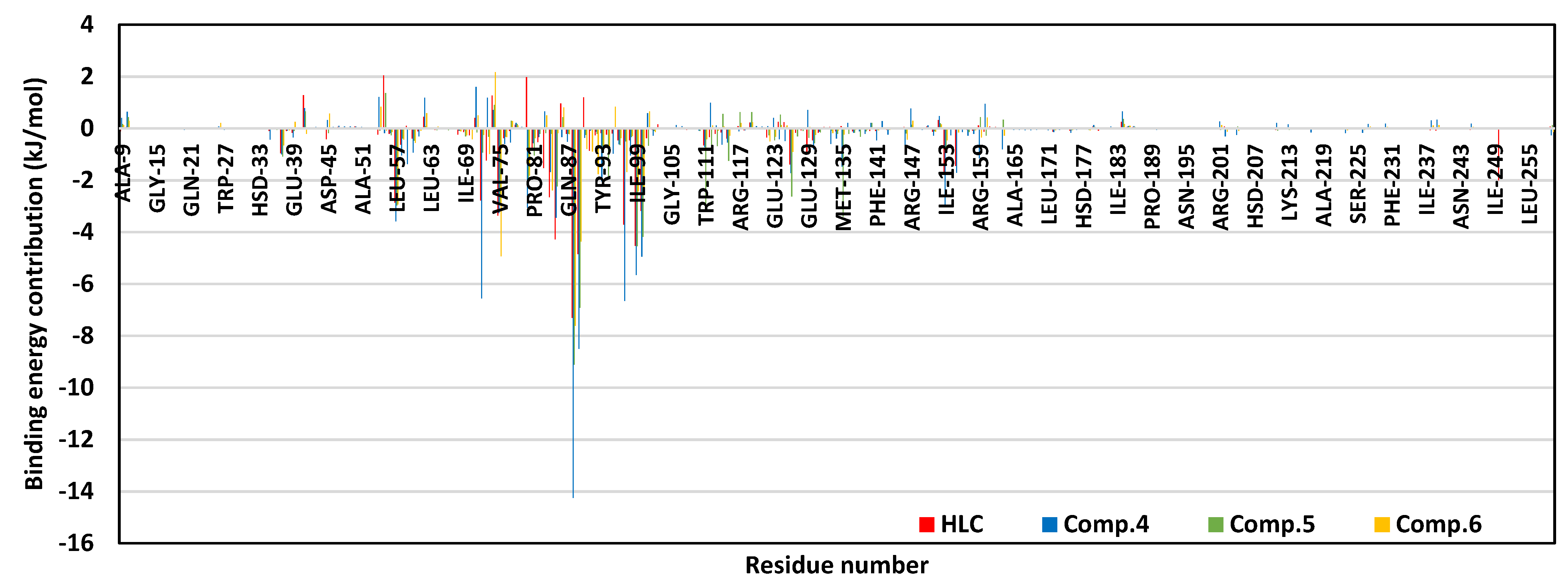
2.2. Molecular Dynamics Simulation
2.2.1. Analysis of Ligand–QscR P. aeruginosa Complex
2.2.2. Ligand–CviR C. violaceum Complex Analysis
2.3. Determination of Minimum Inhibitory Concentration (MIC) of Terazosin against C. violaceum and P. aeruginosa
2.4. Terazosin Inhibited the Violacein Production
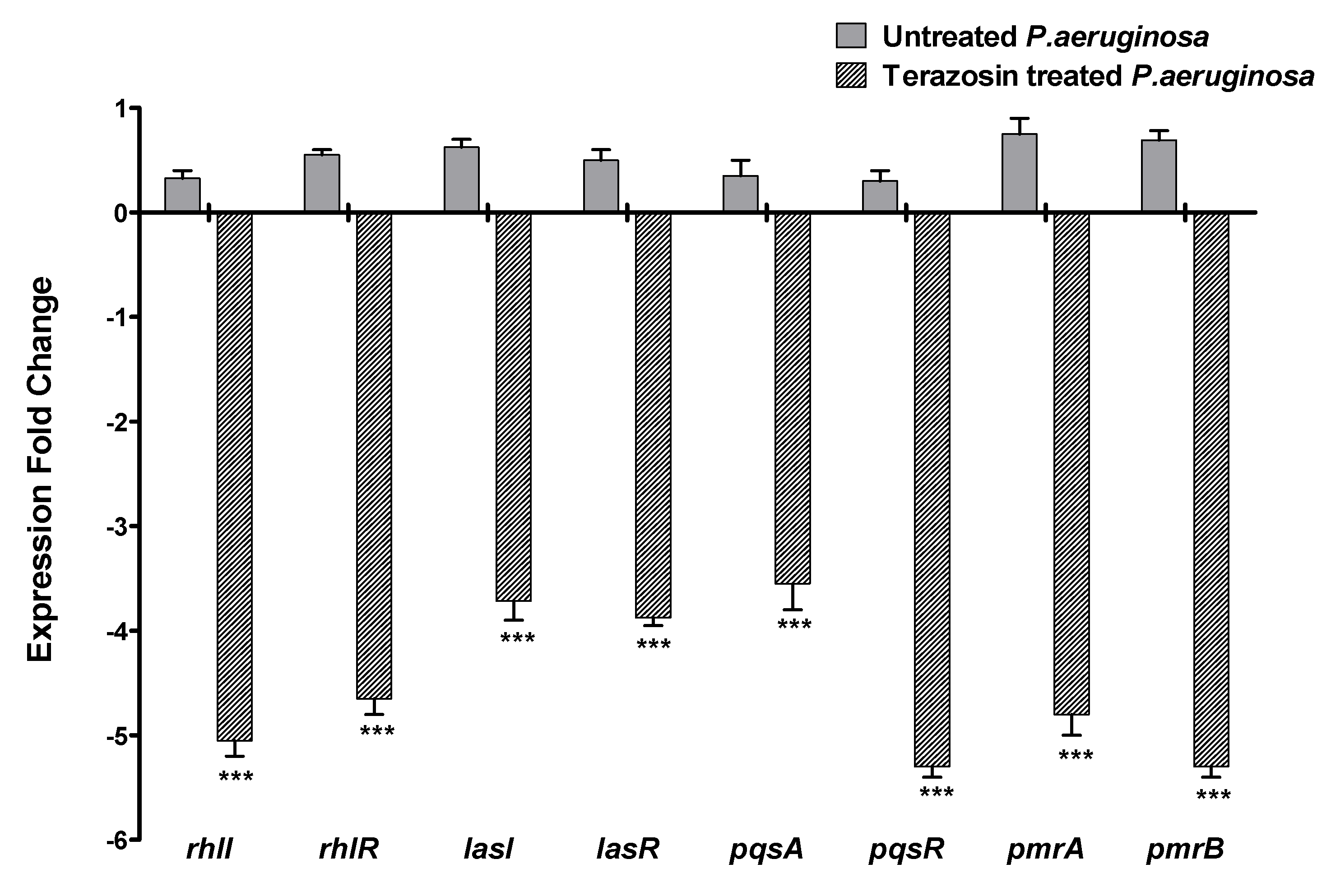
2.5. Terazosin Downregulated the P. aeruginosa Virulence and QS-Encoding Genes
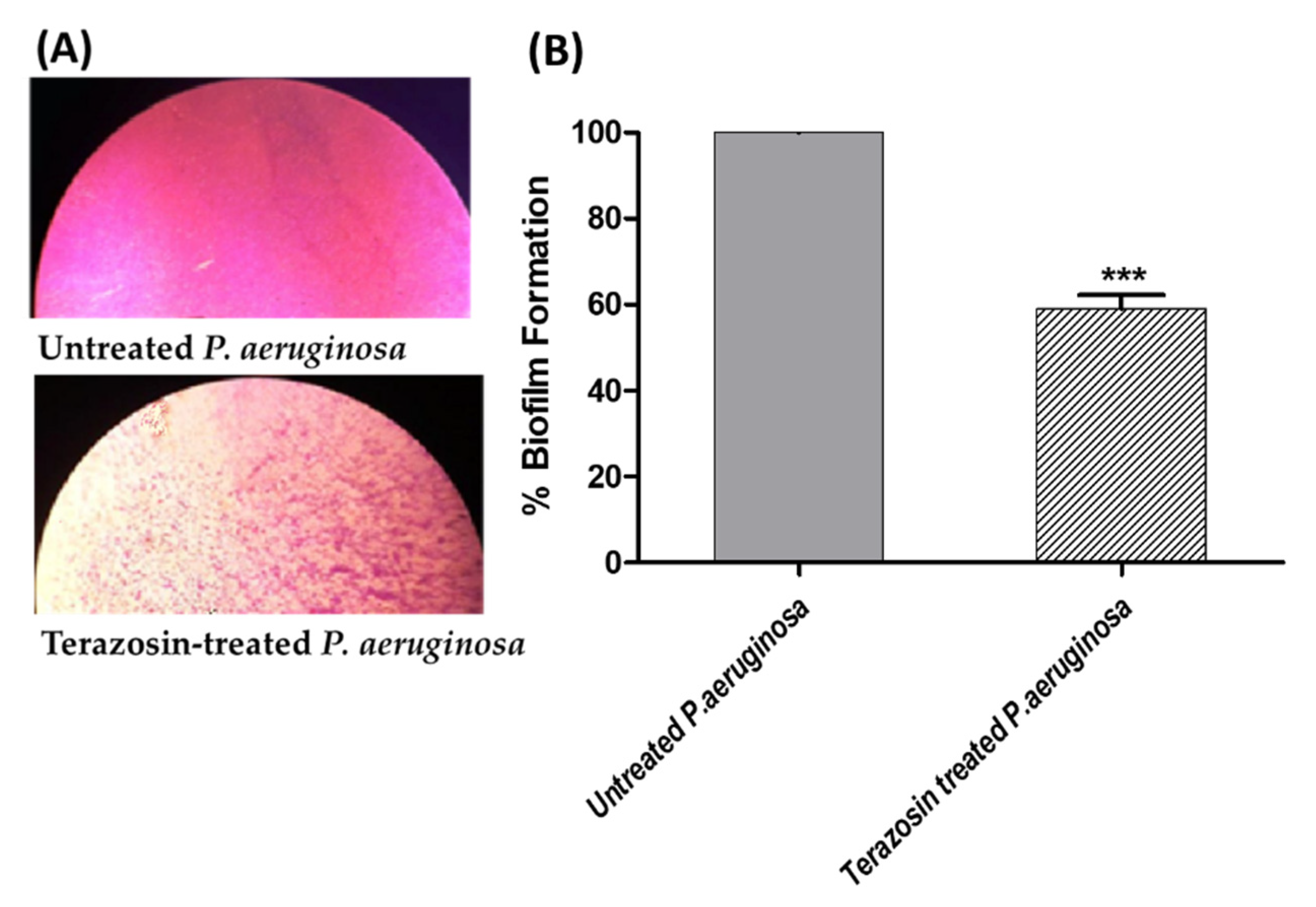
2.6. Terazosin Diminished the Biofilm Formation
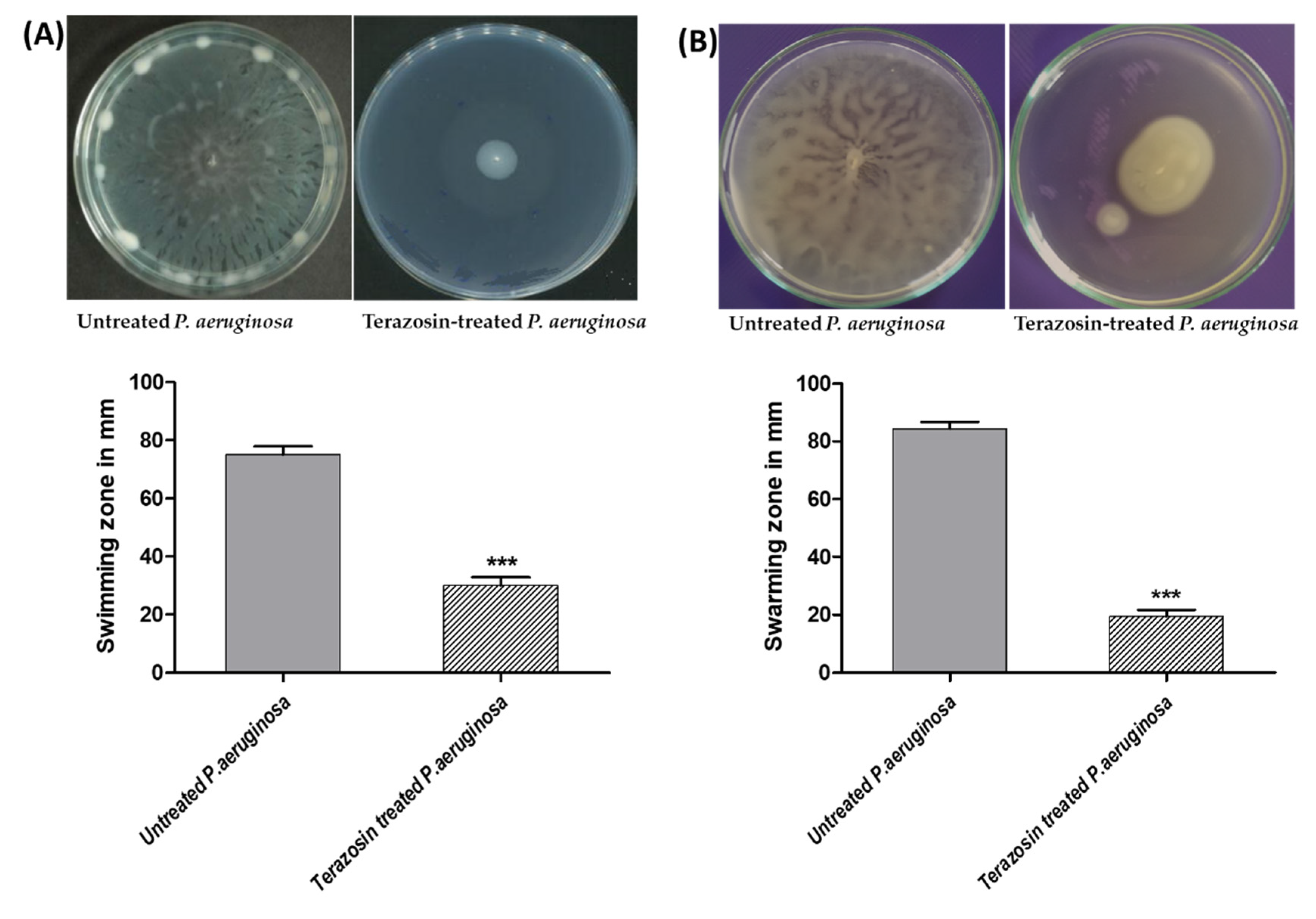
2.7. Terazosin Diminished the P. aeruginosa Motility
2.8. Terazosin Decreased the P. aeruginosa Virulence
2.9. Terazosin Protected Mice against P. aeruginosa
3. Discussion
4. Materials and Methods
4.1. Structural Preparation of QS Targets and Building Ligands for In Silico Investigation
4.2. Double-Staged Multiple Biological Target Docking Investigation
4.3. Molecular Dynamics Simulations
4.4. Bacterial Strains Chemicals and Microbiological Media
4.5. Determination of MIC of Terazosin and Its Effect at Sub-MIC on the Growth of Bacteria
4.6. Evaluation of Violacein Production in C. violaceum
4.7. Quantitative RT-PCR of P. aeruginosa Virulence Involved and QS-Encoding Genes
4.8. Evaluation of P. aeruginosa Biofilm Formation
4.9. Evaluation of P. aeruginosa Motilities
4.10. Evaluation of P. aeruginosa Hemolysins
4.11. Evaluation of P. aeruginosa Protease
4.12. Evaluation of P. aeruginosa Elastase
4.13. Evaluation of P. aeruginosa Pigment Pyocyanin
4.14. Evaluation of P. aeruginosa Resistance to Oxidative Stress
4.15. In Vivo Protection Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Abbas, H.A.; Hegazy, W.A.H. Repurposing anti-diabetic drug “Sitagliptin” as a novel virulence attenuating agent in Serratia marcescens. PLoS ONE 2020, 15, e0231625. [Google Scholar]
- Hegazy, W.A.H.; Khayat, M.T.; Ibrahim, T.S.; Nassar, M.S.; Bakhrebah, M.A.; Abdulaal, W.H.; Alhakamy, N.A.; Bendary, M.M. Repurposing Anti-diabetic Drugs to Cripple Quorum Sensing in Pseudomonas aeruginosa. Microorganisms 2020, 8, 1285. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Z.; Zhi, D.G. Ligand-protein inverse docking and its potential use in the computer search of protein targets of a small molecule. Proteins 2001, 43, 217–226. [Google Scholar] [CrossRef]
- Meng, X.-Y.; Zhang, H.-X.; Mezei, M.; Cui, M. Molecular docking: A powerful approach for structure-based drug discovery. Curr. Comput. Aided Drug Des. 2011, 7, 146–157. [Google Scholar] [CrossRef]
- Huang, H.; Zhang, G.; Zhou, Y.; Lin, C.; Chen, S.; Lin, Y.; Mai, S.; Huang, Z. Reverse Screening Methods to Search for the Protein Targets of Chemopreventive Compounds. Front. Chem. 2018, 6, 138. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Geng, Q.; Pan, Z.; Li, X.; Tie, L.; Pan, Y.; Li, X. Clarifying off-target effects for torcetrapib using network pharmacology and reverse docking approach. BMC Syst. Biol. 2012, 6, 152. [Google Scholar] [CrossRef] [Green Version]
- Sertkaya, A.; Wong, H.-H.; Jessup, A.; Beleche, T. Key cost drivers of pharmaceutical clinical trials in the United States. Clin. Trials 2016, 13, 117–126. [Google Scholar] [CrossRef]
- Abd El-Hamid, M.I.; ES, Y.E.-N.; T, M.K.; Hegazy, W.A.H.; Mosbah, R.A.; Nassar, M.S.; Bakhrebah, M.A.; Abdulaal, W.H.; Alhakamy, N.A.; Bendary, M.M. Promising Antibiofilm Agents: Recent Breakthrough against Biofilm Producing Methicillin-Resistant Staphylococcus aureus. Antibiotics 2020, 9, 667. [Google Scholar] [CrossRef]
- Bendary, M.M.; Ibrahim, D.; Mosbah, R.A.; Mosallam, F.; Hegazy, W.A.H.; Awad, N.F.S.; Alshareef, W.A.; Alomar, S.Y.; Zaitone, S.A.; Abd El-Hamid, M.I. Thymol Nanoemulsion: A New Therapeutic Option for Extensively Drug Resistant Foodborne Pathogens. Antibiotics 2020, 10, 25. [Google Scholar] [CrossRef]
- Hegazy, W.A.H.; Khayat, M.T.; Ibrahim, T.S.; Youns, M.; Mosbah, R.; Soliman, W.E. Repurposing of antidiabetics as Serratia marcescens virulence inhibitors. Braz. J. Microbiol. 2021, 52, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Khayyat, A.N.; Abbas, H.A.; Mohamed, M.F.A.; Asfour, H.Z.; Khayat, M.T.; Ibrahim, T.S.; Youns, M.; Khafagy, E.-S.; Abu Lila, A.S.; Safo, M.K.; et al. Not Only Antimicrobial: Metronidazole Mitigates the Virulence of Proteus mirabilis Isolated from Macerated Diabetic Foot Ulcer. Appl. Sci. 2021, 11, 6847. [Google Scholar] [CrossRef]
- Hentzer, M.; Wu, H.; Andersen, J.B.; Riedel, K.; Rasmussen, T.B.; Bagge, N.; Kumar, N.; Schembri, M.A.; Song, Z.; Kristoffersen, P.; et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 2003, 22, 3803–3815. [Google Scholar] [CrossRef] [PubMed]
- Rasko, D.A.; Sperandio, V. Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 2010, 9, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Hegazy, W.A.H.; Rajab, A.A.H.; Abu Lila, A.S.; Abbas, H.A. Anti-diabetics and antimicrobials: Harmony of mutual interplay. World J. Diabetes 2021, 12, 1832–1855. [Google Scholar] [CrossRef]
- Blair, J.M.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Muhlen, S.; Dersch, P. Anti-virulence Strategies to Target Bacterial Infections. Curr. Top. Microbiol. Immunol. 2016, 398, 147–183. [Google Scholar]
- Stevens, A.M.; Dolan, K.M.; Greenberg, E.P. Synergistic binding of the Vibrio fischeri LuxR transcriptional activator domain and RNA polymerase to the lux promoter region. Proc. Natl. Acad. Sci. USA 1994, 91, 12619–12623. [Google Scholar] [CrossRef] [Green Version]
- Askoura, M.; Hegazy, W.A.H. Ciprofloxacin interferes with Salmonella Typhimurium intracellular survival and host virulence through repression of Salmonella pathogenicity island-2 (SPI-2) genes expression. Pathog. Dis. 2020, 78, ftaa011. [Google Scholar] [CrossRef]
- Vishwa, B.; Moin, A.; Gowda, D.V.; Rizvi, S.M.D.; Hegazy, W.A.H.; Abu Lila, A.S.; Khafagy, E.S.; Allam, A.N. Pulmonary Targeting of Inhalable Moxifloxacin Microspheres for Effective Management of Tuberculosis. Pharmaceutics 2021, 13, 79. [Google Scholar] [CrossRef]
- Al Saqr, A.; Khafagy, E.S.; Alalaiwe, A.; Aldawsari, M.F.; Alshahrani, S.M.; Anwer, M.K.; Khan, S.; Lila, A.S.A.; Arab, H.H.; Hegazy, W.A.H. Synthesis of Gold Nanoparticles by Using Green Machinery: Characterization and In Vitro Toxicity. Nanomaterials 2021, 11, 808. [Google Scholar] [CrossRef] [PubMed]
- Hegazy, W.A.H. Diclofenac inhibits virulence of Proteus mirabilis isolated from diabetic foot ulcer. Afr. J. Microbiol. Res. 2016, 10, 733–743. [Google Scholar]
- Khayyat, A.N.; Abbas, H.A.; Khayat, M.T.; Shaldam, M.A.; Askoura, M.; Asfour, H.Z.; Khafagy, E.-S.; Abu Lila, A.S.; Allam, A.N.; Hegazy, W.A.H. Secnidazole Is a Promising Imidazole Mitigator of Serratia marcescens Virulence. Microorganisms 2021, 9, 2333. [Google Scholar] [CrossRef] [PubMed]
- Khayyat, A.N.; Hegazy, W.A.H.; Shaldam, M.A.; Mosbah, R.; Almalki, A.J.; Ibrahim, T.S.; Khayat, M.T.; Khafagy, E.S.; Soliman, W.E.; Abbas, H.A. Xylitol Inhibits Growth and Blocks Virulence in Serratia marcescens. Microorganisms 2021, 9, 1083. [Google Scholar] [CrossRef]
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef]
- LaSarre, B.; Federle, M.J. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol. Mol. Biol. Rev. 2013, 77, 73–111. [Google Scholar] [CrossRef] [Green Version]
- Askoura, M.; Youns, M.; Halim Hegazy, W.A. Investigating the influence of iron on Campylobacter jejuni transcriptome in response to acid stress. Microb. Pathog. 2020, 138, 103777. [Google Scholar] [CrossRef]
- Hegazy, W.A.H.; Henaway, M. Hepatitis C virus pathogenesis: Serum IL-33 level indicates liver damage. Afr. J. Microbiol. Res. 2015, 9, 1386–1393. [Google Scholar]
- Abbas, H.A.; Hegazy, W.A.H. Targeting the virulence factors of Serratia marcescens by ambroxol. Roum. Arch. Microbiol. Immunol. 2017, 76, 27–32. [Google Scholar]
- Saqr, A.A.; Aldawsari, M.F.; Khafagy, E.-S.; Shaldam, M.A.; Hegazy, W.A.H.; Abbas, H.A. A Novel Use of Allopurinol as A Quorum-Sensing Inhibitor in Pseudomonas aeruginosa. Antibiotics 2021, 10, 1385. [Google Scholar] [CrossRef]
- García-Contreras, R. Is Quorum Sensing Interference a Viable Alternative to Treat Pseudomonas aeruginosa Infections? Front. Microbiol. 2016, 7, 1454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maeda, T.; García-Contreras, R.; Pu, M.; Sheng, L.; Garcia, L.R.; Tomás, M.; Wood, T.K. Quorum quenching quandary: Resistance to antivirulence compounds. ISME J. 2012, 6, 493–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Contreras, R.; Peréz-Eretza, B.; Jasso-Chávez, R.; Lira-Silva, E.; Roldán-Sánchez, J.A.; González-Valdez, A.; Soberón-Chávez, G.; Coria-Jiménez, R.; Martínez-Vázquez, M.; Alcaraz, L.D.; et al. High variability in quorum quenching and growth inhibition by furanone C-30 in Pseudomonas aeruginosa clinical isolates from cystic fibrosis patients. Pathog. Dis. 2015, 73, ftv040. [Google Scholar] [CrossRef] [Green Version]
- Abisado, R.G.; Benomar, S.; Klaus, J.R.; Dandekar, A.A.; Chandler, J.R. Bacterial Quorum Sensing and Microbial Community Interactions. mBio 2018, 9, e02331-17. [Google Scholar] [CrossRef] [Green Version]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Hughes, D.T.; Sperandio, V. Inter-kingdom signalling: Communication between bacteria and their hosts. Nat. Rev. Microbiol. 2008, 6, 111–120. [Google Scholar] [CrossRef] [Green Version]
- Kendall, M.M.; Sperandio, V. What a Dinner Party! Mechanisms and Functions of Interkingdom Signaling in Host-Pathogen Associations. mBio 2016, 7, e01748. [Google Scholar] [CrossRef] [Green Version]
- Karavolos, M.H.; Winzer, K.; Williams, P.; Khan, C.M. Pathogen espionage: Multiple bacterial adrenergic sensors eavesdrop on host communication systems. Mol. Microbiol. 2013, 87, 455–465. [Google Scholar] [CrossRef]
- Flierl, M.A.; Rittirsch, D.; Nadeau, B.A.; Sarma, J.V.; Day, D.E.; Lentsch, A.B.; Huber-Lang, M.S.; Ward, P.A. Upregulation of phagocyte-derived catecholamines augments the acute inflammatory response. PLoS ONE 2009, 4, e4414. [Google Scholar] [CrossRef] [Green Version]
- Lintz, M.J.; Oinuma, K.; Wysoczynski, C.L.; Greenberg, E.P.; Churchill, M.E. Crystal structure of QscR, a Pseudomonas aeruginosa quorum sensing signal receptor. Proc. Natl. Acad. Sci. USA 2011, 108, 15763–15768. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Swem, L.R.; Swem, D.L.; Stauff, D.L.; O’Loughlin, C.T.; Jeffrey, P.D.; Bassler, B.L.; Hughson, F.M. A strategy for antagonizing quorum sensing. Mol. Cell 2011, 42, 199–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Negi, A.; Bhandari, N.; Shyamlal, B.R.K.; Chaudhary, S. Inverse docking based screening and identification of protein targets for Cassiarin alkaloids against Plasmodium falciparum. Saudi Pharm. J. 2018, 26, 546–567. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Negi, A.; Mirallai, S.I.; Warta, R.; Herold-Mende, C.; Carty, M.P.; Ye, X.-S.; Murphy, P.V. N-Alkyl-1,5-dideoxy-1,5-imino-l-fucitols as fucosidase inhibitors: Synthesis, molecular modelling and activity against cancer cell lines. Bioorganic Chem. 2019, 84, 418–433. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef] [Green Version]
- De Souza, A.S.; Pacheco, B.D.C.; Pinheiro, S.; Muri, E.M.F.; Dias, L.R.S.; Lima, C.H.S.; Garrett, R.; de Moraes, M.B.M.; de Souza, B.E.G.; Puzer, L. 3-Acyltetramic acids as a novel class of inhibitors for human kallikreins 5 and 7. Bioorganic Med. Chem. Lett. 2019, 29, 1094–1098. [Google Scholar] [CrossRef] [PubMed]
- Albuquerque, S.O.; Barros, T.G.; Dias, L.R.S.; Lima, C.; Azevedo, P.; Flores-Junior, L.A.P.; Dos Santos, E.G.; Loponte, H.F.; Pinheiro, S.; Dias, W.B.; et al. Biological evaluation and molecular modeling of peptidomimetic compounds as inhibitors for O-GlcNAc transferase (OGT). Eur. J. Pharm. Sci. Off. J. Eur. Fed. Pharm. Sci. 2020, 154, 105510. [Google Scholar] [CrossRef]
- Cavasotto, C.N. Binding Free Energy Calculation Using Quantum Mechanics Aimed for Drug Lead Optimization. Methods Mol. Biol. 2020, 2114, 257–268. [Google Scholar]
- Kumari, R.; Kumar, R.; Lynn, A. g_mmpbsa—A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Modeling 2014, 54, 1951–1962. [Google Scholar] [CrossRef]
- Aldawsari, M.F.; Khafagy, E.S.; Saqr, A.A.; Alalaiwe, A.; Abbas, H.A.; Shaldam, M.A.; Hegazy, W.A.H.; Goda, R.M. Tackling Virulence of Pseudomonas aeruginosa by the Natural Furanone Sotolon. Antibiotics 2021, 10, 871. [Google Scholar] [CrossRef]
- García-Contreras, R.; Nuñez-López, L.; Jasso-Chávez, R.; Kwan, B.W.; Belmont, J.A.; Rangel-Vega, A.; Maeda, T.; Wood, T.K. Quorum sensing enhancement of the stress response promotes resistance to quorum quenching and prevents social cheating. ISME J. 2015, 9, 115–125. [Google Scholar] [CrossRef]
- Swem, L.R.; Swem, D.L.; O’Loughlin, C.T.; Gatmaitan, R.; Zhao, B.; Ulrich, S.M.; Bassler, B.L. A quorum-sensing antagonist targets both membrane-bound and cytoplasmic receptors and controls bacterial pathogenicity. Mol. Cell 2009, 35, 143–153. [Google Scholar] [CrossRef] [Green Version]
- Geske, G.D.; O’Neill, J.C.; Miller, D.M.; Mattmann, M.E.; Blackwell, H.E. Modulation of bacterial quorum sensing with synthetic ligands: Systematic evaluation of N-acylated homoserine lactones in multiple species and new insights into their mechanisms of action. J. Am. Chem. Soc. 2007, 129, 13613–13625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kontoyianni, M.; McClellan, L.M.; Sokol, G.S. Evaluation of Docking Performance: Comparative Data on Docking Algorithms. J. Med. Chem. 2004, 47, 558–565. [Google Scholar] [CrossRef] [PubMed]
- Askoura, M.; Almalki, A.J.; Lila, A.S.A.; Almansour, K.; Alshammari, F.; Khafagy, E.-S.; Ibrahim, T.S.; Hegazy, W.A.H. Alteration of Salmonella enterica Virulence and Host Pathogenesis through Targeting sdiA by Using the CRISPR-Cas9 System. Microorganisms 2021, 9, 2564. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Tong, X.; Sun, P.; Bi, L.; Lin, K. Virtual screening and biological evaluation of biofilm inhibitors on dual targets in quorum sensing system. Future Med. Chem. 2017, 9, 1983–1994. [Google Scholar] [CrossRef] [PubMed]
- Sadiq, S.; Rana, N.F.; Zahid, M.A.; Zargaham, M.K.; Tanweer, T.; Batool, A.; Naeem, A.; Nawaz, A.; Rizwan Ur, R.; Muneer, Z.; et al. Virtual Screening of FDA-Approved Drugs against LasR of Pseudomonas aeruginosa for Antibiofilm Potential. Molecules 2020, 25, 3723. [Google Scholar] [CrossRef]
- Vetrivel, A.; Natchimuthu, S.; Subramanian, V.; Murugesan, R. High-Throughput Virtual Screening for a New Class of Antagonist Targeting LasR of Pseudomonas aeruginosa. ACS Omega 2021, 6, 18314–18324. [Google Scholar] [CrossRef]
- Martins, F.G.; Melo, A.; Sousa, S.F. Identification of New Potential Inhibitors of Quorum Sensing through a Specialized Multi-Level Computational Approach. Molecules 2021, 26, 2600. [Google Scholar] [CrossRef]
- Ravichandran, V.; Zhong, L.; Wang, H.; Yu, G.; Zhang, Y.; Li, A. Virtual Screening and Biomolecular Interactions of CviR-Based Quorum Sensing Inhibitors Against Chromobacterium violaceum. Front. Cell. Infect. Microbiol. 2018, 8, 292. [Google Scholar] [CrossRef]
- Herrera-Arizmendi, J.L.; Curiel-Quesada, E.; Correa-Basurto, J.; Bello, M.; Reyes-Arellano, A. Effect of New Analogs of Hexyloxy Phenyl Imidazoline on Quorum Sensing in Chromobacterium violaceum and In Silico Analysis of Ligand-Receptor Interactions. J. Chem. 2020, 2020, 8735190. [Google Scholar] [CrossRef] [Green Version]
- Venkatramanan, M.; Sankar Ganesh, P.; Senthil, R.; Akshay, J.; Veera Ravi, A.; Langeswaran, K.; Vadivelu, J.; Nagarajan, S.; Rajendran, K.; Shankar, E.M. Inhibition of Quorum Sensing and Biofilm Formation in Chromobacterium violaceum by Fruit Extracts of Passiflora edulis. ACS Omega 2020, 5, 25605–25616. [Google Scholar] [CrossRef] [PubMed]
- Arnittali, M.; Rissanou, A.N.; Harmandaris, V. Structure Of Biomolecules Through Molecular Dynamics Simulations. Procedia Comput. Sci. 2019, 156, 69–78. [Google Scholar] [CrossRef]
- Benson, N.C.; Daggett, V. A comparison of multiscale methods for the analysis of molecular dynamics simulations. J. Phys. Chem. B 2012, 116, 8722–8731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.G.; Pappas, K.M.; Brace, J.L.; Miller, P.C.; Oulmassov, T.; Molyneaux, J.M.; Anderson, J.C.; Bashkin, J.K.; Winans, S.C.; Joachimiak, A. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature 2002, 417, 971–974. [Google Scholar] [CrossRef]
- Mellini, M.; Di Muzio, E.; D’Angelo, F.; Baldelli, V.; Ferrillo, S.; Visca, P.; Leoni, L.; Polticelli, F.; Rampioni, G. In silico Selection and Experimental Validation of FDA-Approved Drugs as Anti-quorum Sensing Agents. Front. Microbiol. 2019, 10, 2355. [Google Scholar] [CrossRef]
- Passos da Silva, D.; Patel, H.K.; González, J.F.; Devescovi, G.; Meng, X.; Covaceuszach, S.; Lamba, D.; Subramoni, S.; Venturi, V. Studies on synthetic LuxR solo hybrids. Front. Cell. Infect. Microbiol. 2015, 5, 52. [Google Scholar] [CrossRef] [Green Version]
- McCready, A.R.; Paczkowski, J.E.; Henke, B.R.; Bassler, B.L. Structural determinants driving homoserine lactone ligand selection in the Pseudomonas aeruginosa LasR quorum-sensing receptor. Proc. Natl. Acad. Sci. USA 2019, 116, 245. [Google Scholar]
- Zhao, X.; Yu, Z.; Ding, T. Quorum-Sensing Regulation of Antimicrobial Resistance in Bacteria. Microorganisms 2020, 8, 425. [Google Scholar] [CrossRef] [Green Version]
- Fong, J.; Zhang, C.; Yang, R.; Boo, Z.Z.; Tan, S.K.; Nielsen, T.E.; Givskov, M.; Liu, X.-W.; Bin, W.; Su, H.; et al. Combination Therapy Strategy of Quorum Quenching Enzyme and Quorum Sensing Inhibitor in Suppressing Multiple Quorum Sensing Pathways of P. aeruginosa. Sci. Rep. 2018, 8, 1155. [Google Scholar] [CrossRef] [Green Version]
- Harrison, A.M.; Soby, S.D. Reclassification of Chromobacterium violaceum ATCC 31532 and its quorum biosensor mutant CV026 to Chromobacterium subtsugae. AMB Express 2020, 10, 202. [Google Scholar] [CrossRef]
- Horcajada, J.P.; Montero, M.; Oliver, A.; Sorli, L.; Luque, S.; Gomez-Zorrilla, S.; Benito, N.; Grau, S. Epidemiology and Treatment of Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa Infections. Clin. Microbiol. Rev. 2019, 32, e00031-19. [Google Scholar] [CrossRef] [PubMed]
- Moradali, M.F.; Ghods, S.; Rehm, B.H. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front. Cell Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, G.; He, J.; Rahme, L.G. Mutation analysis of the Pseudomonas aeruginosa mvfR and pqsABCDE gene promoters demonstrates complex quorum-sensing circuitry. Microbiology 2006, 152 Pt 6, 1679–1686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pacheco, A.R.; Sperandio, V. Inter-kingdom signaling: Chemical language between bacteria and host. Curr. Opin. Microbiol. 2009, 12, 192–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreira, C.G.; Sperandio, V. Interplay between the QseC and QseE bacterial adrenergic sensor kinases in Salmonella enterica serovar Typhimurium pathogenesis. Infect. Immun. 2012, 80, 4344–4353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmer, B.M. Cell-to-cell signalling in Escherichia coli and Salmonella enterica. Mol. Microbiol. 2004, 52, 933–945. [Google Scholar] [CrossRef]
- Rasko, D.A.; Moreira, C.G.; Li de, R.; Reading, N.C.; Ritchie, J.M.; Waldor, M.K.; Williams, N.; Taussig, R.; Wei, S.; Roth, M.; et al. Targeting QseC signaling and virulence for antibiotic development. Science 2008, 321, 1078–1080. [Google Scholar] [CrossRef] [Green Version]
- Okada, B.K.; Li, A.; Seyedsayamdost, M.R. Identification of the Hypertension Drug Guanfacine as an Antivirulence Agent in Pseudomonas aeruginosa. Chembiochem 2019, 20, 2005–2011. [Google Scholar] [CrossRef]
- Francis, V.I.; Stevenson, E.C.; Porter, S.L. Two-component systems required for virulence in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef]
- Moskowitz, S.M.; Ernst, R.K.; Miller, S.I. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J. Bacteriol. 2004, 186, 575–579. [Google Scholar] [CrossRef] [Green Version]
- Medzhitov, R. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 2001, 1, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Aldawsari, M.F.; Alalaiwe, A.; Khafagy, E.S.; Al Saqr, A.; Alshahrani, S.M.; Alsulays, B.B.; Alshehri, S.; Abu Lila, A.S.; Danish Rizvi, S.M.; Hegazy, W.A.H. Efficacy of SPG-ODN 1826 Nanovehicles in Inducing M1 Phenotype through TLR-9 Activation in Murine Alveolar J774A.1 Cells: Plausible Nano-Immunotherapy for Lung Carcinoma. Int. J. Mol. Sci. 2021, 22, 6833. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.W.; Zhou, D.; Haddad, G.G. Antimicrobial peptides increase tolerance to oxidant stress in Drosophila melanogaster. J. Biol. Chem. 2011, 286, 6211–6218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denton, M.; Kerr, K.; Mooney, L.; Keer, V.; Rajgopal, A.; Brownlee, K.; Arundel, P.; Conway, S. Transmission of colistin-resistant Pseudomonas aeruginosa between patients attending a pediatric cystic fibrosis center. Pediatr. Pulmonol. 2002, 34, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Turnidge, J.; Milne, R.; Nation, R.L.; Coulthard, K. In vitro pharmacodynamic properties of colistin and colistin methanesulfonate against Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Antimicrob. Agents Chemother. 2001, 45, 781–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castro, D.; Cordeiro, I.B.; Taquita, P.; Eberlin, M.N.; Garcia, J.S.; Souza, G.H.M.F.; Arruda, M.A.Z.; Andrade, E.V.; Filho, S.A.; Crainey, J.L.; et al. Proteomic analysis of Chromobacterium violaceum and its adaptability to stress. BMC Microbiol. 2015, 15, 272. [Google Scholar] [CrossRef] [Green Version]
- Lin, Q.; Pilewski, J.M.; Di, Y.P. Acidic Microenvironment Determines Antibiotic Susceptibility and Biofilm Formation of Pseudomonas aeruginosa. Front. Microbiol. 2021, 12, 3459. [Google Scholar] [CrossRef]
- Wadie, M.A.; Kishk, S.M.; Darwish, K.M.; Mostafa, S.M.; Elgawish, M.S. Simultaneous Determination of Losartan and Rosuvastatin in Rat Plasma Using Liquid Chromatography–Tandem Mass Spectrometric Technique for Application into Pharmacokinetic and Drug–Drug Interaction Studies. Chromatographia 2020, 83, 1477–1494. [Google Scholar] [CrossRef]
- Malebari, A.; Ibrahim, T.; Salem, I.; Salama, I.; Khayyat, A.; Mostafa, S.; El-Sabbagh, O.; Darwish, K. The Anticancer Activity for the Bumetanide-Based Analogs via Targeting the Tumor-Associated Membrane Bound Human Carbonic Anhydrase-IX Enzyme. Pharmaceuticals 2020, 13, 252. [Google Scholar] [CrossRef]
- El Raey, M.A.; El-Hagrassi, A.M.; Osman, A.F.; Darwish, K.M.; Emam, M. Acalypha wilkesiana flowers: Phenolic profiling, cytotoxic activity of their biosynthesized silver nanoparticles and molecular docking study for its constituents as Topoisomerase-I inhibitors. Biocatal. Agric. Biotechnol. 2019, 20, 101243. [Google Scholar] [CrossRef]
- Saenz-Méndez, P.; Eriksson, M.; Eriksson, L.A. Ligand Selectivity between the ADP-Ribosylating Toxins: An Inverse-Docking Study for Multitarget Drug Discovery. ACS Omega 2017, 2, 1710–1719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ban, T.; Ohue, M.; Akiyama, Y. Multiple grid arrangement improves ligand docking with unknown binding sites: Application to the inverse docking problem. Comput. Biol. Chem. 2018, 73, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Soga, S.; Shirai, H.; Kobori, M.; Hirayama, N. Use of amino acid composition to predict ligand-binding sites. J. Chem. Inf. Modeling 2007, 47, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Edelsbrunner, H.; Woodward, C. Anatomy of protein pockets and cavities: Measurement of binding site geometry and implications for ligand design. Protein Sci. Publ. Protein Soc. 1998, 7, 1884–1897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitchen, D.B.; Decornez, H.; Furr, J.R.; Bajorath, J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat. Reviews.Drug Discov. 2004, 3, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Wojciechowski, M.; Lesyng, B. Generalized Born Model: Analysis, Refinement, and Applications to Proteins. J. Phys. Chem. B 2004, 108, 18368–18376. [Google Scholar] [CrossRef]
- Labute, P. The generalized Born/volume integral implicit solvent model: Estimation of the free energy of hydration using London dispersion instead of atomic surface area. J. Comput. Chem. 2008, 29, 1693–1698. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System, 2.0.6; Schrödinger, LLC: New York, NY, USA; Impact, Schrödinger, LLC: New York, NY, USA, 2016; Prime, Schrödinger, LLC: New York, NY, USA, 2020.
- Elhady, S.S.; Abdelhameed, R.F.A.; Malatani, R.T.; Alahdal, A.M.; Bogari, H.A.; Almalki, A.J.; Mohammad, K.A.; Ahmed, S.A.; Khedr, A.I.M.; Darwish, K.M. Molecular Docking and Dynamics Simulation Study of Hyrtios erectus Isolated Scalarane Sesterterpenes as Potential SARS-CoV-2 Dual Target Inhibitors. Biology 2021, 10, 389. [Google Scholar] [CrossRef]
- Páll, S.; Abraham, M.J.; Kutzner, C.; Hess, B.; Lindahl, E. Tackling Exascale Software Challenges in Molecular Dynamics Simulations with GROMACS; Solving Software Challenges for Exascale; Markidis, S., Laure, E., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 3–27. [Google Scholar]
- Ross, G.A.; Rustenburg, A.S.; Grinaway, P.B.; Fass, J.; Chodera, J.D. Biomolecular Simulations under Realistic Macroscopic Salt Conditions. J. Phys. Chem. B 2018, 122, 5466–5486. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Páll, S.; Hess, B. A flexible algorithm for calculating pair interactions on SIMD architectures. Comput. Phys. Commun. 2013, 184, 2641–2650. [Google Scholar] [CrossRef] [Green Version]
- Youns, M.; Askoura, M.; Abbas, H.A.; Attia, G.H.; Khayyat, A.N.; Goda, R.M.; Almalki, A.J.; Khafagy, E.S.; Hegazy, W.A.H. Celastrol Modulates Multiple Signaling Pathways to Inhibit Proliferation of Pancreatic Cancer via DDIT3 and ATF3 Up-Regulation and RRM2 and MCM4 Down-Regulation. Onco. Targets Ther. 2021, 14, 3849–3860. [Google Scholar] [CrossRef]
- Hegazy, W.A.H.; Abbas, H.A. Evaluation of the role of SsaV ‘Salmonella pathogenicity island-2 dependent type III secretion system components on the virulence behavior of Salmonella enterica serovar Typhimurium. Afr. J. Biotechnol. 2017, 16, 718–726. [Google Scholar]
- Johnson, L.; Mulcahy, H.; Kanevets, U.; Shi, Y.; Lewenza, S. Surface-localized spermidine protects the Pseudomonas aeruginosa outer membrane from antibiotic treatment and oxidative stress. J. Bacteriol. 2012, 194, 813–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanz-Garcia, F.; Hernando-Amado, S.; Martinez, J.L. Mutational Evolution of Pseudomonas aeruginosa Resistance to Ribosome-Targeting Antibiotics. Front. Genet. 2018, 9, 451. [Google Scholar] [CrossRef] [Green Version]








| Compound | 2D Structure | Nomenclature | Docking Binding Energy (Kcal/mol) a | |
|---|---|---|---|---|
| 3SZT | 3QP5 | |||
| 1 |  | Phenoxybenzamine | −6.8144 | −6.4353 |
| 2 |  | Phentolamine | −4.7680 | −5.5767 |
| 3 | 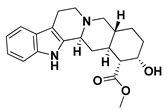 | Yohimbine | −4.2864 | −4.0237 |
| 4 |  | Tamsulosin | −6.9369 | −7.8553 |
| 5 |  | Terazosin | −7.4416 | −7.5163 |
| 6 | 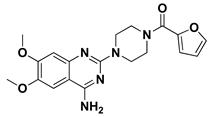 | Prazosin | −7.5679 | −7.2600 |
| 7 | 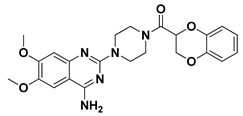 | Doxazosin | −6.6578 | −7.0163 |
| 3SZT Reference |  | O-C12-HSL | −7.5547 | - |
| 3QP5 Reference | 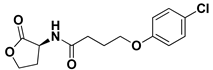 | HLC | −7.6488 | −7.2051 |
| Compound | Docking Binding Energy (kcal/mol) a | H-Bond (Polar) Interactions | Hydrophobic Contacts | π-Interactions | van der Waal with C-Side Chain | |
|---|---|---|---|---|---|---|
| Preliminary (Rigid) | Secondary (Induced-Fit) | |||||
| Terazosin | −7.4416 | −8.0143 | Ser38, Tyr58, Trp62, Met127, Ser129 | Ala41, Tyr52, His53, Tyr58, Trp62, Tyr66, Ile77, Val78, Trp90, Phe101, Trp102, Ile110, Ile125, Met127, Val131 | Phe54 (π–H) Trp102 (π–H) | Arg42 (Cβ) |
| Prazosin | −7.5679 | −8.1023 | Ser38, Tyr58, Trp62, Met127, Ser129 | Ala41, Tyr52, His53, Tyr58, Trp62, Tyr66, Ile77, Val78, Leu82, Trp90, Phe101, Trp102, Ala105, Ile110, Ile125, Met127, Leu128, Val131 | Phe54 (π–π) Trp102 (π–H) | Arg42 (Cβ) |
| HLC | −7.6488 | −7.9912 | Ser38, Tyr58, Trp62, Tyr66, Asp75 | Phe39, Ala41, Tyr52, Tyr58, Trp62, Ile77, Val78, Phe101, Trp102, Ala105, Ile110, Ile125, Met127 | Phe54 (π–π) Trp90 (π–H) | Arg42 (Cβ) |
| Compound | Docking Binding Energy (kcal/mol) a | H-Bond (Polar) Interactions | Hydrophobic Contacts | Π Interactions | van der Waal with C-Side Chain | |
|---|---|---|---|---|---|---|
| Preliminary (Rigid) | Secondary (Induced-Fit) | |||||
| Tamsulosin | −7.8553 | −8.7628 | Tyr80, Asp97, Ser115 | Leu57, Leu72, Val75, Trp84, Leu85, Met89, Ile99, Leu100, Trp111, Phe126, Met135, Ile153, Val250, Met253, Met257 | Tyr80 (π–π) Tyr88 (π–H) | Asn92 (Cβ) |
| Terazosin | −7.5163 | −8.3934 | Tyr80, Met89, Trp84, Trp111 | Leu57, Ala59, Leu72, Val75, Trp84, Leu85, Tyr88, Met89, Ala94, Pro98, Ile99, Leu100, Phe115, Phe126, Ala130, Met135, Ile153, Val250, Met253 | Tyr80 (π–H) Trp111 (π–H) | Arg42 (Cβ) |
| Prazosin | −7.2600 | −8.0092 | Leu72, Trp84, Tyr88 | Leu57, Leu72, Val75, Trp84, Leu85, Ala94, Ile99, Leu100, Phe115, Phe126, Ala130, Met135, Ile153, Val250, Met253 | Leu72 (π–H) Tyr80 (π–H) Tyr88 (π–π) Trp111 (π–π) | - |
| HLC | −7.2051 | −8.08374 | Tyr80, Trp84 *, Asp97 | Leu57, Leu72, Val75, Trp84, Leu85, Met89, Ala94, Ile99, Leu100, Phe115, Phe126, Ala130, Met135, Ile153, Val250, Met253 | Tyr80 (π–H) Tyr88 (π–π) Trp111 (π–H) | - |
| Energy (kJ/mol ± SD) | Ligand–Protein Complexes | ||
|---|---|---|---|
| HLC | Comp. 5 | Comp. 6 | |
| ΔGvan der Waal | −122.79 ± 14.13 | −241.58 ± 15.41 | −239.11 ± 2.66 |
| ΔGElectrostatics | −46.75 ± 2.55 | −56.77 ± 2.86 | −72.40 ± 2.83 |
| ΔGSolvation; Polar | 120.42 ± 1.28 | 217.32 ± 4.88 | 233.61 ± 17.03 |
| ΔGSolvation; Apolar; Only-SASA | −18.75 ± 0.04 | −21.19 ± 0.95 | −22.01 ± 0.97 |
| ΔGTotal binding | −67.87 ± 10.34 | −102.22 ± 6.73 | −99.91 ± 10.58 |
| Energy (kJ/mol ± SD) | Ligand–Protein Complexes | |||
|---|---|---|---|---|
| HLC | Comp. 4 | Comp. 5 | Comp. 6 | |
| ΔGvan der Waal | −170.53 ± 20.47 | −228.70 ± 2.05 | −193.04 ± 10.12 | −120.62 ± 8.69 |
| ΔGElectrostatics | −29.09 ± 11.43 | −84.10 ± 9.68 | −46.61 ± 5.18 | −52.81 ± 15.96 |
| ΔGSolvation; Polar | 153.34 ± 34.91 | 198.05 ± 11.43 | 183.12 ± 12.17 | 120.50 ± 15.88 |
| ΔGSolvation; Apolar; Only-SASA | −20.13 ± 1.36 | −23.05 ± 10.19 | −22.96 ± 0.84 | −18.00 ± 12.27 |
| ΔGTotal binding | −66.41 ± 1.65 | −137.80 ± 17.97 | −79.49 ± 17.95 | −70.93 ± 29.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almalki, A.J.; Ibrahim, T.S.; Elhady, S.S.; Darwish, K.M.; Hegazy, W.A.H. Repurposing α-Adrenoreceptor Blockers as Promising Anti-Virulence Agents in Gram-Negative Bacteria. Antibiotics 2022, 11, 178. https://doi.org/10.3390/antibiotics11020178
Almalki AJ, Ibrahim TS, Elhady SS, Darwish KM, Hegazy WAH. Repurposing α-Adrenoreceptor Blockers as Promising Anti-Virulence Agents in Gram-Negative Bacteria. Antibiotics. 2022; 11(2):178. https://doi.org/10.3390/antibiotics11020178
Chicago/Turabian StyleAlmalki, Ahmad J., Tarek S. Ibrahim, Sameh S. Elhady, Khaled M. Darwish, and Wael A. H. Hegazy. 2022. "Repurposing α-Adrenoreceptor Blockers as Promising Anti-Virulence Agents in Gram-Negative Bacteria" Antibiotics 11, no. 2: 178. https://doi.org/10.3390/antibiotics11020178
APA StyleAlmalki, A. J., Ibrahim, T. S., Elhady, S. S., Darwish, K. M., & Hegazy, W. A. H. (2022). Repurposing α-Adrenoreceptor Blockers as Promising Anti-Virulence Agents in Gram-Negative Bacteria. Antibiotics, 11(2), 178. https://doi.org/10.3390/antibiotics11020178









