Prevalence, Antibiotic Resistance, Toxin-Typing and Genotyping of Clostridium perfringens in Raw Beef Meats Obtained from Qazvin City, Iran
Abstract
:1. Introduction
2. Results
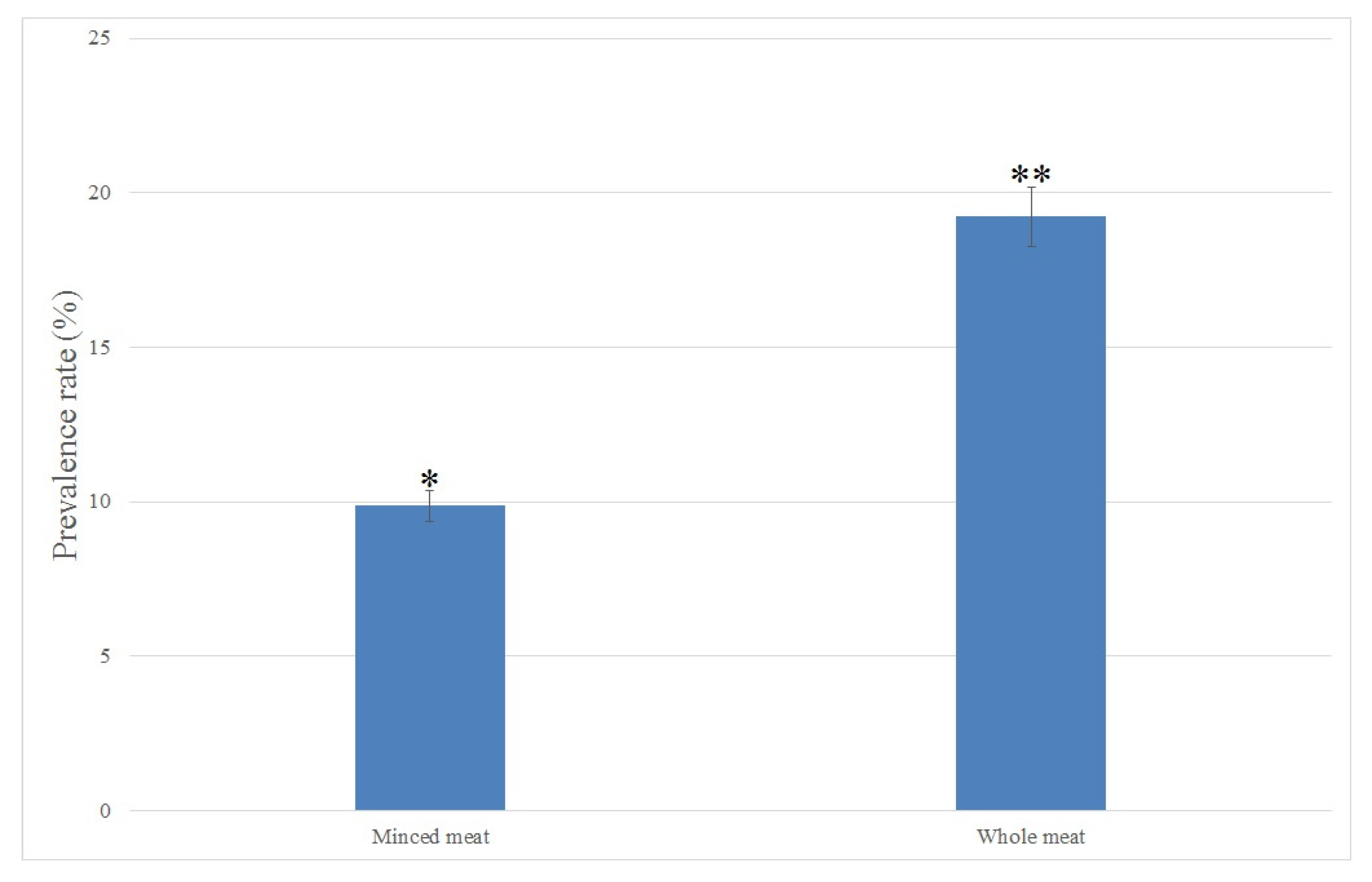
2.1. Isolation and Identification of C. perfringens in Raw Meat Samples
2.2. Antimicrobial Susceptibility Testing of the C. perfringens Isolates
2.3. Toxin-Encoding Genes in C. perfringens Isolates
2.4. Genotyping and Molecular Toxinotyping of the C. perfringens Isolates
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. C. perfringens Isolation
4.3. Antimicrobial Susceptibility Testing
4.4. DNA Extraction
4.5. Identification of Toxin Genes
4.6. Determination of Genetic Diversity
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McClane, B.A.; Robertson, S.L.; Li, J. Clostridium perfringens. In Food Microbiology: Fundamentals and Frontiers; Wiley Publishing: Hoboken, NJ, USA, 2012; pp. 465–489. [Google Scholar]
- Hailegebreal, G. A review on clostridium perfringens food poisoning. Glob. Res. J. Public Health Epidemiol. 2017, 4, 104–109. [Google Scholar]
- Carey, J.; Cole, J.; Venkata, S.L.G.; Hoyt, H.; Mingle, L.; Nicholas, D.; Musser, K.A.; Wolfgang, W.J. Determination of Genomic Epidemiology of Historical Clostridium perfringens Outbreaks in New York State by Use of Two Web-Based Platforms: National Center for Biotechnology Information Pathogen Detection and FDA GalaxyTrakr. J. Clin. Microbiol. 2021, 59, e02200-20. [Google Scholar] [CrossRef] [PubMed]
- Navarro, M.A.; McClane, B.A.; Uzal, F.A. Mechanisms of Action and Cell Death Associated with Clostridium Perfringens Toxins. Toxins 2018, 10, 212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duracova, M.; Klimentova, J.; Fucikova, A.M.; Zidkova, L.; Sheshko, V.; Rehulkova, H.; Dresler, J.; Krocova, Z. Targeted Mass Spectrometry Analysis of Clostridium Perfringens Toxins. Toxins 2019, 11, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rood, J.I.; Adams, V.; Lacey, J.; Lyras, D.; McClane, B.A.; Melville, S.B.; Moore, R.J.; Popoff, M.R.; Sarker, M.R.; Songer, J.G.; et al. Expansion of the Clostridium perfringens toxin-based typing scheme. Anaerobe 2018, 53, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Uzal, F.A.; Freedman, J.C.; Shrestha, A.; Theoret, J.R.; Garcia, J.; Awad, M.M.; Adams, V.; Moore, R.J.; Rood, J.I.; McClane, B.A. Towards an understanding of the role of Clostridium perfringens toxins in human and animal disease. Future Microbiol. 2014, 9, 361–377. [Google Scholar] [CrossRef] [Green Version]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef] [Green Version]
- Sifri, Z.; Chokshi, A.; Cennimo, D.; Horng, H. Global contributors to antibiotic resistance. J. Glob. Infect. Dis. 2019, 11, 36–42. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.J. Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 2018, 42, fux053. [Google Scholar] [CrossRef]
- Adams, V.; Han, X.; Lyras, D.; Rood, J.I. Antibiotic resistance plasmids and mobile genetic elements of Clostridium perfringens. Plasmid 2018, 99, 32–39. [Google Scholar] [CrossRef]
- Bin Zaman, S.; Hussain, M.A.; Nye, R.; Mehta, V.; Mamun, K.T.; Hossain, N. A Review on Antibiotic Resistance: Alarm Bells are Ringing. Cureus 2017, 9, e1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spurgin, L.G.; Richardson, D.S. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc. R. Soc. B Biol. Sci. 2010, 277, 979–988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Y.; Fan, X.; Zhu, L.; Yang, X.; Liu, Y.; Gao, S.; Jin, X.; Liu, D.; Ding, J.; Guo, Y.; et al. Phylogenetic and genomic analysis reveals high genomic openness and genetic diversity of Clostridium perfringens. Microb. Genom. 2020, 6, e000441. [Google Scholar] [CrossRef] [PubMed]
- Xiu, L.; Liu, Y.; Wu, W.; Chen, S.; Zhong, Z.; Wang, H. Prevalence and multilocus sequence typing of Clostridium perfringens isolated from 4 duck farms in Shandong province, China. Poult. Sci. 2020, 99, 5105–5117. [Google Scholar] [CrossRef]
- Chukwu, E.; Nwaokorie, F.; Coker, A.O.; Avila-Campos, M.J.; Solís, R.L.; Llanco, L.; Ogunsola, F.T. Detection of toxigenic Clostridium perfringens and Clostridium botulinum from food sold in Lagos, Nigeria. Anaerobe 2016, 42, 176–181. [Google Scholar] [CrossRef]
- Kahlmeter, G.; Giske, C.G.; Kirn, T.J.; Sharp, S.E. Point-Counterpoint: Differences between the European Committee on Antimicrobial Susceptibility Testing and Clinical and Laboratory Standards Institute Recommendations for Reporting Antimicrobial Susceptibility Results. J. Clin. Microbiol. 2019, 57, e01129-19. [Google Scholar] [CrossRef] [Green Version]
- Humphries, R.; Ambler, J.; Mitchell, S.; Castanheira, M.; Dingle, T.; Hindler, J.; Koeth, L.; Sei, K.; on behalf of the CLSI Methods Development and Standardization Working Group of the Subcommittee on Antimicrobial Susceptibility Testing. CLSI Methods Development and Standardization Working Group best practices for evaluation of antimicrobial susceptibility tests. J. Clin. Microbiol. 2018, 56, e01934-17. [Google Scholar] [CrossRef] [Green Version]
- Chukwu, E.E.; Nwaokorie, F.O.; Coker, A.O.; Avila-Campos, M.J.; Ogunsola, F.T. Genetic variation among Clostridium perfringens isolated from food and faecal specimens in Lagos. Microb. Pathog. 2017, 111, 232–237. [Google Scholar] [CrossRef]
- Pavel, A.B.; Vasile, C.I. PyElph—A software tool for gel images analysis and phylogenetics. BMC Bioinform. 2012, 13, 9. [Google Scholar] [CrossRef] [Green Version]
- NTSYS-PC. Numerical Taxonomy and Multivariate Analyses System: NTSYS-PC 2.2. Available online: http://www.appliedbiostat.com/ntsyspc/ntsyspc.html (accessed on 25 November 2021).
- Ghoneim, N.; Hamza, D. Epidemiological studies on Clostridium perfringens food poisoning in retail foods. Rev. Sci. Tech. 2017, 36, 1025–1032. [Google Scholar] [CrossRef]
- Lee, C.-A.; Labbé, R. Distribution of Enterotoxin- and Epsilon-Positive Clostridium perfringens Spores in U.S. Retail Spices. J. Food Prot. 2018, 81, 394–399. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhou, Y.; Yang, D.; Zhang, S.; Sun, Z.; Wang, Y.; Wang, S.; Wu, C. Prevalence and antimicrobial susceptibility of Clostridium perfringens in chickens and pigs from Beijing and Shanxi, China. Vet. Microbiol. 2020, 252, 108932. [Google Scholar] [CrossRef] [PubMed]
- Kouassi, K.A.; Dadie, A.T.; N’Guessan, K.F.; Dje, K.M.; Loukou, Y.G. Clostridium perfringens and Clostridium difficile in cooked beef sold in Côte d’Ivoire and their antimicrobial susceptibility. Anaerobe 2014, 28, 90–94. [Google Scholar] [CrossRef]
- Maikanov, B.; Mustafina, R.; Auteleyeva, L.; Wiśniewski, J.; Anusz, K.; Grenda, T.; Kwiatek, K.; Goldsztejn, M.; Grabczak, M. Clostridium botulinum and Clostridium perfringens Occurrence in Kazakh Honey Samples. Toxins 2019, 11, 472. [Google Scholar] [CrossRef] [Green Version]
- Stagnitta, P.V.; Micalizzi, B.; de Guzmán, A.M.a.S. Prevalence of enterotoxigenic Clostridium perfringens in meats in San Luis, Argentina. Anaerobe 2002, 8, 253–258. [Google Scholar] [CrossRef]
- Miki, Y.; Miyamoto, K.; Kaneko-Hirano, I.; Fujiuchi, K.; Akimoto, S. Prevalence and Characterization of Enterotoxin Gene-Carrying Clostridium perfringens Isolates from Retail Meat Products in Japan. Appl. Environ. Microbiol. 2008, 74, 5366–5372. [Google Scholar] [CrossRef] [Green Version]
- Aras, Z.; Hadimli, H.H. Detection and molecular typing of Clostridium perfringens isolates from beef, chicken and turkey meats. Anaerobe 2015, 32, 15–17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhang, W.; Ai, D.; Zhang, R.; Lu, Q.; Luo, Q.; Shao, H. Prevalence and characterization of Clostridium perfringens in broiler chickens and retail chicken meat in central China. Anaerobe 2018, 54, 100–103. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.-S.; Kim, D.-H.; Bae, D.; Kim, S.-H.; Kim, H.; Moon, J.-S.; Song, K.-Y.; Chon, J.-W.; Seo, K.-H. Prevalence, toxin-typing, and antimicrobial susceptibility of Clostridium perfringens from retail meats in Seoul, Korea. Anaerobe 2020, 64, 102235. [Google Scholar] [CrossRef]
- Choi, Y.; Kang, J.; Lee, Y.; Seo, Y.; Lee, H.; Kim, S.; Lee, J.; Ha, J.; Oh, H.; Kim, Y.; et al. Quantitative microbial risk assessment for Clostridium perfringens foodborne illness following consumption of kimchi in South Korea. Food Sci. Biotechnol. 2020, 29, 1131–1139. [Google Scholar] [CrossRef]
- Ziomek, M.; Drozd, Ł.; Gondek, M.; Pyz-Łukasik, R.; Pedonese, F.; Florek, M.; Domaradzki, P.; Skałecki, P. Microbiological Changes in Meat and Minced Meat from Beavers (Castor fiber L.) during Refrigerated and Frozen Storage. Foods 2021, 10, 1270. [Google Scholar] [CrossRef]
- Yibar, A.; Cetin, E.; Ata, Z.; Erkose, E.; Tayar, M. Clostridium perfringensContamination in Retail Meat and Meat-Based Products in Bursa, Turkey. Foodborne Pathog. Dis. 2018, 15, 239–245. [Google Scholar] [CrossRef]
- Lerminiaux, N.A.; Cameron, A.D.S. Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 2019, 65, 34–44. [Google Scholar] [CrossRef]
- Martinez, J.L.; Coque, T.M.; Baquero, F. What is a resistance gene? Ranking risk in resistomes. Nat. Rev. Genet. 2014, 13, 116–123. [Google Scholar] [CrossRef]
- Walsh, C.; Fanning, S. Antimicrobial resistance in foodborne pathogens-A cause for concern? Curr. Drug Targets 2008, 9, 808–815. [Google Scholar] [CrossRef]
- Anju, K.; Karthik, K.; Divya, V.; Mala Priyadharshini, M.L.; Sharma, R.K.; Manoharan, S. Toxinotyping and molecular characterization of antimicrobial resistance in Clostridium perfringens isolated from different sources of livestock and poultry. Anaerobe 2020, 67, 102298. [Google Scholar] [CrossRef]
- Park, M.; Rafii, F. The prevalence of plasmid-coded cpe enterotoxin, β2 toxin, tpeL toxin, and tetracycline resistance in Clostridium perfringens strains isolated from different sources. Anaerobe 2019, 56, 124–129. [Google Scholar] [CrossRef]
- Erol, I.; Goncuoglu, M.; Ayaz, N.; Ormancı, F.S.B.; Hildebrandt, G. Molecular typing of Clostridium perfringens isolated from turkey meat by multiplex PCR. Lett. Appl. Microbiol. 2008, 47, 31–34. [Google Scholar] [CrossRef]
- Guran, H.; Oksuztepe, G. Detection and typing of C lostridium perfringens from retail chicken meat parts. Lett. Appl. Microbiol. 2013, 57, 77–82. [Google Scholar] [CrossRef]
- Afshari, A.; Jamshidi, A.; Razmyar, J.; Rad, M. Genotyping of Clostridium perfringens isolated from broiler meat in northeastern of Iran. Vet. Res. Forum Int. 2015, 6, 279–284. [Google Scholar]
- Góra, B.; Gofron, Z.; Grosiak, M.; Aptekorz, M.; Kazek, B.; Kocelak, P.; Radosz-Komoniewska, H.; Chudek, J.; Martirosian, G. Toxin profile of fecal Clostridium perfringens strains isolated from children with autism spectrum disorders. Anaerobe 2018, 51, 73–77. [Google Scholar] [CrossRef]
- Miao, J.; Wang, W.; Xu, W.; Su, J.; Li, L.; Li, B.; Zhang, X.; Xu, Z. The fingerprint mapping and genotyping systems application on methicillin-resistant Staphylococcus aureus. Microb. Pathog. 2018, 125, 246–251. [Google Scholar] [CrossRef]
- Chen, Q.; Xie, S. Genotypes, Enterotoxin Gene Profiles, and Antimicrobial Resistance of Staphylococcus aureus Associated with Foodborne Outbreaks in Hangzhou, China. Toxins 2019, 11, 307. [Google Scholar] [CrossRef] [Green Version]
- Llanco, L.; Nakano, V.; Avila-Campos, M.J. Sialidase Production and Genetic Diversity in Clostridium perfringens Type A Isolated from Chicken with Necrotic Enteritis in Brazil. Curr. Microbiol. 2014, 70, 330–337. [Google Scholar] [CrossRef]
- Hu, W.-S.; Kim, H.; Koo, O.K. Molecular genotyping, biofilm formation and antibiotic resistance of enterotoxigenic Clostridium perfringens isolated from meat supplied to school cafeterias in South Korea. Anaerobe 2018, 52, 115–121. [Google Scholar] [CrossRef]
- Pakbin, B.; Mahmoudi, R.; Mousavi, S.; Allahyari, S.; Amani, Z.; Peymani, A.; Qajarbeygi, P.; Hoseinabadi, Z. Genotypic and antimicrobial resistance characterizations of Cronobacter sakazakii isolated from powdered milk infant formula: A comparison between domestic and imported products. Food Sci. Nutr. 2020, 8, 6708–6717. [Google Scholar] [CrossRef]

| Antibiotic Class | Antibiotic Agent | n (%) | ||
|---|---|---|---|---|
| Whole Meat (n = 10) | Minced Meat (n = 8) | Total (n = 18) | ||
| β-Lactams | imipenem | 4 (40.0) | 1 (12.5) | 5 (27.7) |
| amoxicillin | 7 (70.0) | 4 (50.0) | 11 (61.1) | |
| ampicillin | 7 (70.0) | 6 (75.0) | 13 (72.2) | |
| cefepime | 3 (30.0) | 0 (0.0) | 3 (16.6) | |
| Cephalosporins | ceftriaxone | 4 (40.0) | 0 (0.0) | 4 (22.2) |
| Aminoglycosides | amikacin | 2 (20.0) | 1 (12.5) | 3 (16.6) |
| Fluoroquinolones | ciprofloxacin | 5 (50.0) | 2 (25.0) | 7 (38.8) |
| Phenicols | chloramphenicol | 3 (30.0) | 3 (37.5) | 6 (33.3) |
| Tetracyclines | tetracycline | 7 (70.0) | 5 (62.5) | 12 (66.6) |
| No. Classes of Antibiotics | Patterns of Multidrug Resistance a (No. Isolates in Each Pattern) | No. Total Isolates (%) (n = 18) |
|---|---|---|
| One | βLs (n = 1) | 3 (16.6) |
| TCs (n = 2) | ||
| Two | βLs-TCs (n = 5) | 8 (44.4) |
| βLs-CPs (n = 1) | ||
| βLs-PNs (n = 1) | ||
| QNs-TCs (n = 1) | ||
| Three | βLs-TCs-QNs (n = 1) | 3 (16.6) |
| βLs-PNs-QNs (n = 1) | ||
| βLs-TCs-CPs (n = 1) | ||
| Four | βLs-PNs-QNs-AGs (n = 1) | 2 (11.1) |
| βLs-PNs-QNs-TCs (n = 1) | ||
| Five | βLs-CPs-QNs-TCs-AGs (n = 1) | 1 (5.5) |
| Six | βLs-CPs-QNs-TCs-AGs-PNs (n = 1) | 1 (5.5) |
| No. Sample | Isolate | Source | Resistance Phenotype a | Toxin Genes | Toxin Type | OPA-3 Group |
|---|---|---|---|---|---|---|
| 1 | CPQM19-1 | Whole meat | TET and CIP | cpa+ | A | O1 |
| 2 | CPQM19-2 | Whole meat | IPM, AMX, AMK, AMP, TET, FEP, CHL, CIP, and CRO | cpa+ | A | O1 |
| 3 | CPQM19-3 | Whole meat | AMX, TET, and AMP | cpa+ cpe+ | Ae | O1 |
| 4 | CPQM19-4 | Whole meat | CIP, AMX, AMP, and TET | cpa+ | A | O1 |
| 5 | CPQM19-5 | Whole meat | CRO, FEP, AMX, AMP, and TET | cpa+ etx+ | D | O1 |
| 6 | CPQM19-6 | Whole meat | CRO and AMP | cpa+ | A | O1 |
| 7 | CPQM19-7 | Whole meat | FEP and CHL | cpa+ | A | O2 |
| 8 | CPQM19-8 | Minced meat | TET and AMP | cpa+ etx+ | D | O1 |
| 9 | CPQM19-9 | Minced meat | AMK, CIP, AMX, AMP, and CHL | cpa+ cpe+ | Ae | O2 |
| 10 | CPQM19-10 | Minced meat | AMP | cpa+ cpe+ cpb+ | Ce | O3 |
| 11 | CPQM19-11 | Minced meat | TET | cpa+ cpb+ etx+ | B | O3 |
| 12 | CPQM19-12 | Minced meat | IPM, CIP, AMX, AMP, TET, and CHL | cpa+ etx+ | D | O4 |
| 13 | CPQM19-13 | Minced meat | TET | cpa+ cpe+ | Ae | O3 |
| 14 | CPQM19-14 | Minced meat | AMX, AMP, and TET | cpa+ iap+ | E | O5 |
| 15 | CPQM19-15 | Whole meat | IPM, AMX, AMP, and TET | cpa+ etx+ | D | O2 |
| 16 | CPQM19-16 | Whole meat | IPM, AMX, AMK, AMP, TET, CIP, and CRO | cpa+ | A | O1 |
| 17 | CPQM19-17 | Whole meat | IPM, CIP, AMX, and CHL | cpa+ | A | O2 |
| 18 | CPQM19-18 | Minced meat | FEP, AMX, and TET | cpa+ cpe+ | Ae | O2 |
| Primer | Sequence (5′-3′) | Annealing Temperature (°C) | Amplicon (bp) |
|---|---|---|---|
| Cpa | AGTCTACGCTTGGGATGGAA | 56 | 900 |
| TTTCCTGGGTTGTCCATTTC | |||
| Cpe | GGGGAACCCTCAGTAGTTTCA | 56 | 506 |
| ACCAGCTGGATTTGAGTTTAATG | |||
| Cpb | TCCTTTCTTGAGGGAGGATAAA | 56 | 611 |
| TGAACCTCCTATTTTGTATCCCA | |||
| Etx | TGGGAACTTCGATACAAGCA | 56 | 396 |
| TTAACTCATCTCCCATAACTGCAC | |||
| Iap | AAACGCATTAAAGCTCACACC | 56 | 293 |
| CTGCATAACCTGGAATGGCT | |||
| OPA-3 | AGTCAGCCAC | 42 | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassani, S.; Pakbin, B.; Brück, W.M.; Mahmoudi, R.; Mousavi, S. Prevalence, Antibiotic Resistance, Toxin-Typing and Genotyping of Clostridium perfringens in Raw Beef Meats Obtained from Qazvin City, Iran. Antibiotics 2022, 11, 340. https://doi.org/10.3390/antibiotics11030340
Hassani S, Pakbin B, Brück WM, Mahmoudi R, Mousavi S. Prevalence, Antibiotic Resistance, Toxin-Typing and Genotyping of Clostridium perfringens in Raw Beef Meats Obtained from Qazvin City, Iran. Antibiotics. 2022; 11(3):340. https://doi.org/10.3390/antibiotics11030340
Chicago/Turabian StyleHassani, Samaneh, Babak Pakbin, Wolfram Manuel Brück, Razzagh Mahmoudi, and Shaghayegh Mousavi. 2022. "Prevalence, Antibiotic Resistance, Toxin-Typing and Genotyping of Clostridium perfringens in Raw Beef Meats Obtained from Qazvin City, Iran" Antibiotics 11, no. 3: 340. https://doi.org/10.3390/antibiotics11030340
APA StyleHassani, S., Pakbin, B., Brück, W. M., Mahmoudi, R., & Mousavi, S. (2022). Prevalence, Antibiotic Resistance, Toxin-Typing and Genotyping of Clostridium perfringens in Raw Beef Meats Obtained from Qazvin City, Iran. Antibiotics, 11(3), 340. https://doi.org/10.3390/antibiotics11030340







