Distribution and Genomic Characterization of Third-Generation Cephalosporin-Resistant Escherichia coli Isolated from a Single Family and Home Environment: A 2-Year Longitudinal Study
Abstract
:1. Introduction
2. Results
2.1. Participant Characteristic
2.2. Antimicrobial Susceptibility Profiles
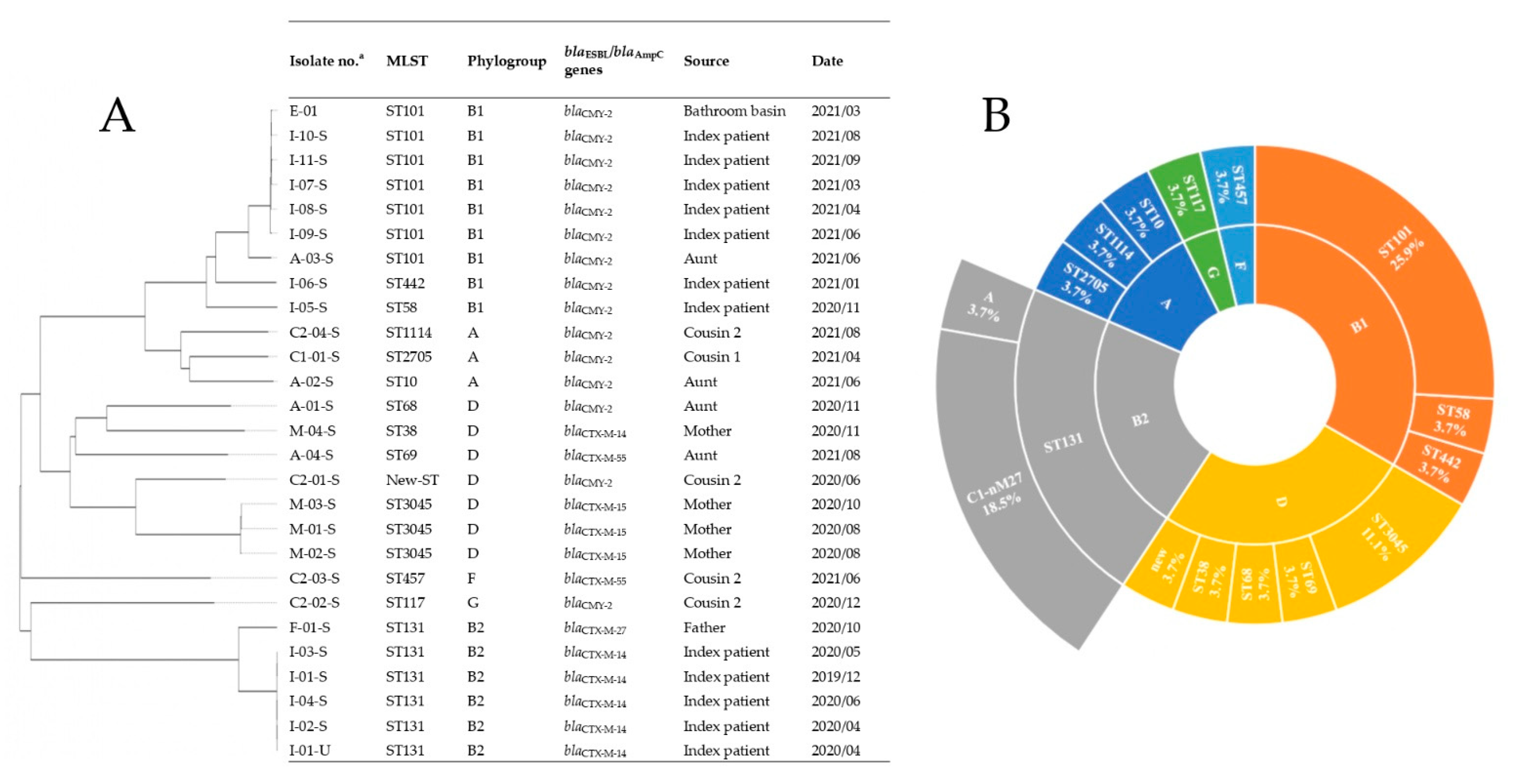
2.3. Genome Sequences, Sequence Types, Serotypes, and Phylogenetic Groups
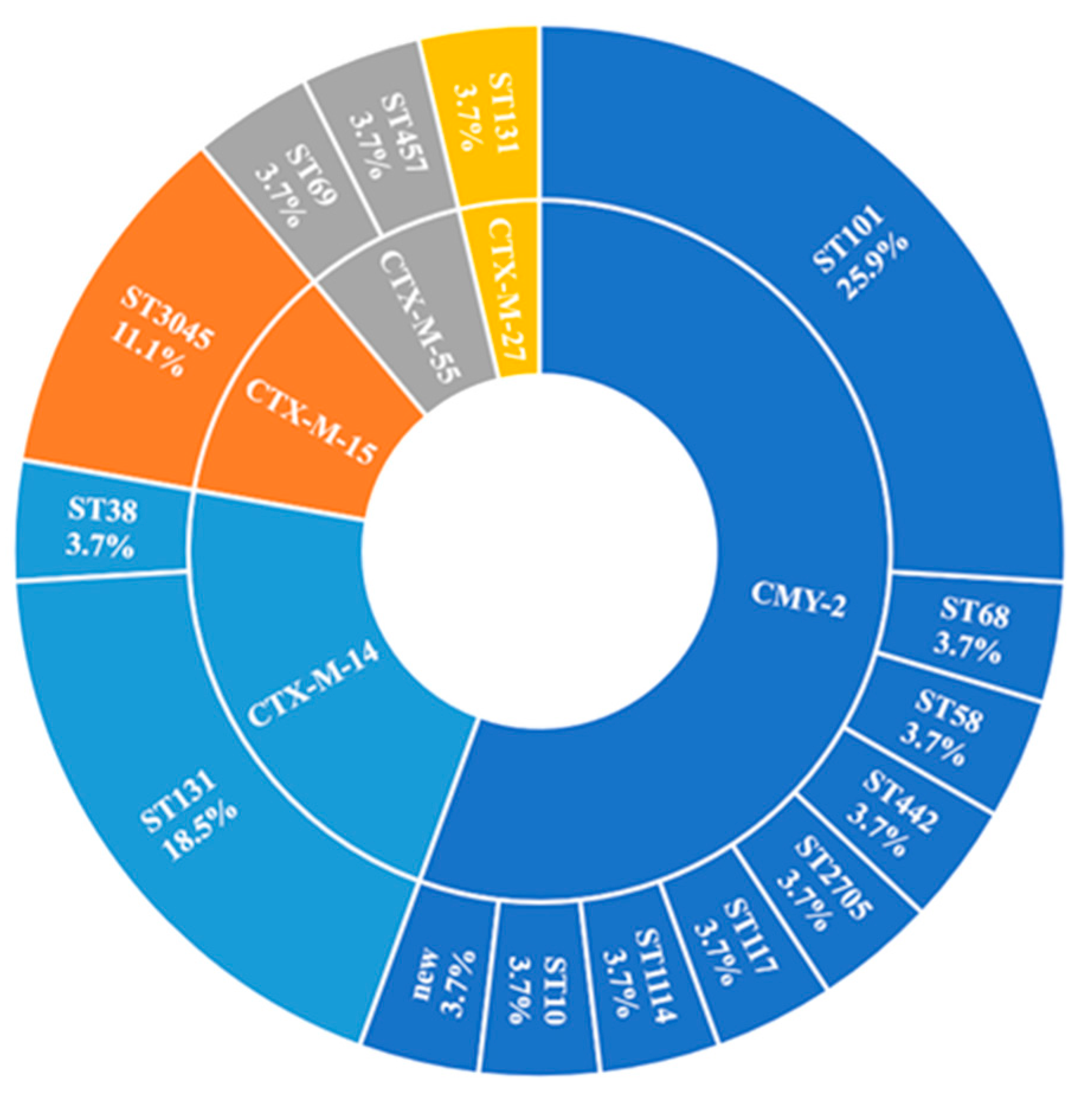
2.4. ARGs
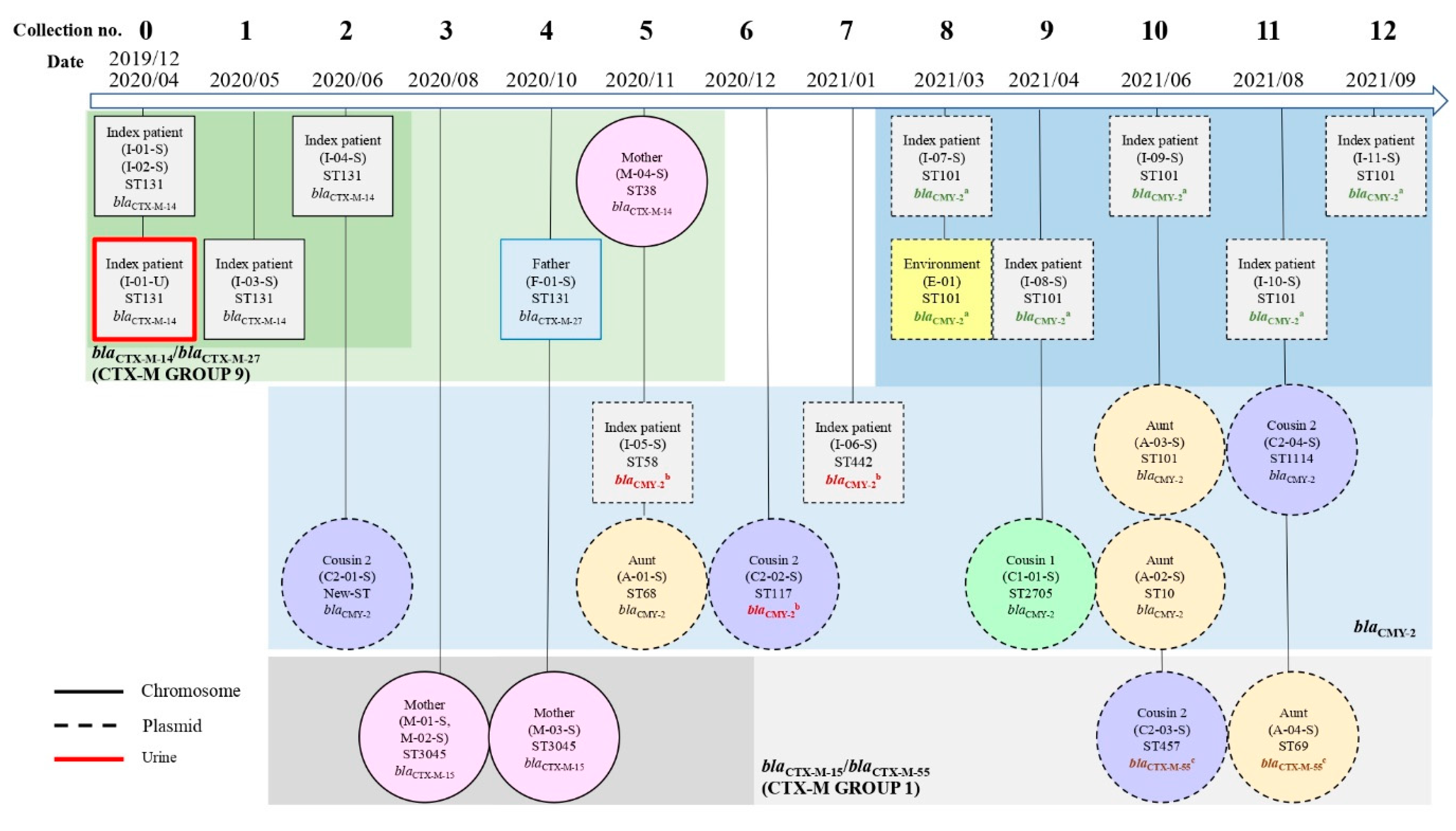
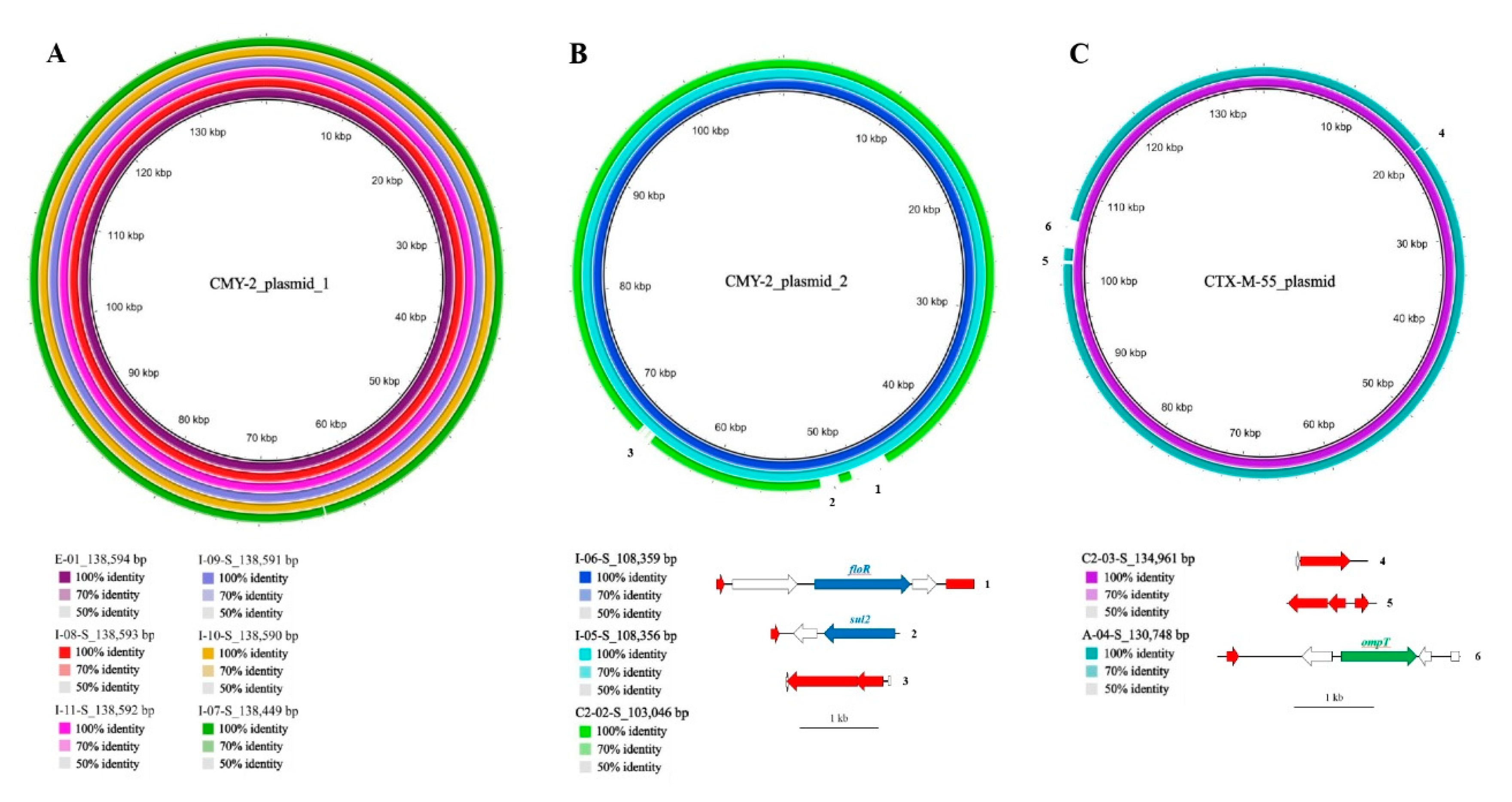
2.4.1. β-Lactamase-Encoding Genes and Household Transmission
2.4.2. ARGs Other Than β-Lactamase Genes
2.5. Virulence Factors (VF)-Encoding Genes
3. Discussion
4. Materials and Methods
4.1. Participants and Sample Collection
4.2. Questionnaire Design
4.3. Microbiological Analysis and Antimicrobial Susceptibility Testing
4.4. Nanopore Sequencing and De Novo Assembly
4.5. Pathogenic Types of E. coli
4.6. Bioinformatics and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Franz, E.; Veenman, C.; van Hoek, A.H.; de Roda Husman, A.; Blaak, H. Pathogenic Escherichia coli producing Extended Spectrum β-Lactamases isolated from surface water and wastewater. Sci. Rep. 2015, 5, 14372. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Zhao, X.; Li, R.; Han, Y.; Hao, G.; Wang, G.; Sun, S. Companion Animals as Potential Reservoirs of Antibiotic Resistant Diarrheagenic Escherichia coli in Shandong, China. Antibiotics 2022, 11, 828. [Google Scholar] [CrossRef]
- Imre, K.; Ban-Cucerzan, A.; Herman, V.; Sallam, K.I.; Cristina, R.T.; Abd-Elghany, S.M.; Morar, D.; Popa, S.A.; Imre, M.; Morar, A. Occurrence, Pathogenic Potential and Antimicrobial Resistance of Escherichia coli Isolated from Raw Milk Cheese Commercialized in Banat Region, Romania. Antibiotics 2022, 11, 721. [Google Scholar] [CrossRef] [PubMed]
- Smet, A.; Martel, A.; Persoons, D.; Dewulf, J.; Heyndrickx, M.; Herman, L.; Haesebrouck, F.; Butaye, P. Broad-spectrum β-lactamases among Enterobacteriaceae of animal origin: Molecular aspects, mobility and impact on public health. FEMS Microbiol. Rev. 2010, 34, 295–316. [Google Scholar] [CrossRef]
- Sheng, W.H.; Badal, R.E.; Hsueh, P.R. Distribution of extended-spectrum β-lactamases, AmpC β-lactamases, and carbapenemases among Enterobacteriaceae isolates causing intra-abdominal infections in the Asia-Pacific region: Results of the study for Monitoring Antimicrobial Resistance Trends (SMART). Antimicrob. Agents Chemother. 2013, 57, 2981–2988. [Google Scholar] [CrossRef] [PubMed]
- Bezabih, Y.M.; Sabiiti, W.; Alamneh, E.; Bezabih, A.; Peterson, G.M.; Bezabhe, W.M.; Roujeinikova, A. The global prevalence and trend of human intestinal carriage of ESBL-producing Escherichia coli in the community. J. Antimicrob. Chemother. 2021, 76, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.F.; Chen, W.L.; Hung, W.Y.; Huang, I.F.; Chiou, Y.H.; Chen, Y.S.; Lee, S.S.; Hung, C.H.; Wang, J.L. Emergence of extended spectrum-beta-lactamase-producing Escherichia coli O25b-ST131: A major community-acquired uropathogen in infants. Pediatr. Infect. Dis. J. 2015, 34, 469–475. [Google Scholar] [CrossRef]
- Huang, I.F.; Lee, W.Y.; Wang, J.L.; Hung, C.H.; Hu, H.H.; Hung, W.Y.; Hung, Y.J.; Chen, W.C.; Shen, Y.T.; Cheng, M.F. Fecal carriage of multidrug-resistant Escherichia coli by community children in southern Taiwan. BMC Gastroenterol. 2018, 18, 86. [Google Scholar] [CrossRef]
- Tsai, W.L.; Hung, C.H.; Chen, H.A.; Wang, J.L.; Huang, I.F.; Chiou, Y.H.; Chen, Y.S.; Lee, S.S.; Hung, W.Y.; Cheng, M.F. Extended-spectrum beta-lactamase-producing Escherichia coli bacteremia: Comparison of pediatric and adult populations. J. Microbiol. Immunol. Infect. 2018, 51, 723–731. [Google Scholar] [CrossRef]
- Chen, W.C.; Hung, C.H.; Chen, Y.S.; Cheng, J.S.; Lee, S.S.; Tseng, F.C.; Cheng, M.F.; Wang, J.L. Bloodstream Infections Caused by Extended-Spectrum Beta-Lactamase-Producing Escherichia coli in Patients with Liver Cirrhosis. Pathogens 2021, 10, 37. [Google Scholar] [CrossRef]
- Cheng, M.F.; Chen, W.L.; Huang, I.F.; Chen, J.R.; Chiou, Y.H.; Chen, Y.S.; Lee, S.S.; Hung, W.Y.; Hung, C.H.; Wang, J.L. Urinary tract infection in infants caused by extended-spectrum beta-lactamase-producing Escherichia coli: Comparison between urban and rural hospitals. Pediatr. Nephrol. 2016, 31, 1305–1312. [Google Scholar] [CrossRef] [PubMed]
- von Wintersdorff, C.J.H.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.M.; Wolffs, P.F.G. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef] [PubMed]
- Zalewska, M.; Błażejewska, A.; Czapko, A.; Popowska, M. Antibiotics and Antibiotic Resistance Genes in Animal Manure—Consequences of Its Application in Agriculture. Front. Microbiol. 2021, 12, 610656. [Google Scholar] [CrossRef]
- Reuland, E.A.; Halaby, T.; Hays, J.P.; de Jongh, D.M.; Snetselaar, H.D.; van Keulen, M.; Elders, P.J.; Savelkoul, P.H.; Vandenbroucke-Grauls, C.M.; Al Naiemi, N. Plasmid-mediated AmpC: Prevalence in community-acquired isolates in Amsterdam, the Netherlands, and risk factors for carriage. PLoS ONE 2015, 10, e0113033. [Google Scholar] [CrossRef]
- Mandal, D.K.; Sah, S.K.; Mishra, S.K.; Sharma, S.; Kattel, H.P.; Pandit, S.; Yadav, P.K.; Laghu, U.; Lama, R.; Sah, N.P.; et al. Carriage of Extended-Spectrum-β-Lactamase- and AmpC-β-Lactamase-Producing Enterobacteriaceae (ESBL-PE) in Healthy Community and Outpatient Department (OPD) Patients in Nepal. Can. J. Infect. Dis. Med. Microbiol. 2020, 2020, 5154217. [Google Scholar] [CrossRef] [PubMed]
- Latorre-Pérez, A.; Villalba-Bermell, P.; Pascual, J.; Vilanova, C. Assembly methods for nanopore-based metagenomic sequencing: A comparative study. Sci. Rep. 2020, 10, 13588. [Google Scholar] [CrossRef]
- Huang, Y.-T.; Liu, P.-Y.; Shih, P.-W. Homopolish: A method for the removal of systematic errors in nanopore sequencing by homologous polishing. Genome Biol. 2021, 22, 95. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Tetzschner, A.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and Easy In Silico Serotyping of Escherichia coli Isolates by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2015, 53, 2410–2426. [Google Scholar] [CrossRef]
- Wu, P.C.; Wang, J.L.; Hsueh, P.R.; Lin, P.H.; Cheng, M.F.; Huang, I.F.; Chen, Y.S.; Lee, S.S.; Guang-Yuan, M.; Yu, H.C.; et al. Prevalence and risk factors for colonization by extended-spectrum beta-lactamase-producing or ST 131 Escherichia coli among asymptomatic adults in community settings in Southern Taiwan. Infect. Drug Resist. 2019, 12, 1063–1071. [Google Scholar] [CrossRef] [Green Version]
- Riley, L.W. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin. Microbiol. Infect. 2014, 20, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Ewers, C.; Bethe, A.; Semmler, T.; Guenther, S.; Wieler, L.H. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: A global perspective. Clin. Microbiol. Infect. 2012, 18, 646–655. [Google Scholar] [CrossRef] [PubMed]
- Valenza, G.; Nickel, S.; Pfeifer, Y.; Eller, C.; Krupa, E.; Lehner-Reindl, V.; Höller, C. Extended-spectrum-β-lactamase-producing Escherichia coli as intestinal colonizers in the German community. Antimicrob. Agents Chemother. 2014, 58, 1228–1230. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, R.L.; Nielsen, J.B.; Friis-Møller, A.; Fjeldsøe-Nielsen, H.; Schønning, K. Prevalence and molecular characterization of clinical isolates of Escherichia coli expressing an AmpC phenotype. J. Antimicrob. Chemother. 2010, 65, 460–464. [Google Scholar] [CrossRef] [PubMed]
- Jean, S.-S.; Hsueh, P.-R. Distribution of ESBLs, AmpC β-lactamases and carbapenemases among Enterobacteriaceae isolates causing intra-abdominal and urinary tract infections in the Asia-Pacific region during 2008–14: Results from the Study for Monitoring Antimicrobial Resistance Tr. J. Antimicrob. Chemother. 2017, 72, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.-J.; Hong, C.-Y.; Ko, W.-C.; Chen, Y.-J.; Tsai, S.-H.; Chuang, C.-L.; Wu, J.-J. Dissemination of bla CMY-2 among Escherichia coli Isolates from Food Animals, Retail Ground Meats, and Humans in Southern Taiwan. Antimicrob. Agents Chemother. 2004, 48, 1353–1356. [Google Scholar] [CrossRef]
- Alonso, N.; Miró, E.; Pascual, V.; Rivera, A.; Simó, M.; Garcia, M.C.; Xercavins, M.; Morera, M.A.; Espejo, E.; Gurguí, M.; et al. Molecular characterisation of acquired and overproduced chromosomal blaAmpC in Escherichia coli clinical isolates. Int. J. Antimicrob. Agents 2016, 47, 62–68. [Google Scholar] [CrossRef]
- van Hoek, A.H.; Schouls, L.; van Santen, M.G.; Florijn, A.; de Greeff, S.C.; van Duijkeren, E. Molecular characteristics of extended-spectrum cephalosporin-resistant Enterobacteriaceae from humans in the community. PLoS ONE 2015, 10, e0129085. [Google Scholar] [CrossRef]
- Karaiskos, I.; Giamarellou, H. Carbapenem-Sparing Strategies for ESBL Producers: When and How. Antibiotics 2020, 9, 61. [Google Scholar] [CrossRef]
- Rizi, K.S.; Mosavat, A.; Youssefi, M.; Jamehdar, S.A.; Ghazvini, K.; Safdari, H.; Amini, Y.; Farsiani, H. High prevalence of bla(CMY) AmpC beta-lactamase in ESBL co-producing Escherichia coli and Klebsiella spp. clinical isolates in the northeast of Iran. J. Glob. Antimicrob. Resist. 2020, 22, 477–482. [Google Scholar] [CrossRef]
- Drinkovic, D.; Morris, A.J.; Dyet, K.; Bakker, S.; Heffernan, H. Plasmid-mediated AmpC beta-lactamase-producing Escherichia coli causing urinary tract infection in the Auckland community likely to be resistant to commonly prescribed antimicrobials. N. Z. Med. J. 2015, 128, 50–59. [Google Scholar] [PubMed]
- Ramphal, R.; Ambrose, P.G. Extended-Spectrum β-Lactamases and Clinical Outcomes: Current Data. Clin. Infect. Dis. 2006, 42, S164–S172. [Google Scholar] [CrossRef] [PubMed]
- Chia, J.H.; Siu, L.K.; Su, L.H.; Lin, H.S.; Kuo, A.J.; Lee, M.H.; Wu, T.L. Emergence of carbapenem-resistant Escherichia coli in Taiwan: Resistance due to combined CMY-2 production and porin deficiency. J. Chemother. 2009, 21, 621–626. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.C.; Cheng, M.F.; Chen, W.L.; Hung, W.Y.; Wang, J.L.; Hung, C.H. Risk Factors and Prevalence of mcr-1-Positive Escherichia coli in Fecal Carriages Among Community Children in Southern Taiwan. Front. Microbiol. 2021, 12, 748525. [Google Scholar] [CrossRef] [PubMed]
- Lautenbach, E.; Bilker, W.B.; Tolomeo, P.; Maslow, J.N. Impact of diversity of colonizing strains on strategies for sampling Escherichia coli from fecal specimens. J. Clin. Microbiol. 2008, 46, 3094–3096. [Google Scholar] [CrossRef]
- Smati, M.; Clermont, O.; Le Gal, F.; Schichmanoff, O.; Jauréguy, F.; Eddi, A.; Denamur, E.; Picard, B. Real-time PCR for quantitative analysis of human commensal Escherichia coli populations reveals a high frequency of subdominant phylogroups. Appl. Environ. Microbiol. 2013, 79, 5005–5012. [Google Scholar] [CrossRef]
- Weinstein, M.P.; Kim, T.J., Jr.; Lewis, J.S., II.; Limbago, B.; Bobenchik, A.M.; Mathers, A.J.; Campeau, S.; Mazzulli, T.; Cullen, S.K.; Satlin, M.; et al. Performance Standards for Antimicrobial Testing; CLSI Supplement M100, 30th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- FDA-Recognized Antimicrobial Susceptibility Test Interpretive Criteria: Tigecycline. Available online: https://www.fda.gov/drugs/development-resources/tigecycline-injection-products (accessed on 1 October 2021).
- Khurana, S.; Malhotra, R.; Mathur, P. Evaluation of Vitek®2 performance for colistin susceptibility testing for Gram-negative isolates. JAC-Antimicrob. Resist. 2020, 2, dlaa101. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Herschleb, J.; Ananiev, G.; Schwartz, D.C. Pulsed-field gel electrophoresis. Nat. Protocols 2007, 2, 677–684. [Google Scholar] [CrossRef]
- Liao, Y.-C.; Cheng, H.-W.; Wu, H.-C.; Kuo, S.-C.; Lauderdale, T.-L.Y.; Chen, F.-J. Completing Circular Bacterial Genomes With Assembly Complexity by Using a Sampling Strategy From a Single MinION Run With Barcoding. Front. Microbiol. 2019, 10, 2068. [Google Scholar] [CrossRef] [Green Version]
- Malberg Tetzschner, A.M.; Johnson, J.R.; Johnston, B.D.; Lund, O.; Scheutz, F. In Silico Genotyping of Escherichia coli Isolates for Extraintestinal Virulence Genes by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2020, 58, e01269-20. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, R.L.; Pouseele, H.; Chen, J.C.; Strockbine, N.A.; Carleton, H.A. Implementation of Whole Genome Sequencing (WGS) for Identification and Characterization of Shiga Toxin-Producing Escherichia coli (STEC) in the United States. Front. Microbiol. 2016, 7, 766. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Porter, S.; Johnston, B.; Kuskowski, M.A.; Spurbeck, R.R.; Mobley, H.L.T.; Williamson, D.A. Host Characteristics and Bacterial Traits Predict Experimental Virulence for Escherichia coli Bloodstream Isolates From Patients With Urosepsis. Open Forum Infect. Dis. 2015, 2, ofv083. [Google Scholar] [CrossRef] [PubMed]
- Spurbeck, R.R.; Dinh, P.C., Jr.; Walk, S.T.; Stapleton, A.E.; Hooton, T.M.; Nolan, L.K.; Kim, K.S.; Johnson, J.R.; Mobley, H.L. Escherichia coli isolates that carry vat, fyuA, chuA, and yfcV efficiently colonize the urinary tract. Infect. Immun. 2012, 80, 4115–4122. [Google Scholar] [CrossRef]
- Johnson, T.J.; Wannemuehler, Y.; Doetkott, C.; Johnson, S.J.; Rosenberger, S.C.; Nolan, L.K. Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. J. Clin. Microbiol. 2008, 46, 3987–3996. [Google Scholar] [CrossRef]
- Center for Genomic Epidemiology. Available online: http://www.genomicepidemiology.org/services/ (accessed on 8 March 2022).
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2021, 50, D801–D807. [Google Scholar] [CrossRef]
- Alikhan, N.-F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genomics 2011, 12, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Variables | Index Patient | Mother | Father | Aunt | Uncle | Cousin 1 | Cousin 2 |
|---|---|---|---|---|---|---|---|
| Demographic data | |||||||
| Age (years) | 0.3 | 35 | 37 | 46 | 42 | 8 | 6 |
| Male/Female | M | F | M | F | M | F | F |
| Travel abroad in the past 12 months | - | - | - | Japan | Japan China | Japan | Japan |
| Postal code | 81146, Nanzi District, live in the same house but different floors | ||||||
| Daily animal contact | Dog | ||||||
| Diet habit | |||||||
| Feed water source | RO water | RO water | RO water | RO water | RO water | RO water | RO water |
| Boiled water use | + | + | + | + | + | + | + |
| Vegetarian | Breastmilk or formula milk | - | - | - | - | - | - |
| Consumption of meats, consumption days per week | |||||||
| Pork | 0 | 6 | 7 | 5 | 6 | 6 | 5 |
| Beef | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| Sheep | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Chicken | 0 | 4 | 2 | 4 | 4 | 4 | 4 |
| Duck | 0 | 1 | 2 | 1 | 1 | 1 | 1 |
| Goose | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Fish | 0 | 4 | 7 | 4 | 4 | 4 | 4 |
| Family cleaning habits | |||||||
| Clean toilet bowl every month | + | + | + | + | + | + | + |
| Clean bathroom sink every month | + | + | + | + | + | + | + |
| Clean bathroom floor every month | + | + | + | + | + | + | + |
| Clean children’s bedrooms every month | + | + | + | + | + | + | + |
| Clean kitchen floor every month | + | + | + | + | + | + | + |
| Clean living room floor every month | + | + | + | + | + | + | + |
| Clean master bedroom every month | + | + | + | + | + | + | + |
| Clean toys every year | + | + | + | + | + | + | + |
| Medical history | |||||||
| Previous hospitalization in the past 12 months | 2019/12 UTI 2020/04 UTI | 2019/12 Labor | - | - | - | 2019/5 UTI | 2019/07 Pneumonia |
| Previous antimicrobial treatment in the past 12 months | + | + | - | - | - | + | + |
| Previous probiotics use in the past 12 months, consumption days per week | 0 | 0 | 0 | 1 | 0 | 7 | 7 |
| Antimicrobial Resistance Phenotype | Ampicillin/ Sulbactam | Piperacillin | Piperacillin/ Tazobactam | Cefazolin | Cefoxitin | Cefixime b | Ceftazidime b | Ceftriaxone b | Cefepime | Ertapenem | Imipenem | Amikacin | Gentamicin | Ciprofloxacin | Minocycline | Tigecycline | Colistin | Trimethoprim/ Sulfamethoxazole | Chloramphenicol | Azithromycin | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolate No. a | Source | Date | blaESBL/blaAmpC Genes | ||||||||||||||||||||
| I-01-S | Index case | 2019/12 | blaCTX-M-14 | ND c | ND d | ND d | |||||||||||||||||
| I-01-U | Index case | 2020/04 | blaCTX-M-14 | ||||||||||||||||||||
| I-02-S | Index case | 2020/04 | blaCTX-M-14 | ||||||||||||||||||||
| I-03-S | Index case | 2020/05 | blaCTX-M-14 | ||||||||||||||||||||
| I-04-S | Index case | 2020/06 | blaCTX-M-14 | ||||||||||||||||||||
| I-05-S | Index case | 2020/11 | blaCMY-2 | ||||||||||||||||||||
| I-06-S | Index case | 2021/01 | blaCMY-2 | ||||||||||||||||||||
| I-07-S | Index case | 2021/03 | blaCMY-2 | ||||||||||||||||||||
| I-08-S | Index case | 2021/04 | blaCMY-2 | ||||||||||||||||||||
| I-09-S | Index case | 2021/06 | blaCMY-2 | ||||||||||||||||||||
| I-10-S | Index case | 2021/08 | blaCMY-2 | ||||||||||||||||||||
| I-11-S | Index case | 2021/09 | blaCMY-2 | ||||||||||||||||||||
| F-01-S | Father | 2020/10 | blaCTX-M-27 | ||||||||||||||||||||
| M-01-S | Mother | 2020/08 | blaCTX-M-15 | ||||||||||||||||||||
| M-02-S | Mother | 2020/08 | blaCTX-M-15 | ||||||||||||||||||||
| M-03-S | Mother | 2020/10 | blaCTX-M-15 | ||||||||||||||||||||
| M-04-S | Mother | 2020/11 | blaCTX-M-14 | ||||||||||||||||||||
| A-01-S | Aunt | 2020/11 | blaCMY-2 | ||||||||||||||||||||
| A-02-S | Aunt | 2021/06 | blaCMY-2 | ||||||||||||||||||||
| A-03-S | Aunt | 2021/06 | blaCMY-2 | ||||||||||||||||||||
| A-04-S | Aunt | 2021/08 | blaCTX-M-55 | ||||||||||||||||||||
| C1-01-S | Cousin 1 | 2021/04 | blaCMY-2 | ||||||||||||||||||||
| C2-01-S | Cousin 2 | 2020/06 | blaCMY-2 | ||||||||||||||||||||
| C2-02-S | Cousin 2 | 2020/12 | blaCMY-2 | ||||||||||||||||||||
| C2-03-S | Cousin 2 | 2021/06 | blaCTX-M-55 | ||||||||||||||||||||
| C2-04-S | Cousin 2 | 2021/08 | blaCMY-2 | ||||||||||||||||||||
| E-01 | Bathroom basin | 2021/03 | blaCMY-2 | ||||||||||||||||||||
| Percentage of non-susceptibility | 92.6% | 100.0% | 0 | 100.0% | 55.6% | 100.0% | 55.6% | 100.0% | 7.4% | 0.0% | 0.0% | 0.0% | 59.3% | 63.0% | 63.0% | 0.0% | NA | 55.6% | NA | NA | |||
| Antimicrobial Resistance Genes | β-lactam | Aminoglycoside | Quinolone | Tetracycline | Colistin | Trimethoprim/Sulfamethoxazole | Amphenicol | Macrolide | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolate No. a | Source | Date | blaTEM | blaCTX-Mb | blaAmpC b | dfrA | sul | ||||||||||||||||||||||
| TEM-1B | TEM-135 | CTX-M-14 | CTX-M-15 | CTX-M-27 | CTX-M-55 | CMY-2 | aac(3)-IId | aac(3)-VIa | gyrA c | qnrS1 | qnrS13 | tet(A) | tet(B) | tet(M) | mcr-1.1 | dfrA5 | dfrA12 | dfrA14 | dfrA17 | sul1 | sul2 | sul3 | cmlA1 | catA1 | floR | mph(A) | |||
| I-01-S | Index case | 2019/12 | |||||||||||||||||||||||||||
| I-01-U | Index case | 2020/04 | |||||||||||||||||||||||||||
| I-02-S | Index case | 2020/04 | |||||||||||||||||||||||||||
| I-03-S | Index case | 2020/05 | |||||||||||||||||||||||||||
| I-04-S | Index case | 2020/06 | |||||||||||||||||||||||||||
| I-05-S | Index case | 2020/11 | |||||||||||||||||||||||||||
| I-06-S | Index case | 2021/01 | |||||||||||||||||||||||||||
| I-07-S | Index case | 2021/03 | |||||||||||||||||||||||||||
| I-08-S | Index case | 2021/04 | |||||||||||||||||||||||||||
| I-09-S | Index case | 2021/06 | |||||||||||||||||||||||||||
| I-10-S | Index case | 2021/08 | |||||||||||||||||||||||||||
| I-11-S | Index case | 2021/09 | |||||||||||||||||||||||||||
| F-01-S | Father | 2020/10 | |||||||||||||||||||||||||||
| M-01-S | Mother | 2020/08 | |||||||||||||||||||||||||||
| M-02-S | Mother | 2020/08 | |||||||||||||||||||||||||||
| M-03-S | Mother | 2020/10 | |||||||||||||||||||||||||||
| M-04-S | Mother | 2020/11 | |||||||||||||||||||||||||||
| A-01-S | Aunt | 2020/11 | |||||||||||||||||||||||||||
| A-02-S | Aunt | 2021/06 | |||||||||||||||||||||||||||
| A-03-S | Aunt | 2021/06 | |||||||||||||||||||||||||||
| A-04-S | Aunt | 2021/08 | |||||||||||||||||||||||||||
| C1-01-S | Cousin 1 | 2021/04 | |||||||||||||||||||||||||||
| C2-01-S | Cousin 2 | 2020/06 | |||||||||||||||||||||||||||
| C2-02-S | Cousin 2 | 2020/12 | |||||||||||||||||||||||||||
| C2-03-S | Cousin 2 | 2021/06 | |||||||||||||||||||||||||||
| C2-04-S | Cousin 2 | 2021/08 | |||||||||||||||||||||||||||
| E-01 | Bathroom basin | 2021/03 | |||||||||||||||||||||||||||
| Percentage of positive by either of one gene | 20 (74.1%) | 12 (44.4%) | 15 (55.6%) | 16 (59.2%) | 19 (70.3%) | 21 (77.8%) | 1 (3.7%) | 15 (55.6%) | 17 (63%) | 16 (59.2%) | 8 (29.6%) | ||||||||||||||||||
| Virulence Factor (VF)-Encoding Genes | Adherence | Capsule | Iron Uptake | Toxin | Miscellaneous | Definition | ||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolate No. a | Source | Date | afaA | afaB | afaC | afaD | focC | hra | iha | lpfA | papA | papC | sfaD | yfcV | kpsE | kpsM II | kpsM III | chuA | fyuA | iroN | irp2 | iucC | iutA | sitA | air | astA | ccI | cea | cib | cma | cnf1 | hlyF | pic | sat | senB | tsh | vat | cvaC | eilA | etsC | gad | iss | katP | mcbA | mchF | ompT | terC | traT | usp | ExPEC | UPEC | APEC |
| I-01-S | Index patient | 2019/12 |  | |||||||||||||||||||||||||||||||||||||||||||||||||
| I-01-U | Index patient | 2020/04 |  | |||||||||||||||||||||||||||||||||||||||||||||||||
| I-02-S | Index patient | 2020/04 |  | |||||||||||||||||||||||||||||||||||||||||||||||||
| I-03-S | Index patient | 2020/05 |  | |||||||||||||||||||||||||||||||||||||||||||||||||
| I-04-S | Index patient | 2020/06 |  | |||||||||||||||||||||||||||||||||||||||||||||||||
| I-05-S | Index patient | 2020/11 |  | |||||||||||||||||||||||||||||||||||||||||||||||||
| I-06-S | Index patient | 2021/01 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| I-07-S | Index patient | 2021/03 |  | |||||||||||||||||||||||||||||||||||||||||||||||||
| I-08-S | Index patient | 2021/04 |  | |||||||||||||||||||||||||||||||||||||||||||||||||
| I-09-S | Index patient | 2021/06 |  | |||||||||||||||||||||||||||||||||||||||||||||||||
| I-10-S | Index patient | 2021/08 |  | |||||||||||||||||||||||||||||||||||||||||||||||||
| I-11-S | Index patient | 2021/09 |  | |||||||||||||||||||||||||||||||||||||||||||||||||
| F-01-S | Father | 2020/10 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| M-01-S | Mother | 2020/08 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| M-02-S | Mother | 2020/08 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| M-03-S | Mother | 2020/10 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| M-04-S | Mother | 2020/11 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| A-01-S | Aunt | 2020/11 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| A-02-S | Aunt | 2021/06 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| A-03-S | Aunt | 2021/06 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| A-04-S | Aunt | 2021/08 |  | |||||||||||||||||||||||||||||||||||||||||||||||||
| C1-01-S | Cousin 1 | 2021/04 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| C2-01-S | Cousin 2 | 2020/06 |  | |||||||||||||||||||||||||||||||||||||||||||||||||
| C2-02-S | Cousin 2 | 2020/12 |  | |||||||||||||||||||||||||||||||||||||||||||||||||
| C2-03-S | Cousin 2 | 2021/06 |  | |||||||||||||||||||||||||||||||||||||||||||||||||
| C2-04-S | Cousin 2 | 2021/08 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| E-01 | Bathroom basin | 2021/03 |  | |||||||||||||||||||||||||||||||||||||||||||||||||
| ExPEC:≧2 of 5 markers, including papAH and/or papC, sfa/focDE, afa/draBC, kpsM II, and iutA | ||||||||||||||||||||||||||||||||||||||||||||||||||||
| UPEC:≧3 of 4 markers, including chuA, fyuA, vat, and yfcV | ||||||||||||||||||||||||||||||||||||||||||||||||||||
| APEC:≧4 of 5 markers, including hlyF, iutA, iroN, iss, and ompT | ||||||||||||||||||||||||||||||||||||||||||||||||||||
| Presence of predicted VF-encoding genes | ||||||||||||||||||||||||||||||||||||||||||||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, Y.-C.; Liou, C.-H.; Ng, W.V.; Chen, F.-J.; Hung, C.-H.; Liu, P.-Y.; Liao, Y.-C.; Wu, H.-C.; Cheng, M.-F. Distribution and Genomic Characterization of Third-Generation Cephalosporin-Resistant Escherichia coli Isolated from a Single Family and Home Environment: A 2-Year Longitudinal Study. Antibiotics 2022, 11, 1152. https://doi.org/10.3390/antibiotics11091152
Feng Y-C, Liou C-H, Ng WV, Chen F-J, Hung C-H, Liu P-Y, Liao Y-C, Wu H-C, Cheng M-F. Distribution and Genomic Characterization of Third-Generation Cephalosporin-Resistant Escherichia coli Isolated from a Single Family and Home Environment: A 2-Year Longitudinal Study. Antibiotics. 2022; 11(9):1152. https://doi.org/10.3390/antibiotics11091152
Chicago/Turabian StyleFeng, Yin-Chih, Ci-Hong Liou, Wailap Victor Ng, Feng-Jui Chen, Chih-Hsin Hung, Po-Yen Liu, Yu-Chieh Liao, Han-Chieh Wu, and Ming-Fang Cheng. 2022. "Distribution and Genomic Characterization of Third-Generation Cephalosporin-Resistant Escherichia coli Isolated from a Single Family and Home Environment: A 2-Year Longitudinal Study" Antibiotics 11, no. 9: 1152. https://doi.org/10.3390/antibiotics11091152
APA StyleFeng, Y.-C., Liou, C.-H., Ng, W. V., Chen, F.-J., Hung, C.-H., Liu, P.-Y., Liao, Y.-C., Wu, H.-C., & Cheng, M.-F. (2022). Distribution and Genomic Characterization of Third-Generation Cephalosporin-Resistant Escherichia coli Isolated from a Single Family and Home Environment: A 2-Year Longitudinal Study. Antibiotics, 11(9), 1152. https://doi.org/10.3390/antibiotics11091152






