New Characterization of Multi-Drug Resistance of Streptococcus suis and Biofilm Formation from Swine in Heilongjiang Province of China
Abstract
:1. Introduction
2. Results
2.1. Isolation and Identification of S. suis
2.2. Antimicrobial Susceptibility Testing
2.3. Detection of Antimicrobial Resistance Genes
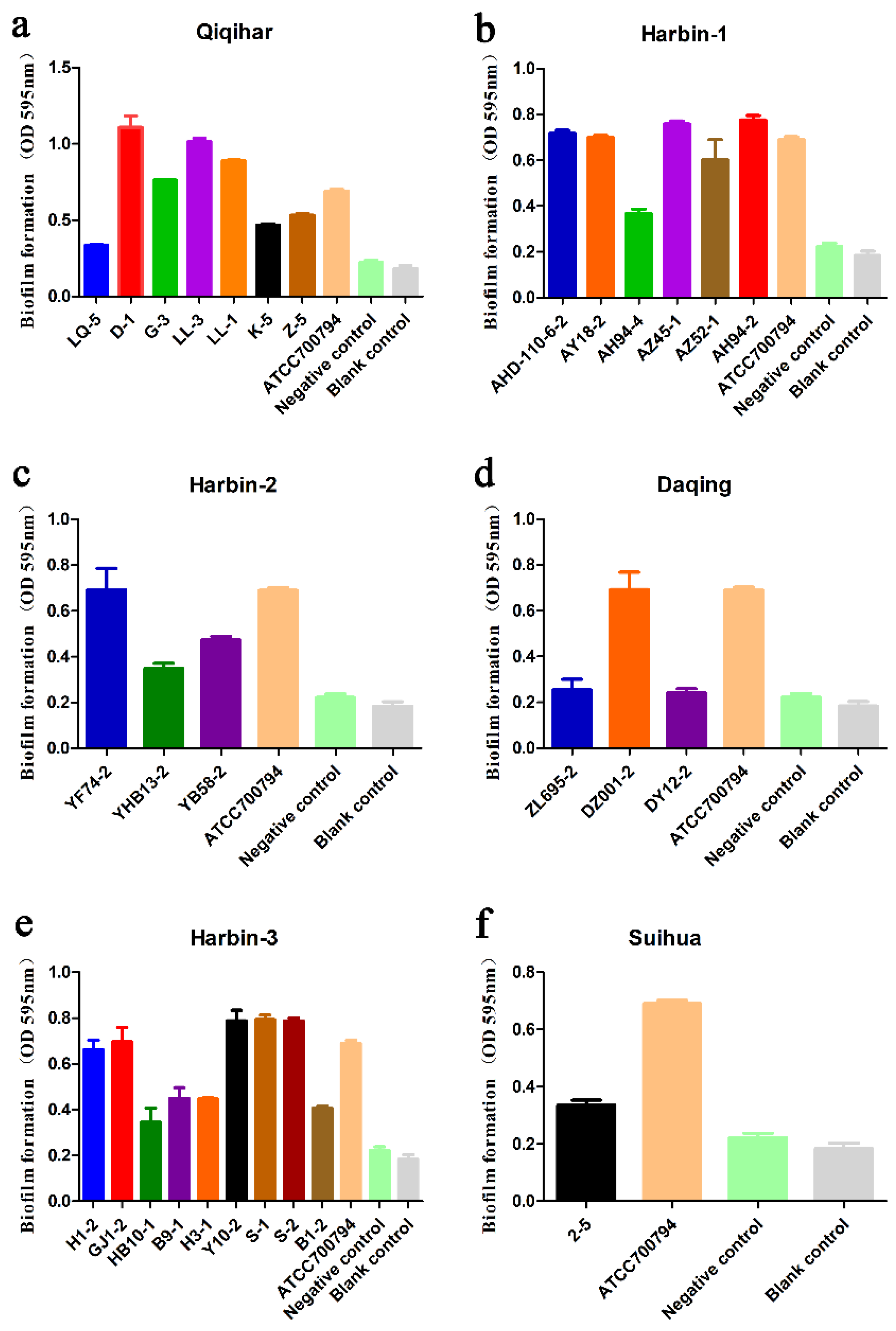
2.4. Biofilm Formation Analysis
3. Discussion
4. Materials and Methods
4.1. Samples
4.2. S. suis Isolation and Serotypes Identification
4.3. MIC Testing
4.4. Detection of Antimicrobial Resistance Genes
4.5. Biofilm Formation
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jiang, H.; Liu, J.; Wu, T.; Liu, M.; Sun, Q.; Wang, J.; Zhu, R.; Qu, G.; Li, S.; Liu, H.; et al. Proteomics analysis of important molecules in serum from meningitic piglets caused by Streptococcus suis serotype 2. J. Infect. Dev. Ctries. 2020, 14, 502–510. [Google Scholar] [CrossRef] [PubMed]
- Okura, M.; Takamatsu, D.; Maruyama, F.; Nozawa, T.; Nakagawa, I.; Osaki, M.; Sekizaki, T.; Gottschalk, M.; Kumagai, Y.; Hamada, S. Genetic analysis of capsular polysaccharide synthesis gene clusters from all serotypes of Streptococcus suis: Potential mechanisms for generation of capsular variation. Appl. Environ. Microbiol. 2013, 79, 2796–2806. [Google Scholar] [CrossRef] [Green Version]
- Gurung, M.; Tamang, M.D.; Moon, D.C.; Kim, S.R.; Jeong, J.H.; Jang, G.C.; Jung, S.C.; Park, Y.H.; Lim, S.K. Molecular Basis of Resistance to Selected Antimicrobial Agents in the Emerging Zoonotic Pathogen Streptococcus suis. J. Clin. Microbiol. 2015, 53, 2332–2336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.; Chen, B.; Zhang, Q.; Liu, L.; Zhang, A.; Yang, Y.; Huang, K.; Yan, S.; Yu, J.; Sun, X.; et al. Streptococcus suis 2 Transcriptional Regulator TstS Stimulates Cytokine Production and Bacteremia to Promote Streptococcal Toxic Shock-Like Syndrome. Front. Microbiol. 2018, 9, 1309. [Google Scholar] [CrossRef] [Green Version]
- Yongkiettrakul, S.; Maneerat, K.; Arechanajan, B.; Malila, Y.; Srimanote, P.; Gottschalk, M.; Visessanguan, W. Antimicrobial susceptibility of Streptococcus suis isolated from diseased pigs, asymptomatic pigs, and human patients in Thailand. BMC Vet. Res. 2019, 15, 5. [Google Scholar] [CrossRef] [Green Version]
- Tan, M.F.; Tan, J.; Zeng, Y.B.; Li, H.Q.; Yang, Q.; Zhou, R. Antimicrobial resistance phenotypes and genotypes of Streptococcus suis isolated from clinically healthy pigs from 2017 to 2019 in Jiangxi Province, China. J. Appl. Microbiol. 2021, 130, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, C.; Varaldo, P.E.; Facinelli, B. Streptococcus suis, an Emerging Drug-Resistant Animal and Human Pathogen. Front. Microbiol. 2011, 2, 235. [Google Scholar] [CrossRef] [Green Version]
- Segura, M.; Aragon, V.; Brockmeier, S.L.; Gebhart, C.; Greeff, A.; Kerdsin, A.; O’Dea, M.A.; Okura, M.; Salery, M.; Schultsz, C.; et al. Update on Streptococcus suis Research and Prevention in the Era of Antimicrobial Restriction: 4th International Workshop on S. suis. Pathogens 2020, 9, 374. [Google Scholar] [CrossRef] [PubMed]
- Rajkhowa, S.; Rajesh, J.B. Virulence associated gene profiling and antimicrobial resistance pattern of Streptococcus suis isolated from clinically healthy pigs from North East India. Lett. Appl. Microbiol. 2021, 73, 392–397. [Google Scholar] [CrossRef]
- Varela, N.P.; Gadbois, P.; Thibault, C.; Gottschalk, M.; Dick, P.; Wilson, J. Antimicrobial resistance and prudent drug use for Streptococcus suis. Anim. Health Res. Rev. 2013, 14, 68–77. [Google Scholar] [CrossRef]
- Huang, J.; Ma, J.; Shang, K.; Hu, X.; Liang, Y.; Li, D.; Wu, Z.; Dai, L.; Chen, L.; Wang, L. Evolution and Diversity of the Antimicrobial Resistance Associated Mobilome in Streptococcus suis: A Probable Mobile Genetic Elements Reservoir for Other Streptococci. Front. Cell. Infect. Microbiol. 2016, 6, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seitz, M.; Valentin-Weigand, P.; Willenborg, J. Use of Antibiotics and Antimicrobial Resistance in Veterinary Medicine as Exemplified by the Swine Pathogen Streptococcus suis. Curr. Top. Microbiol. Immunol. 2016, 398, 103–121. [Google Scholar] [CrossRef]
- Yao, J.; Shang, K.; Huang, J.; Ran, W.; Kashif, J.; Wang, L. Overexpression of an ABC transporter and mutations of GyrA, GyrB, and ParC in contributing to high-level ciprofloxacin resistance in Streptococcus suis type 2. Biosci. Trends 2014, 8, 84–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, Y.W.; Cheung, T.K.; Chu, M.Y.; Tsang, V.Y.; Fung, J.T.; Kam, K.M.; Lo, J.Y. Resistance to tetracycline, erythromycin and clindamycin in Streptococcus suis serotype 2 in Hong Kong. Int. J. Antimicrob. Agents 2009, 34, 181–182. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Wu, J.; Xia, Y.; Yang, M.; Xiao, J.; Yu, J. Molecular dynamics simulation of the complex PBP-2x with drug cefuroxime to explore the drug resistance mechanism of Streptococcus suis R61. PLoS ONE 2012, 7, e35941. [Google Scholar] [CrossRef] [Green Version]
- Hauschild, T.; Stepanović, S.; Vuković, D.; Dakić, I.; Schwarz, S. Occurrence of chloramphenicol resistance and corresponding resistance genes in members of the Staphylococcus sciuri group. Int. J. Antimicrob. Agents 2009, 33, 383–384. [Google Scholar] [CrossRef]
- Dechene-Tempier, M.; Marois-Crehan, C.; Libante, V.; Jouy, E.; Leblond-Bourget, N.; Payot, S. Update on the Mechanisms of Antibiotic Resistance and the Mobile Resistome in the Emerging Zoonotic Pathogen Streptococcus suis. Microorganisms 2021, 9, 1765. [Google Scholar] [CrossRef]
- Pinheiro, S.; Radhouani, H.; Coelho, C.; Gonçalves, A.; Carvalho, E.; Carvalho, J.A.; Ruiz-Larrea, F.; Torres, C.; Igrejas, G.; Poeta, P. Prevalence and mechanisms of erythromycin resistance in Streptococcus agalactiae from healthy pregnant women. Microb. Drug. Resist. 2009, 15, 121–124. [Google Scholar] [CrossRef]
- Van der Linden, M.; Al-Lahham, A.; Haupts, S.; Reinert, R.R. Clonal spread of mef-positive macrolide-resistant Streptococcus pneumoniae isolates causing invasive disease in adults in Germany. Antimicrob. Agents. Chemother. 2007, 51, 1830–1834. [Google Scholar] [CrossRef] [Green Version]
- Martel, A.; Baele, M.; Devriese, L.A.; Goossens, H.; Wisselink, H.J.; Decostere, A.; Haesebrouck, F. Prevalence and mechanism of resistance against macrolides and lincosamides in Streptococcus suis isolates. Vet. Microbiol. 2001, 83, 287–297. [Google Scholar] [CrossRef]
- Tenson, T.; Lovmar, M.; Ehrenberg, M. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J. Mol. Biol. 2003, 330, 1005–1014. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Liu, J.; Zhang, Y.; Chen, S.; Ma, J.; Dong, W.; Wu, Z.; Yao, H. A novel integrative conjugative element mediates transfer of multi-drug resistance between Streptococcus suis strains of different serotypes. Vet. Microbiol. 2019, 229, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Holmer, I.; Salomonsen, C.M.; Jorsal, S.E.; Astrup, L.B.; Jensen, V.F.; Hog, B.B.; Pedersen, K. Antibiotic resistance in porcine pathogenic bacteria and relation to antibiotic usage. BMC Vet. Res. 2019, 15, 449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jørgensen, K.M.; Wassermann, T.; Jensen, P.; Hengzuang, W.; Molin, S.; Høiby, N.; Ciofu, O. Sublethal ciprofloxacin treatment leads to rapid development of high-level ciprofloxacin resistance during long-term experimental evolution of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2013, 57, 4215–4221. [Google Scholar] [CrossRef] [Green Version]
- Kaplan, J.B. Antibiotic-induced biofilm formation. Int. J. Artif. Organs 2011, 34, 737–751. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S. Antimicrobial Tolerance in Biofilms. Microbiol. Spectr. 2015, 3, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brauner, A.; Fridman, O.; Gefen, O.; Balaban, N.Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 2016, 14, 320–330. [Google Scholar] [CrossRef]
- Yan, J.; Bassler, B.L. Surviving as a Community: Antibiotic Tolerance and Persistence in Bacterial Biofilms. Cell Host Microbe 2019, 26, 15–21. [Google Scholar] [CrossRef]
- Crabbe, A.; Jensen, P.O.; Bjarnsholt, T.; Coenye, T. Antimicrobial Tolerance and Metabolic Adaptations in Microbial Biofilms. Trends Microbiol. 2019, 27, 850–863. [Google Scholar] [CrossRef]
- Wang, S.J.; Lei, L.C.; Min, X.U.; Sun, C.J.; Cheng-Jun, L.I.; Cai, X.H.; Liu, Y.G.; Zhang, Q.; Liu, D.Q.; Shi, W.D. Isolation, identifiation and epidemiological analysis of swine Streptococcus in the northeast region of China. Chin. J. Vet. Sci. 2009, 12, 877–881. [Google Scholar]
- Goyette-Desjardins, G.; Auger, J.P.; Xu, J.; Segura, M.; Gottschalk, M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg. Microbes Infect. 2014, 3, e45. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Yanxiu, N.I.; Zhu, H.; Wang, D.; Guan, L.; Zhengyu, Y.U.; Zhou, J.; Lixin, L.; Wang, C.; Kongwang, H.E. Study on Biological Characteristics of Streptococcus suis Type 4 Isolates. China. Anim. Husb. Vet. Med. 2019, 46, 80–88. [Google Scholar]
- Wang, J.; Liu, Q.; Zhou, R.Y.; Jia, A.Q.; Wang, G.P. The Streptococcus suis epidemiological analysis of swine in Guangdong province. Chin. J. Prev. Vet. Med. 2017, 39, 621–623. [Google Scholar]
- Wang, S.; Gao, M.; An, T.; Liu, Y.; Jin, J.; Wang, G.; Jiang, C.; Tu, Y.; Hu, S.; Li, J.; et al. Genetic diversity and virulence of novel sequence types of Streptococcus suis from diseased and healthy pigs in China. Front. Microbiol. 2015, 6, 173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Ku, X.; Yu, X.; Sun, Q.; Wu, H.; Chen, F.; Zhang, X.; Guo, L.; Tang, X.; He, Q. Prevalence and antimicrobial susceptibilities of bacterial pathogens in Chinese pig farms from 2013 to 2017. Sci. Rep. 2019, 9, 9908. [Google Scholar] [CrossRef] [Green Version]
- Haenni, M.; Lupo, A.; Madec, J.Y. Antimicrobial Resistance in Streptococcus spp. Microbiol. Spectr. 2018, 6, 2–6. [Google Scholar] [CrossRef] [PubMed]
- Cai, T.; Luo, X.; Yindi, X.U.; Wei, D.; Dandan, H.E.; Liu, J.; Yuan, L.; Gongzheng, H.U. Research on the resistance of Streptococcus suis to macrolide antibiotics. J. Henan. Agric. Univ. 2019, 1, 73–81. [Google Scholar]
- Huang, J.H.; Li, Y.X.; Shang, K.X.; Kashif, J.; Qian, X.L.; Wang, L.P. Efflux Pump, Methylation and Mutations in the 23S rRNA Genes Contributing to the Development of Macrolide Resistance in Streptococcus suis Isolated from Infected Human and Swine in China. Pak. Vet. J. 2014, 34, 82–86. [Google Scholar]
- Martel, A.; Decostere, A.; Leener, E.D.; Marien, M.; Graef, E.D.; Heyndrickx, M.; Goossens, H.; Lammens, C.; Devriese, L.A.; Haesebrouck, F. Comparison and transferability of the erm (B) genes between human and farm animal streptococci. Microb. Drug. Resist. 2005, 11, 295–302. [Google Scholar] [CrossRef]
- Cheng-gang, J.; Yan-hua, L.; Xue-hui, C.; Hua-ji, Q.; Heng-min, T.; Di-qiu, L.; Chang-jiang, S. Erythromycin Resistance of Streptococcus suis Isolates In Vivo. Agric. Sci. China 2009, 8, 502–507. [Google Scholar]
- Huang, J.; Liang, Y.; Guo, D.; Shang, K.; Ge, L.; Kashif, J.; Wang, L. Comparative Genomic Analysis of the ICESa2603 Family ICEs and Spread of erm(B)- and tet(O)-Carrying Transferable 89K-Subtype ICEs in Swine and Bovine Isolates in China. Front. Microbiol. 2016, 7, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Wang, Y.; Sun, L.; Grenier, D.; Yi, L. Streptococcus suis biofilm: Regulation, drug-resistance mechanisms, and disinfection strategies. Appl. Microbiol. Biotechnol. 2018, 102, 9121–9129. [Google Scholar] [CrossRef] [PubMed]
- Waack, U.; Nicholson, T.L. Subinhibitory Concentrations of Amoxicillin, Lincomycin, and Oxytetracycline Commonly Used to Treat Swine Increase Streptococcus suis Biofilm Formation. Front. Microbiol. 2018, 9, 2707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Liu, B.; Li, J.; Gong, S.; Dong, X.; Mao, C.; Yi, L. LuxS/AI-2 system is involved in fluoroquinolones susceptibility in Streptococcus suis through overexpression of efflux pump SatAB. Vet. Microbiol. 2019, 233, 154–158. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.; Jin, M.; Li, J.; Grenier, D.; Wang, Y. Antibiotic resistance related to biofilm formation in Streptococcus suis. Appl. Microbiol. Biotechnol. 2020, 104, 1–12. [Google Scholar] [CrossRef]
- Bonifait, L.; Grignon, L.; Grenier, D. Fibrinogen induces biofilm formation by Streptococcus suis and enhances its antibiotic resistance. Appl. Environ. Microbiol. 2008, 74, 4969–4972. [Google Scholar] [CrossRef] [PubMed]
- Grenier, D.; Grignon, L.; Gottschalk, M. Characterisation of biofilm formation by a Streptococcus suis meningitis isolate. Vet. J. 2009, 179, 292–295. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Yi, L.; Yu, N.; Wang, G.; Ma, Z.; Lin, H.; Fan, H. Streptococcus suis Serotype 2 Biofilms Inhibit the Formation of Neutrophil Extracellular Traps. Front. Cell. Infect. Microbiol. 2017, 7, 86. [Google Scholar] [CrossRef] [Green Version]
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef] [Green Version]
- Sindeldecker, D.; Stoodley, P. The many antibiotic resistance and tolerance strategies of Pseudomonas aeruginosa. Biofilm 2021, 3, 100056. [Google Scholar] [CrossRef]
- Fridman, O.; Goldberg, A.; Ronin, I.; Shoresh, N.; Balaban, N.Q. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 2014, 513, 418–421. [Google Scholar] [CrossRef] [PubMed]
- Maneerat, K.; Yongkiettrakul, S.; Kramomtong, I.; Tongtawe, P.; Tapchaisri, P.; Luangsuk, P.; Chaicumpa, W.; Gottschalk, M.; Srimanote, P. Virulence genes and genetic diversity of Streptococcus suis serotype 2 isolates from Thailand. Transbound. Emerg. Dis. 2013, 60 (Suppl. 2), 69–79. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zheng, H.; Gottschalk, M.; Bai, X.; Lan, R.; Ji, S.; Liu, H.; Xu, J. Development of multiplex PCR assays for the identification of the 33 serotypes of Streptococcus suis. PLoS ONE 2013, 8, e72070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callens, B.F.; Haesebrouck, F.; Maes, D.; Butaye, P.; Dewulf, J.; Boyen, F. Clinical resistance and decreased susceptibility in Streptococcus suis isolates from clinically healthy fattening pigs. Microb. Drug Resist. 2013, 19, 146–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Wang, C.; Gao, L.; Cai, H.; Zhou, Y.; Yang, Y.; Xu, C.; Ding, W.; Chen, J.; Muhammad, I.; et al. Rutin Inhibits Streptococcus suis Biofilm Formation by Affecting CPS Biosynthesis. Front. Pharm. 2017, 8, 379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stepanovic, S.; Vukovic, D.; Dakic, I.; Savic, B.; Svabic-Vlahovic, M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods. 2000, 40, 175–179. [Google Scholar] [CrossRef]
- Stepanović, S.; Vuković, D.; Hola, V.; Di Bonaventura, G.; Djukić, S.; Cirković, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS. Acta Pathol. Microbiol. Immunol. Scand. 2007, 115, 891–899. [Google Scholar] [CrossRef]


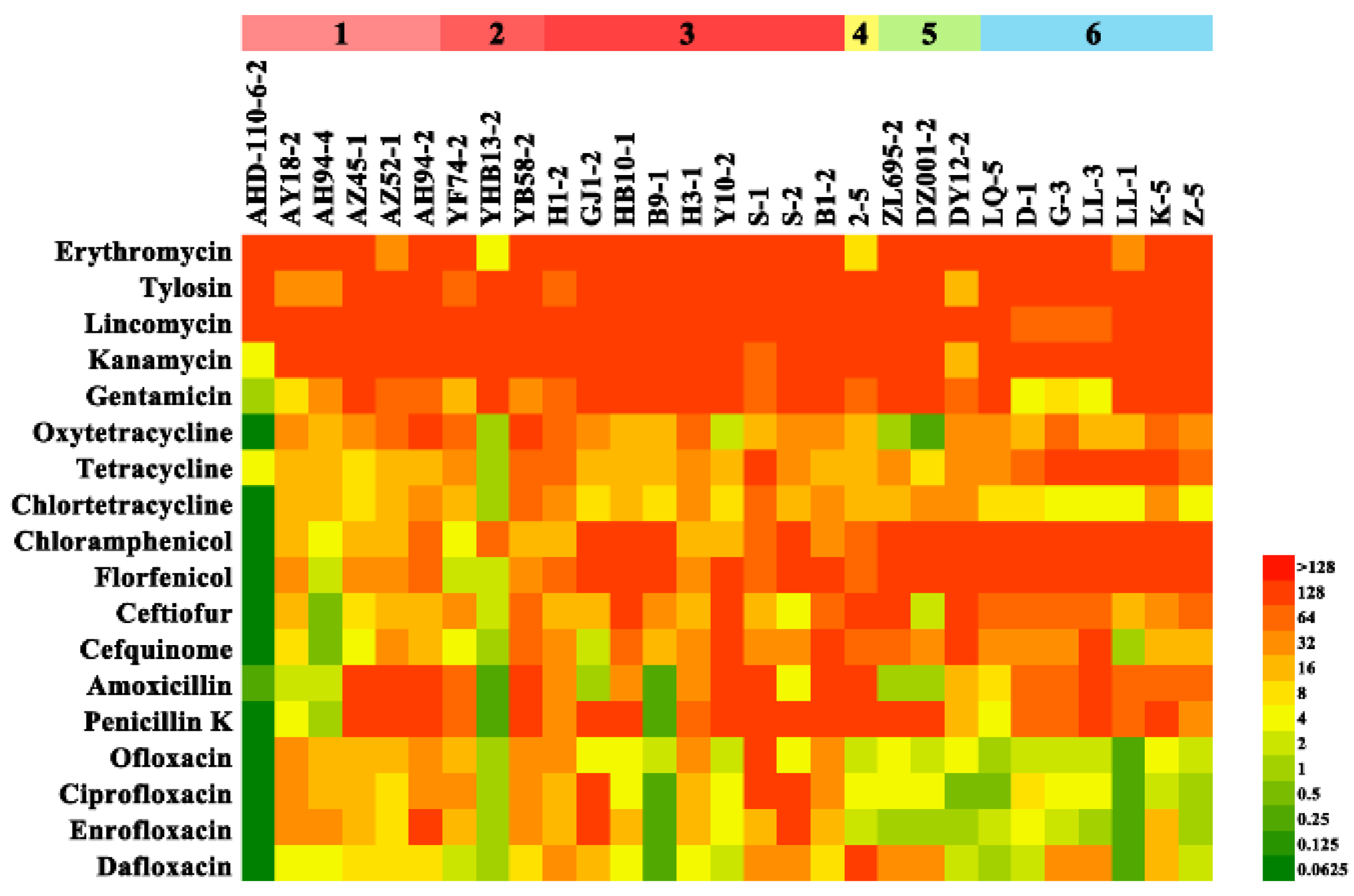

| Serial Number | Serotype | Antimicrobial Resistant Types * | No. of Antimicrobials |
|---|---|---|---|
| AHD-110-6-2 | 4 | ERY-TET | 2 |
| AY18-2 | 4 | CHL-CEF-ENR-ERY-PK-FFL-TET | 7 |
| AH94-4 | 4 | ENR-ERY-GEN-TET | 4 |
| AZ45-1 | 2 | CHL-CEF-ENR-ERY-GEN-PK-FFL-TET | 8 |
| AZ52-1 | 2 | CHL-CEF-ENR-ERY-GEN-PK-FFL-TET | 8 |
| AH94-2 | 4 | CHL-CEF-ENR-ERY-GEN-PK-FFL-TET | 8 |
| YF74-2 | unidentified | CEF-ENR-ERY-GEN-PK-TET | 6 |
| YHB13-2 | 2 | ENR-ERY-GEN | 3 |
| YB58-2 | 9 | CHL-CEF-ENR-ERY-GEN-PK-FFL-TET | 8 |
| H1-2 | 4 | CHL-CEF-ENR-ERY-GEN-PK-FFL-TET | 8 |
| GJ1-2 | 4 | CHL-CEF-ENR-ERY-GEN-PK-FFL-TET | 8 |
| HB10-1 | 4 | CHL-CEF-ENR-ERY-GEN-PK-FFL-TET | 8 |
| B9-1 | 4 | CHL-CEF-ERY-GEN-FFL-TET | 6 |
| H3-1 | 4 | CHL-CEF-ENR-ERY-GEN-PK-FFL-TET | 8 |
| Y10-2 | 4 | CHL-CEF-ENR-ERY-GEN-PK-FFL-TET | 8 |
| S-1 | 4 | CHL-CEF-ENR-ERY-GEN-PK-FFL-TET | 8 |
| S-2 | 4 | CHL-ENR-ERY-GEN-PK-FFL-TET | 7 |
| B1-2 | 4 | CHL-CEF-ENR-ERY-GEN-PK-FFL-TET | 8 |
| 2-5 | unidentified | CHL-CEF-ENR-ERY-GEN-PK-FFL-TET | 8 |
| ZL695-2 | unidentified | CHL-CEF-ERY-GEN-PK-FFL-TET | 7 |
| DZ001-2 | 4 | CHL-ERY-GEN-PK-FFL-TET | 6 |
| DY12-2 | 4 | CHL-CEF-ERY-GEN-PK-FFL-TET | 7 |
| LQ-5 | 2 | CHL-CEF-ENR-ERY-GEN-PK-FFL-TET | 8 |
| D-1 | 2 | CHL-CEF-ENR-ERY-PK-FFL-TET | 7 |
| G-3 | 2 | CHL-CEF-ENR-ERY-PK-FFL-TET | 7 |
| LL-3 | 2 | CHL-CEF-ERY-PK-FFL-TET | 6 |
| LL-1 | 2 | CHL-CEF-ERY-GEN-PK-FFL-TET | 7 |
| K-5 | 2 | CHL-CEF-ENR-ERY-GEN-PK-FFL-TET | 8 |
| Z-5 | 2 | CHL-CEF-ERY-GEN-PK-FFL-TET | 7 |
| Isolation Site | Number of Samples | Isolation S. suis |
|---|---|---|
| Harbin-1 | 40 | 6 |
| Harbin-2 | 26 | 3 |
| Harbin-3 | 44 | 9 |
| Suihua | 4 | 1 |
| Daqing | 40 | 3 |
| Qiqihar | 178 | 7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, C.-L.; Che, R.-X.; Wu, T.; Qu, Q.-W.; Chen, M.; Zheng, S.-D.; Cai, X.-H.; Wang, G.; Li, Y.-H. New Characterization of Multi-Drug Resistance of Streptococcus suis and Biofilm Formation from Swine in Heilongjiang Province of China. Antibiotics 2023, 12, 132. https://doi.org/10.3390/antibiotics12010132
Dong C-L, Che R-X, Wu T, Qu Q-W, Chen M, Zheng S-D, Cai X-H, Wang G, Li Y-H. New Characterization of Multi-Drug Resistance of Streptococcus suis and Biofilm Formation from Swine in Heilongjiang Province of China. Antibiotics. 2023; 12(1):132. https://doi.org/10.3390/antibiotics12010132
Chicago/Turabian StyleDong, Chun-Liu, Rui-Xiang Che, Tong Wu, Qian-Wei Qu, Mo Chen, Si-Di Zheng, Xue-Hui Cai, Gang Wang, and Yan-Hua Li. 2023. "New Characterization of Multi-Drug Resistance of Streptococcus suis and Biofilm Formation from Swine in Heilongjiang Province of China" Antibiotics 12, no. 1: 132. https://doi.org/10.3390/antibiotics12010132
APA StyleDong, C.-L., Che, R.-X., Wu, T., Qu, Q.-W., Chen, M., Zheng, S.-D., Cai, X.-H., Wang, G., & Li, Y.-H. (2023). New Characterization of Multi-Drug Resistance of Streptococcus suis and Biofilm Formation from Swine in Heilongjiang Province of China. Antibiotics, 12(1), 132. https://doi.org/10.3390/antibiotics12010132





