Evaluation of Antimicrobial Resistance of Different Phylogroups of Escherichia coli Isolates from Feces of Breeding and Laying Hens
Abstract
:1. Introduction
2. Results
2.1. Quantification of β-Glucoronidase Positive E. coli from Hens Fecal Samples
2.2. Determination of the E. coli Phylogenetic and Patotypes Groups
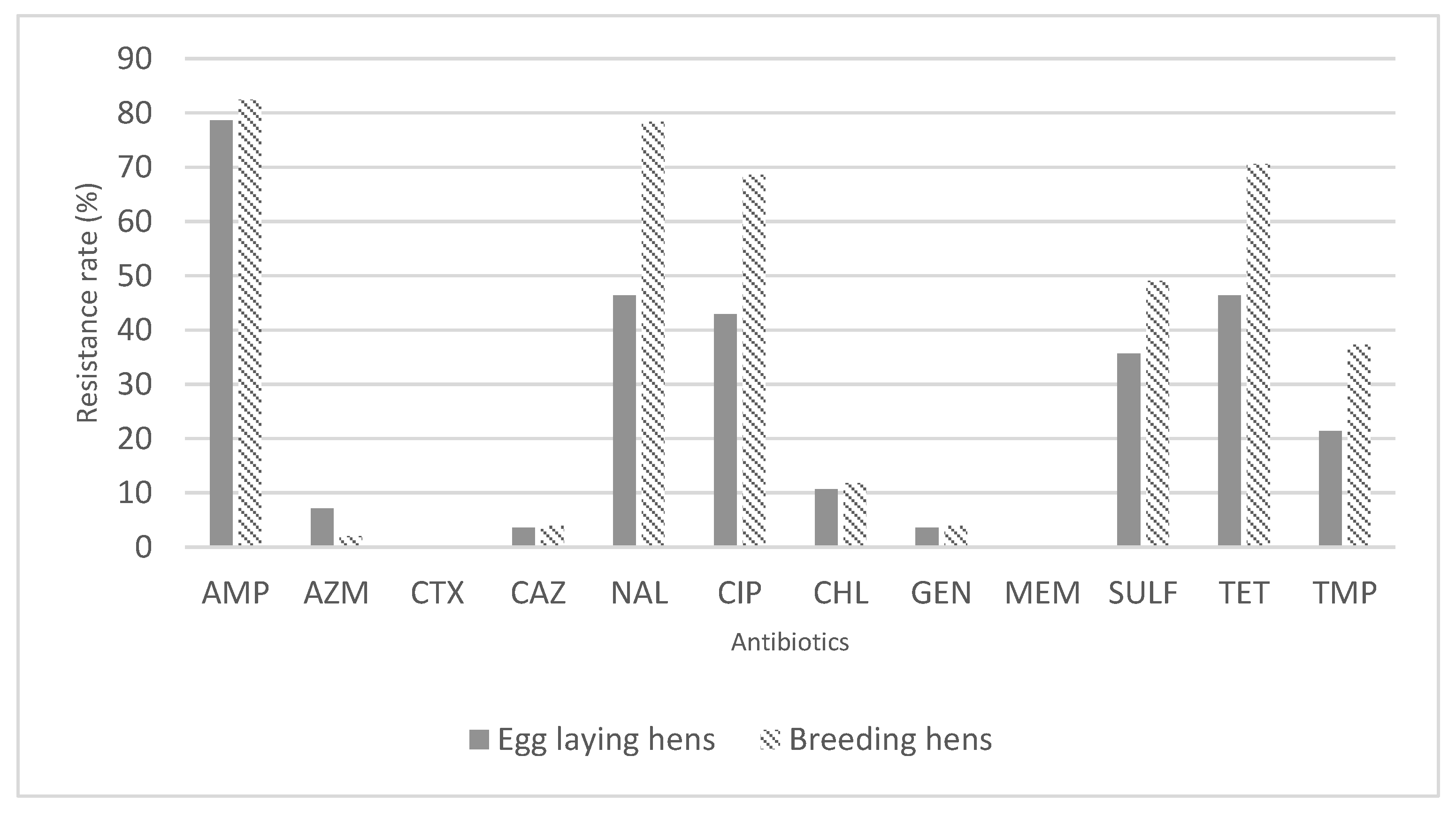
2.3. Susceptibility to Antimicrobials
2.4. Multiresistant Isolates
2.5. Detection of ESBL Resistance Genes
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Sampling and Bacterial Isolation
5.2. Phylogenetic Grouping and Determination of E. coli Pathotypes
5.3. Antimicrobial Susceptibility of E. coli Isolates
5.4. Multiresistant Isolates of E. coli Assessment
5.5. Detection of Extended-Spectrum β-Lactamase Resistance Genes
5.6. Data Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McAfee, A.J.; McSorley, E.M.; Cuskelly, G.J.; Moss, B.W.; Wallace, J.M.W.; Bonham, M.P.; Fearon, A.M. Red meat consumption: An overview of the risks and benefits. Meat Sci. 2010, 84, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Abebe, E.; Gugsa, G.; Ahmed, M. Review on Major Food-Borne Zoonotic Bacterial Pathogens. J. Trop Med. 2020, 2020, 4674235. [Google Scholar] [CrossRef]
- Hughes, C.; Gillespie, I.A.; O’Brien, S.J. Foodborne transmission of infectious intestinal disease in England and Wales, 1992–2003. Food Control 2007, 18, 766–772. [Google Scholar] [CrossRef]
- Algammal, A.M.; Hetta, H.F.; Batiha, G.E.; Hozzein, W.N.; El Kazzaz, W.M.; Hashem, H.R. Virulence-determinants and antibiotic-resistance genes of MDR-E. coli isolated from secondary infections following FMD-outbreak in cattle. Sci. Rep. 2020, 10, 19779. [Google Scholar] [CrossRef] [PubMed]
- Tyson, G.H.; Li, C.; Hsu, C.H.; Jones, S.B.; McDermott, P.F. Diverse fluoroquinolone resistance plasmids from retail meat E. coli in the United States. Front. Microbiol. 2019, 10, 2826. [Google Scholar] [CrossRef] [Green Version]
- Poole, T.L.; Callaway, T.R.; Norman, K.N.; Scott, H.M.; Loneragan, G.H.; Ison, S.A. Transferability of antimicrobial resistance from multidrug-resistant Escherichia coli isolated from cattle in the USA to E. coli and Salmonella Newport recipients. J. Glob. Antimicrob. Resist. 2017, 11, 123–132. [Google Scholar] [CrossRef]
- Tadesse, D.A.; Zhao, S.; Tong, E.; Ayers, S.; Singh, A.; Bartholomew, M.J.; McDermott, P.F. Antimicrobial Drug Resistance in Escherichia coli from humans and food animals, United States, 1950–2002. Emerg. Infect. Dis. 2012, 18, 741–749. [Google Scholar] [CrossRef]
- van Elsas, J.; Semenov, A.; Costa, R. Survival of Escherichia coli in the environment: Fundamental and public health aspects. ISME J. 2011, 5, 173–183. [Google Scholar] [CrossRef] [Green Version]
- Adzitey, F. Incidence and antimicrobial susceptibility of Escherichia coli isolated from beef (meat muscle, liver and kidney) samples in Wa Abattoir, Ghana. Cogent. Food Agric. 2020, 6, 2–10. [Google Scholar] [CrossRef]
- Gupta, P.; Adhikari, A. Novel approaches to environmental monitoring and control of Listeria monocytogenes in food production facilities. Foods 2022, 11, 1760. [Google Scholar] [CrossRef]
- Caudry, S.D.; Stanisich, V.A. Incidence of antibiotic-resistant Escherichia coli associated with frozen chicken carcasses and characterization of conjugative R plasmids derived from such strains. Antimicrob. Agents Chemother. 1979, 16, 701–709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nazer, A.H. Transmissible drug resistance in Escherichia coli isolated from poultry and their carcasses in Iran. Cornell Vet. 1980, 70, 365–371. [Google Scholar] [PubMed]
- Bensink, J.C.; Botham, F.P. Antibiotic resistant coliform bacilli, isolated from freshly slaughtered poultry and from chilled poultry at retail outlets. Aust. Vet. J. 1983, 60, 80–83. [Google Scholar] [CrossRef] [PubMed]
- Ojeniyi, A.A. Direct transmission of Escherichia coli from poultry to humans. Epidemiol. Infect. 1989, 103, 513–522. [Google Scholar] [CrossRef] [Green Version]
- Chaslus-Dancla, E.; Lafont, J.P. IncH plasmids in Escherichia coli strains isolated from broiler chicken carcasses. Appl. Environ. Microbiol. 1985, 49, 1016–1018. [Google Scholar] [CrossRef] [Green Version]
- Jayaratne, A.H.; Collins-Thompson, D.L.; Trevors, J.T. Occurrence of aminoglycoside phosphotransferase subclass I and II structural genes among Enterobacteriaceae spp. Isolated from meat samples. Appl. Microbiol. Biotechnol. 1990, 33, 547–552. [Google Scholar] [CrossRef]
- Turtura, G.C.; Massa, S.; Ghazvinizadeh, H. Antibiotic resistance among coliform bacteria isolated from carcasses of commercially slaughtered chickens. Int. J. Food Microbiol. 1990, 11, 351–354. [Google Scholar] [CrossRef]
- Lakhotia, R.L.; Stephens, J.F. Drug resistance and R factors among enterobacteria isolated from eggs. Poult. Sci. 1973, 52, 1955–1962. [Google Scholar] [CrossRef]
- van den Bogaard, A.E.; London, N.; Driessen, C.; Stobberingh, E.E. Antibiotic resistance of faecal Escherichia coli in poultry, poultry farmers and poultry slaughterers. J. Antimicrob. Chemother. 2001, 47, 763–771. [Google Scholar] [CrossRef]
- Kaneko, K.I.; Hayashidani, H.; Ohtomo, Y. Bacterial contamination of ready-to-eat foods and fresh products in retail shops and food factories. J. Food Prot. 1999, 62, 644–649. [Google Scholar] [CrossRef]
- Guzmán-Gómez, G.; Valdovinos, M.; Díaz, E.; Montaño, J.; Valle, M.; Vitela, M.; Quezada, S. Frequency of Salmonella and Listeria monocytogenes in Five Commercial Brands of Chicken Eggs Using a Combined Method of Enrichment and Nested-PCR. J. Food Prot. 2013, 76, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Gharbi, M.; Béjaoui, A.; Ben Hamda, C.; Alaya, N.; Hamrouni, S.; Bessoussa, G.; Ghram, A.; Maaroufi, A. Campylobacter spp. In Eggs and Laying Hens in the North-East of Tunisia: High Prevalence and Multidrug-Resistance Phenotypes. Vet. Sci. 2022, 9, 108. [Google Scholar] [CrossRef] [PubMed]
- Kapena, M.S.; Muma, J.B.; Mubita, C.M.; Munyeme, M. Antimicrobial resistance of Escherichia coli and Salmonella in raw retail table eggs in Lusaka, Zambia. Vet. World 2020, 13, 2528–2533. [Google Scholar] [CrossRef] [PubMed]
- Mehdi, Y.; Montminy, M.P.L.; Gaucher, M.L.; Chorfi, Y.; Suresh, G.; Rouissi, T.; Brar, S.K.; Côté, C.; Ramirez, A.A.; Godbout, S. Use of antibiotics in broiler production: Global impacts and alternatives. Anim. Nutr. 2018, 4, 170–178. [Google Scholar] [CrossRef]
- Singh, P.; Karimi, A.; Devendra, K.; Waldroup, P.W.; Cho, K.K.; Kwon, Y.M. Influence of penicillin on microbial diversity of the cecal microbiota in broiler chickens. Poult. Sci. 2013, 92, 272–276. [Google Scholar] [CrossRef]
- O’Brien, T.F. Emergence, spread, and environmental effect of antimicrobial resistance: How use of an antimicrobial anywhere can increase resistance to any antimicrobial anywhere else. Clin. Infect. Dis. 2002, 34, S78–S84. [Google Scholar] [CrossRef] [Green Version]
- Wolny-Koładka, K.; Zdaniewicz, M. Antibiotic Resistance of Escherichia coli Isolated from Processing of Brewery Waste with the Addition of Bulking Agents. Sustainability 2021, 13, 10174. [Google Scholar] [CrossRef]
- Kluytmans, J.A.J.W.; Overdevest, I.T.M.A. Extended-spectrum beta-lactamase-producing Escherichia coli from retail chicken meat and humans: Comparison of strains, plasmids, resistance genes, and virulence factors. Clin. Infect. Dis. 2013, 56, 478–487. [Google Scholar] [CrossRef] [Green Version]
- Zachary, R.; Stromberg, Z.R.; Johnson, J.R.; Fairbrother, J.M.; Kilbourne, J.; Van Goor, A.; Curtiss, R.; Mellata, M. Evaluation of Escherichia coli isolates from healthy chickens to determine their potential risk to poultry and human health. PloS ONE 2017, 12, e0180599. [Google Scholar] [CrossRef] [Green Version]
- Apata, D.F. Antibiotic Resistance in Poultry. Int. J. Poult. Sci. 2009, 8, 404–408. [Google Scholar] [CrossRef]
- Barton, M.D. Antibiotic use in animal feed and its impact on human healt. Nutr. Res. Rev. 2000, 13, 279–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bond, V.; Jewell, J. The Impacts of Antibiotic Use in Animals on Human Health and Animal Welfare; Investor Briefing No. 17; Business Benchmark on Farm Animal Welfare, 2014; pp. 1–9. Available online: https://www.bbfaw.com/media/1070/briefing-17-impacts-of-antibiotic-use-in-animals-on-human-health-and-animal-welfare.pdf (accessed on 23 November 2022).
- DGAV. Available online: https://www.dgav.pt/wp-content/uploads/2021/09/ESVAC-RELATORIO-2017.pdf (accessed on 23 November 2022).
- Tenaillon, O.; Skurnik, D.; Picard, B.; Denamur, E. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 2010, 8, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doumith, M.; Day, M.J.; Hope, R.; Wain, J.N.; Woodford, N. Improved multiplex PCR strategy for rapid assignment of the four major Escherichia coli phylogenetic groups. J. Clin. Microbiol. 2012, 50, 3108–3110. [Google Scholar] [CrossRef] [Green Version]
- Katongole, P.; Kisawuzi, D.B.; Bbosa, H.K.; Kateete, D.P.; Najjuka, C.F. Phylogenetic groups and antimicrobial susceptibility patterns of uropathogenic Escherichia coli clinical isolates from patients at Mulago National Referral Hospital, Kampala, Uganda. F1000Research 2019, 8, 1828. [Google Scholar] [CrossRef]
- Clermont, O.; Christenson, J.K.; Denamur, E. The Clermont Escherichia coli phylotyping method revisited: Improvement of specificity and detection of new phylogroups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]
- Clermont, O.; Gordon, D.; Denamur, E. Guide to the various phylogenetic classification schemes for Escherichia coli and the correspondence among schemes. Microbiology 2015, 161, 980–988. [Google Scholar] [CrossRef]
- Santos, Y.P.A.; Flores, M.E.B.; Camacho, S.P.D.; Beltrán, M.J.U.; Campos, C.A.E.; Unda, J.R.P. Association of phylogenetic distribution and presence of integrons with multidrug resistance in Escherichia coli clinical isolates from children with diarrhoea. J. Infect. Public Health 2020, 13, 767–772. [Google Scholar] [CrossRef]
- Barzan, M.; Rad, M.; Tabar, G.R.H.; Azizzadeh, M. Phylogenetic analysis of Escherichia coli isolates from healthy and diarrhoeic calves in Mashhad, Iran. Bulg. J. Vet. Med. 2017, 20, 11–18. [Google Scholar] [CrossRef]
- Bélanger, L.A.; Garenaux, J.; Harel, M.; Boulianne, E.; Dozois, C. Escherichia coli from animal reservoirs as a potential source of human extraintestinal pathogenic Escherichia coli. FEMS Immunol. Med. Microbiol. 2011, 62, 1–10. [Google Scholar] [CrossRef]
- Croxall, G.; Hale, J.; Weston, V.; Manning, G.; Cheetham, P.; Achtman, M.; McNally, A. Molecular epidemiology of extraintestinal pathogenic Escherichia coli isolates from a regional cohort of elderly patients highlights the prevalence of ST131 strains with increased antimicrobial resistance in both community and hospital care settings. J. Antimicrob. Chemother. 2011, 66, 2501–2508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L.T. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Bekal, S.; Brousseau, R.; Masson, L.; Préfontaine, G.; Fairbrother, J.M.; Harel, J. Rapid identification of Escherichia coli pathotypes by virulence gene detection with DNA microarray. J. Clin. Microbiol. 2003, 41, 2113–2125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caine, L.A.; Nwodo, U.U.; Okoh, A.I.; Ndip, R.N.; Green, E. Occurrence of Virulence Genes Associated with Diarrheagenic Escherichia coli Isolated from Raw Cow’s Milk from Two Commercial Dairy Farms in the Eastern Cape Province, South Africa. Int. J. Environ. Res. Public Health 2014, 11, 11950–11963. [Google Scholar] [CrossRef] [PubMed]
- Murase, T.; Ozaki, H. Relationship between phylogenetic groups of Escherichia coli and Pathogenicity among Isolates from chickens with Colibacillosis and healthy chickens. Poult Sci. 2022, 101, 102007. [Google Scholar] [CrossRef] [PubMed]
- Pavlovic, M.; Luze, A.; Konrad, R.; Berger, A.; Sing, A.; Busch, U.; Huber, I. Development of a duplex real-time PCR for differentiation between E. coli and Shigella spp. J. Appl. Microbiol. 2011, 110, 1245–1251. [Google Scholar] [CrossRef] [PubMed]
- Zenati, F.; Barguigua, A.; Nayme, K.; Benbelaïd, F.; Khadir, A.; Bellahsene, C.; Bendahou, M.; Hafida, H.; Timinouni, M. Characterization of uropathogenic ESBL-producing Escherichia coli isolated from hospitalized patients in western Algeria. J. Infect. Dev. Ctries. 2019, 13, 291–302. [Google Scholar] [CrossRef]
- Alonso, C.A.; Zarazaga, M.; Ben Sallem, R.; Jouini, A.; Ben Slama, K.; Torres, C. Antibiotic resistance in Escherichia coli in husbandry animals: The African perspective. Lett. Appl. Microbiol. 2007, 64, 318–334. [Google Scholar] [CrossRef] [Green Version]
- Riaño, I.; Moreno, M.A.; Teshager, T.; Sáenz, Y.; Dominguez, L.; Torres, C. Detection and characterization of extended-spectrum beta-lactamases in Salmonella enterica strains of healthy food animals in Spain. J. Antimicrob. Chemother. 2006, 58, 844–847. [Google Scholar] [CrossRef] [Green Version]
- Meguenni, N.; Le Devendec, L.; Jouy, E.; Le Corvec, M.; Bounar-Kechih, S.; Bakour, R.; Kempf, I. First description of an extended-spectrum cephalosporin- and fluoroquinolone- resistant avian pathogenic Escherichia coli clone in Algeria. Avian Dis. 2015, 59, 20–23. [Google Scholar] [CrossRef]
- Belmahdi, M.; Bakour, S.; Al Bayssari, C.; Touati, A.; Rolain, J.M. Molecular Characterization of extended-spectrum β-lactamase- and plasmid AmpC-producing Escherichia coli strains isolated from broilers in Béjaïa, Algeria. J. Glob. Antimicrob. Resist. 2016, 6, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Maamar, E.; Hammami, S.; Alonso, C.A.; Dakhli, N.; Abbassi, M.S.; Ferjani, S.; Hamzaoui, Z.; Saidani, M.; Torres, C.; Boutiba-Ben Boubaker, I. High prevalence of extended-spectrum and plasmidic AmpC beta-lactamase-producing Escherichia coli from poultry in Tunisia. Int. J. Food Microbiol. 2016, 231, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Casella, T.; Nogueira, M.C.L.; Saras, E.; Haenni, M.; Madec, J.Y. High prevalence of ESBLs in retail chicken meat despite reduced use of antimicrobials in chicken production, France. Int. J. Food Microbiol. 2017, 257, 271–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 12.0. 2022. Available online: http://www.eucast.org (accessed on 10 October 2022).
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 32nd ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2022; ISSN1 978-1-68440-134-5. ISSN2 978-1-68440-135-2. [Google Scholar]
- Clermont, O.; Dixit, O.; Vangchhia, B.; Condamine, B.; Dion, S.; Bridier-Nahmias, A.; Denamur, E.; Gordon, D. Characterization and rapid identification of phy- logroup G in Escherichia coli, a lineage with high virulence and antibiotic resistance potential. Environ. Microbiol 2019, 21, 3107–3117. [Google Scholar] [CrossRef]
- Bingen, E.; Picard, B.; Brahimi, N.; Mathy, S.; Desjardins, P.; Elion, J.; Denamur, E. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J. Infect. Dis. 1998, 177, 642–650. [Google Scholar] [CrossRef] [Green Version]
- Picard, B.; Garcia, J.S.; Gouriou, S.; Duriez, P.; Brahimi, N.; Bingen, E.; Elion, J.; Denamur, E. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 1999, 67, 546–553. [Google Scholar] [CrossRef] [Green Version]
- Obeng, A.S.; Rickard, H.; Ndi, O.; Sexton, M.; Barton, M. Antibiotic resistance, phylogenetic grouping and virulence potential of Escherichia coli isolated from the faeces of intensively farmed and free range poultry. Vet. Microbiol. 2012, 154, 305–315. [Google Scholar] [CrossRef]
- Hayashi, W.; Ohsaki, Y.; Taniguchi, Y.; Koide, S.; Kawamura, K.; Suzuki, M.; Kimura, K.; Wachino, J.; Nagano, Y.; Arakawa, Y.; et al. High prevalence of blaCTX-M-14 among genetically diverse Escherichia coli recovered from retail raw chicken meat portions in Japan, Int. J. Food Microbiol. 2018, 284, 98–104. [Google Scholar] [CrossRef]
- Projahn, M.; Daehre, K.; Roesler, U.; Friese, A. Extended-spectrum-beta-lactamase- and plasmid-encoded cephamycinase-producing Enterobacteria in the broiler hatchery as a potential mode of pseudo-vertical transmission. Appl. Environ. Microbiol. 2016, 83, e02364-16. [Google Scholar] [CrossRef] [Green Version]
- Adefioye, O.J.; Weinreich, J.; Rödiger, S.; Schierack, P.; Olowe, O.A. Phylogenetic Characterization and Multilocus Sequence Typing of Extended-Spectrum Beta Lactamase-Producing Escherichia coli from Food-Producing Animals, Beef, and Humans in Southwest Nigeria. Microb. Drug Resist. 2021, 27, 111–120. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G.J. Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 2018, 42, fux053. [Google Scholar] [CrossRef] [PubMed]
- Hossain, T.; Rafiq, K.; Islam, Z.; Chowdhury, S.; Islam, P.; Haque, Z.; Samad, M.A.; Sani, A.A.; Ferdous, M.R.A.; Islam, R.; et al. A survey on knowledge, attitude, and practices of large-animal farmers towards antimicrobial use, resistance, and residues in Mymensingh division of Bangladesh. Antibiotics 2022, 11, 442. [Google Scholar] [CrossRef] [PubMed]
- Javadi, A.; Khatibi, S.A. Effect of commercial probiotic (Protexin®) on growth, survival and microbial quality of shrimp (Litopenaeus vannamei). Nutr. Food Sci. 2017, 47, 204–216. [Google Scholar] [CrossRef]
- Hailu, W.; Helmy, Y.A.; Carney-Knisely, G.; Kauffman, M.; Fraga, D.; Rajashekara, G. Prevalence and Antimicrobial Resistance Profiles of Foodborne Pathogens Isolated from Dairy Cattle and Poultry Manure Amended Farms in Northeastern Ohio, the United States. Antibiotics 2021, 10, 1450. [Google Scholar] [CrossRef] [PubMed]
- Langata, L.M.; Maingi, J.M.; Musonye, H.A.; Kiiru, J.; Nyamache, A.K. Antimicrobial resistance genes in Salmonella and Escherichia coli isolates from chicken droppings in Nairobi, Kenya. BMC Res. Notes 2019, 12, 22. [Google Scholar] [CrossRef] [Green Version]
- Abbassi, M.S.; Kilani, H.; Abid, I.; Sáenz, Y.; Hynds, P.; Lengliz, S.; Chehida, N.B.; Boubaker, I.B. Genetic background of antimicrobial resistance in multiantimicrobial-resistant Escherichia coli isolates from feces of healthy broiler chickens in Tunisia. BioMed Res. Int. 2021, 2021, 1269849. [Google Scholar] [CrossRef]
- European Food Safety Authority. Scientific Opinion on the public health risks of bacterial strains producing extended-spectrum β-lactamases and/or AmpC β-lactamases in food and food-producing animals. EFSA J. 2011, 9, 2322. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Wang, P.; Dai, Y.; Liu, Y.; Song, Y.; Yu, L.; Feng, C.; Liu, M.; Xie, Z.; Shang, Y.; et al. Longitudinal monitoring of multidrug resistance in Escherichia coli on broiler chicken fattening farms in Shandong, China. Poult. Sci. 2021, 100, 100887. [Google Scholar] [CrossRef]
- Koju, P.; Shrestha, R.; Shrestha, A.; Tamrakar, S.; Rai, A.; Shrestha, P.; Madhup, S.K.; Katuwal, N.; Shrestha, A.; Shrestha, A.; et al. Antimicrobial resistance in E. coli isolated from chicken cecum samples and factors contributing to antimicrobial resistance in Nepal. Trop. Med. Infect. Dis. 2022, 7, 249. [Google Scholar] [CrossRef]
- Huneau-Salaun, A.; Michel, V.; Huonnic, D.; Balaine, L.; Le Bouquin, S. Factors influencing bacterial eggshell contamination in conventional cages, furnished cages and free-range systems for laying hens under commercial conditions. Br. Poult. Sci. 2010, 51, 163–169. [Google Scholar] [CrossRef]
- Egea, P.; López-Cerero, L.; Navarro, M.D. Assessment of the presence of extended-spectrum beta-lactamase-producing Escherichia coli in eggshells and ready-to-eat products. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 1045–1047. [Google Scholar] [CrossRef]
- Machado, E.; Coque, T.M.; Cantón, R.; Sousa, J.C.; Peixe, L. Antibiotic resistance integrons and extended-spectrum {beta}-lactamases among Enterobacteriaceae isolates recovered from chickens and swine in Portugal. J. Antimicrob. Chemother. 2008, 62, 296–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clemente, L.; Leão, C.; Moura, L.; Albuquerque, T.; Amaro, A. Prevalence and characterization of ESBL/AmpC producing Escherichia coli from fresh meat in Portugal. Antibiotics 2021, 10, 1333. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, Z.H.; Kabir, M.H.; Ali, S.; Moniruzzaman, M.; Imran, K.M.; Nafiz, T.N.; Islam, M.S.; Hussain, A.; Hakim, S.A.I.; Worth, M.; et al. Extended-Spectrum Beta-Lactamase-Producing Escherichia coli in Drinking Water Samples from a Forcibly Displaced, Densely Populated Community Setting in Bangladesh. Front. Public Health 2020, 8, 228. [Google Scholar] [CrossRef]
- ISO 16649-2; Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for the Enumeration of β-glucoronidase-positive Escherichia coli- Part 2: Colony—Count Technique at 44 °C Using 5-bromo-4-chloro-3-indolyl β-D-glucuronide. ISO: Geneva, Switzerland, 2001.
- Schmidt, H.; Knop, C.; Franke, S.; Aleksic, S.; Heesemann, J.; Karch, H. Development of PCR for screening of enteroaggregative Escherichia coli. J. Clin. Microbiol. 1995, 33, 701–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aranda, K.R.; Fabbricotti, S.H.; Fagundes-Neto, U.; Scaletsky, I.C. Single multiplex assay to identify simultaneously enteropathogenic, enteroaggregative, enterotoxigenic, enteroinvasive and Shiga toxin-producing Escherichia coli strains in Brazilian children. FEMS Microbiol. Lett. 2007, 267, 145–150. [Google Scholar] [CrossRef] [Green Version]
- ISO/TS 13136; Microbiology of Food and Animal Feed—Real-Time Polymerase Chain Reaction (PCR)-Based Method for the Detection of Food-Borne Pathogens—Horizontal Method for the Detection of Shiga Toxin-Producing Escherichia coli (STEC) and the Determination of O157, O111, O26, O103 and O145 Serogroups. ISO: Geneva, Switzerland, 2012.
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Arlet, G.; Rouveau, M.; Philippon, A. Substitution of alanine for aspartate at position 179 in the SHV-6 extended-spectrum beta-lactamase. FEMS Microbiol. Lett. 1997, 152, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Monstein, H.J.; Ostholm-Balkhed, A.; Nilsson, M.V.; Nilsson, M.; Dornbusch, K.; Nilsson, L.E. Multiplex PCR amplification assay for the detection of blaSHV, blaTEM and blaCTX-M genes in Enterobacteriaceae. APMIS 2007, 115, 1400–1408. [Google Scholar] [CrossRef]
- Dallenne, C.; Da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef]
- Oliveira, R.; Castro, J.; Silva, S.; Oliveira, H.; Saavedra, M.J.; Azevedo, N.F.; Almeida, C. Exploring the Antibiotic Resistance Profile of Clinical Klebsiella pneumoniae Isolates in Portugal. Antibiotics 2022, 11, 1613. [Google Scholar] [CrossRef] [PubMed]
- IBM. IBM SPSS Statistics for Windows 2015; IBM Corperation: Armonk, NY, USA, 2015. [Google Scholar]
| Sample | Samples Sources | Microbiological Load of E. coli (log CFU/g) |
|---|---|---|
| Log_TBX | Egg laying hens | 6.02 a ± 1.43 |
| Breeding hens | 6.03 a ± 0.98 | |
| Log_TBXamp | Egg laying hens | 4.46 a ± 1.65 |
| Breeding hens | 5.06 a ± 1.63 | |
| Log_TBXenro | Egg laying hens | 2.81 a ± 1.12 |
| Breeding hens | 4.12 b ± 1.33 |
| Samples Source | No. of Samples | No. of Isolates | No. of Isolates AMP (R) | No. of Isolates ENR (R) |
|---|---|---|---|---|
| Breeding hens | 31 | 51 | 21 (41.2%) | 30 (58.8%) |
| Egg laying hens | 29 | 28 | 16 (57.1%) | 12 (42.9%) |
| Total | 60 | 79 | 37 (46.8%) | 42 (53.2%) |
| Phylogroup | |||||
|---|---|---|---|---|---|
| A | B1 | D | E | Unknown | |
| No. of isolates of breeding hens (%) | 8 (15.6%) | 42 (82.4%) | 0 (0%) | 1 (2%) | 0 (0%) |
| No. of isolates of egg laying hens (%) | 7 (25%) | 17 (60.7%) | 1 (3.6%) | 2 (7.1%) | 1 (3.6%) |
| Total no. of isolates (%) | 15 (19%) | 59 (74.6%) | 1 (1.3%) | 3 (3.8%) | 1 (1.3%) |
| Antibiotic | No. of Isolates (%S) | No. of Isolates (%R) |
|---|---|---|
| AMP | 15 (19.0) | 64 (81.0) |
| AZM | 76 (96.2) | 3 (3.8) |
| CTX | 79 (100.0) | 0 (0.0) |
| CAZ | 76 (96.2) | 3 (3.8) |
| NAL | 27 (34.2) | 52 (65.8) |
| CIP | 32 (40.5) | 47 (59.5) |
| CHL | 70 (88.6) | 9 (11.4) |
| GEN | 76 (96.2) | 3 (3.8) |
| MEM | 79 (100.0) | 0 (0.0) |
| SULF | 44 (55.7) | 35 (44.3) |
| TET | 20 (38.0) | 49 (62.0) |
| TMP | 54 (68.4) | 25 (31.6) |
| No. of Antimicrobials Classes Per Group | MDR Pattern | No. of Isolates of Breeding Hens | No. of Isolates of Egg Laying Hens | Total No. of Isolates (%) |
|---|---|---|---|---|
| 6 | PEN + CEP + FQs + TETs + MA + SULFs | 1 0 0 | 0 1 1 | 3 (6.7%) |
| PEN + FQs + M + TETs + MA + SULFs | ||||
| PEN + FQs + AMG + TETs + MA + SULFs | ||||
| 5 | PEN + FQs + TETs + MA + SULFs PEN + AMG + TETs + MA + SULFs FQs + MGS + TETs + MA + SULFs | 14 1 1 | 2 0 1 | 19 (42.2%) |
| 4 | PEN + CEP + FQs + TETs | 0 | 1 | 9 (20%) |
| PEN + FQs + TETs + SULFs | 5 | 0 | ||
| PEN + FQs + TETs + MA | 1 | 1 | ||
| PEN + TETs + MA + SULFs | 1 | 0 | ||
| 3 | PEN + CEP + TETs PEN + FQs + SULFs PEN + FQs + TETs PEN + TETs + MA PEN + TETs + SULFs PEN + MA + SULFs FQs + TETs + SULFs | 1 6 1 0 3 0 1 | 0 0 0 1 0 1 0 | 14 (31.1%) |
| Total | 36 | 9 | 45 (100%) | |
| Isolate | Type | Phylogroup | AMR | MDR |
|---|---|---|---|---|
| 5 AMP | Breeding | E | AMP + CAZ + NAL + CIP + SULF + TET + TMP | 6 classes |
| 15 AMP | Breeding | B1 | AMP + CAZ + TET | 4 classes |
| 29 ENRO | Egg laying | B1 | AMP + CZD + NAL + CIP + TET | 3 classes |
| Primer | Target | Sequence (5′-3′) | PCR Product. (bp) | Ref. |
|---|---|---|---|---|
| arpA fwd | arpA | AACGCTATTCGCCAGCTTGC | 400 | [38] |
| arpA rev | TCTCCCCATACCGTACGCTA | |||
| chuaA fwd | chuaA | ATGGTACCGGACGAACCAAC | 288 | [38] |
| chuaA rev | TGCCGCCAGTACCAAAGAC | |||
| yjaA fwd | yjaA | CAAACGTGAAGTGTCAGGAG | 211 | [38] |
| yjaA rev | AATGCGTTCCTCAACCTGTG | |||
| TspE4.C2 | TspE4.C2 | CACTATTCGTAAGGTCATCC | 152 | [38] |
| TspE4.C2 rev | AGTTTATCGCTGCGGGTCGC |
| Phylogroup | Target Gene | |||
|---|---|---|---|---|
| arpA | chuA | yjaA | TspE4.C2 | |
| A | + | - | - | - |
| A or C | + | - | + | - |
| B1 | + | - | - | + |
| B2 | - | + | + | - |
| B2 | - | + | - | + |
| B2 | - | + | + | + |
| E or D | + | + | - | - |
| E or D | + | + | - | + |
| E or Clade I | + | + | + | - |
| F | - | + | - | - |
| (a) | + | - | + | + |
| Primer | Target | Sequence (5′-3′) | PCR Product. (bp) | Ref. |
|---|---|---|---|---|
| trpAgpC.1 fwd | trpA | AGTTTTATGCCCAGTGCGAG | 219 | [38] |
| trpAgpC.1 rev | TCTGCGCCGGTCACGCCC |
| Primer | Target | Sequence (5′-3′) | PCR Product. (bp) | Ref. |
|---|---|---|---|---|
| ArpAgpE fwd | arpA | GATTCCATCTTGTCAAAATATGCC | 301 | [38] |
| ArpAgpE rev | GAAAAGAAAAAGAATTCCCAAGAG |
| Pathotypes | Primer | Target | Sequence (5′-3′) | PCR Product (bp) | Ref. |
|---|---|---|---|---|---|
| ETEC | est (ST) fwd | est (ST) elt (LT) | ATTTTTMTTTCTGTATTRTCTT | 190 450 | [81] |
| est (ST) rev | CACCCGGTACARGCAGGATT | ||||
| elt (LT) fwd elt (LT) rev | GGCGACAGATTATACCGTGC CGGTCTCTATATTCCCTGTT | ||||
| EIEC | ipaH fwd ipaH rev | ipaH | GTTCCTTGACCGCCTTTCCGATACCGTC GCCGGTCAGCCACCCTCTGAGAGTAC | 600 | [81] |
| EAEC | aggr fwd aggr rev cvd432 fwd cvd432 rev | aggr cvd432 | GTATACACAAAAGAAGGAAGC ACAGAATCGTCAGCATCAGC CTGGCGAAAGACTGTATCAT CAATGTATAGAAATCCGCTGTT | 254 630 | [80] |
| EPEC | bfpA fwd bfpA rev eae fwd eae rev | bfpA eae | AATGGTGCTTGCGCTTGCTGC GCCGCTTTATCCAACCTGGTA GACCCGGCACAAGCATAAGC CCACCTGCAGCAACAAGAGG | 326 384 | [81] |
| STEC | stx1 fwd stx1 rev stx2 fwd stx2 rev eae fwd eae rev | stx1 stx2 eae | ATAAATCGCCATTCGTTGACTAC AGAACGCCCACTGAGATCATC CGCACTGTCTGAAACTGCTCC TCGCCAGTTATCTGACATTCTG GACCCGGCACAAGCATAAGC CCACCTGCAGCAACAAGAGG | 180 255 384 | [82] |
| Primer | Primer Concentration | Amplification Conditions |
|---|---|---|
| est (ST) fwd | 0.5 μM 0.5 μM 0.5 μM 0.5 μM | 95 °C, 5 min 35 cycles of 95 °C, 30 s; 55 °C, 60 s; 72 °C, 14 s 72 °C, 5 min |
| est (ST) rev | ||
| elt (LT) fwd elt (LT) rev | ||
| ipaH fwd ipaH rev | 0.2 μM 0.2 μM | 95 °C, 5 min 30 cycles of 95 °C, 30 s; 60 °C, 1 min; 72 °C, 18 s 72 °C, 5 min |
| aggr fwd aggr rev | 0.2 Mm 0.2 Mm | 95 °C, 5 min 30 cycles of 95 °C, 30 s; 60 °C, 1 min; 72 °C, 8 s 72 °C, 5 min |
| cvd432 fwd cvd432 rev | 0.2 μM 0.2 μM | 95 °C, 5 min 10 cycles of 95 °C, 30 s; 55 °C, 1 min; 72 °C, 19 s 20 cycles of 95 °C, 30 s; 60 °C, 1 min; 72 °C, 19 s 72 °C, 5 min |
| bfpA fwd bfpA rev | 0.2 μM 0.2 μM | 95 °C, 5 min 10 cycles of 95 °C, 30 s; 55 °C, 1 min; 72 °C, 10 s 20 cycles of 95 °C, 30 s; 60 °C, 1 min; 72 °C, 10 s 72 °C, 5 min |
| stx1 fwd stx1 rev stx2 fwd stx2 rev eae fwd eae rev | 0.8 μM 0.8 μM 2.4 μM 2.4 μM 0.8 μM 0.8 μM | 95 °C, 5 min 9 cycles of 95 °C, 60 s; 65 °C, 2 min; 72 °C, 90 s 95 °C, 60 s; 64 °C, 2 min; 72 °C, 90 s; 95 °C, 60 s; 63 °C, 2 min; 72 °C, 90 s 95 °C, 62 s; 64 °C, 2 min; 72 °C, 90 s; 95 °C, 60 s; 61 °C, 2 min; 72 °C, 90 s 10 cycles of 95 °C, 60 s; 60 °C, 2 min; 72 °C, 90 s 9 cycles of 95 °C, 60 s; 60 °C, 2 min; 72 °C, 150 s 72 °C, 5 min |
| Primer | Target | Sequence (5′-3′) | PCR Product. (bp) | Ref. |
|---|---|---|---|---|
| blaTEM fwd | blaTEM | CATTTCCGTCGCCCTTATTC | 800 | [84] |
| blaTEM rev | CGTTCATCCATAGTTGCCTGAC | |||
| blaSHV fwd | blaSHV | AGCCGCTTGAGCAAATTAAAC | 713 | [85] |
| blaSHV rev | ATCCCGCAGATAAATCACCAC | |||
| blaCTX-M fwd | blaCTX-M | ATGTGCAGYACCGTAARGTKATGC | 593 | [86] |
| blaCTX-M rev | TGGGTRAARTARGTSACCAGAAYCAGCGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pais, S.; Costa, M.; Barata, A.R.; Rodrigues, L.; Afonso, I.M.; Almeida, G. Evaluation of Antimicrobial Resistance of Different Phylogroups of Escherichia coli Isolates from Feces of Breeding and Laying Hens. Antibiotics 2023, 12, 20. https://doi.org/10.3390/antibiotics12010020
Pais S, Costa M, Barata AR, Rodrigues L, Afonso IM, Almeida G. Evaluation of Antimicrobial Resistance of Different Phylogroups of Escherichia coli Isolates from Feces of Breeding and Laying Hens. Antibiotics. 2023; 12(1):20. https://doi.org/10.3390/antibiotics12010020
Chicago/Turabian StylePais, Sandra, Mariana Costa, Ana Rita Barata, Lígia Rodrigues, Isabel M. Afonso, and Gonçalo Almeida. 2023. "Evaluation of Antimicrobial Resistance of Different Phylogroups of Escherichia coli Isolates from Feces of Breeding and Laying Hens" Antibiotics 12, no. 1: 20. https://doi.org/10.3390/antibiotics12010020
APA StylePais, S., Costa, M., Barata, A. R., Rodrigues, L., Afonso, I. M., & Almeida, G. (2023). Evaluation of Antimicrobial Resistance of Different Phylogroups of Escherichia coli Isolates from Feces of Breeding and Laying Hens. Antibiotics, 12(1), 20. https://doi.org/10.3390/antibiotics12010020







