Teleost Piscidins—In Silico Perspective of Natural Peptide Antibiotics from Marine Sources
Abstract
:1. Introduction
2. UniProt-Reviewed Piscidins
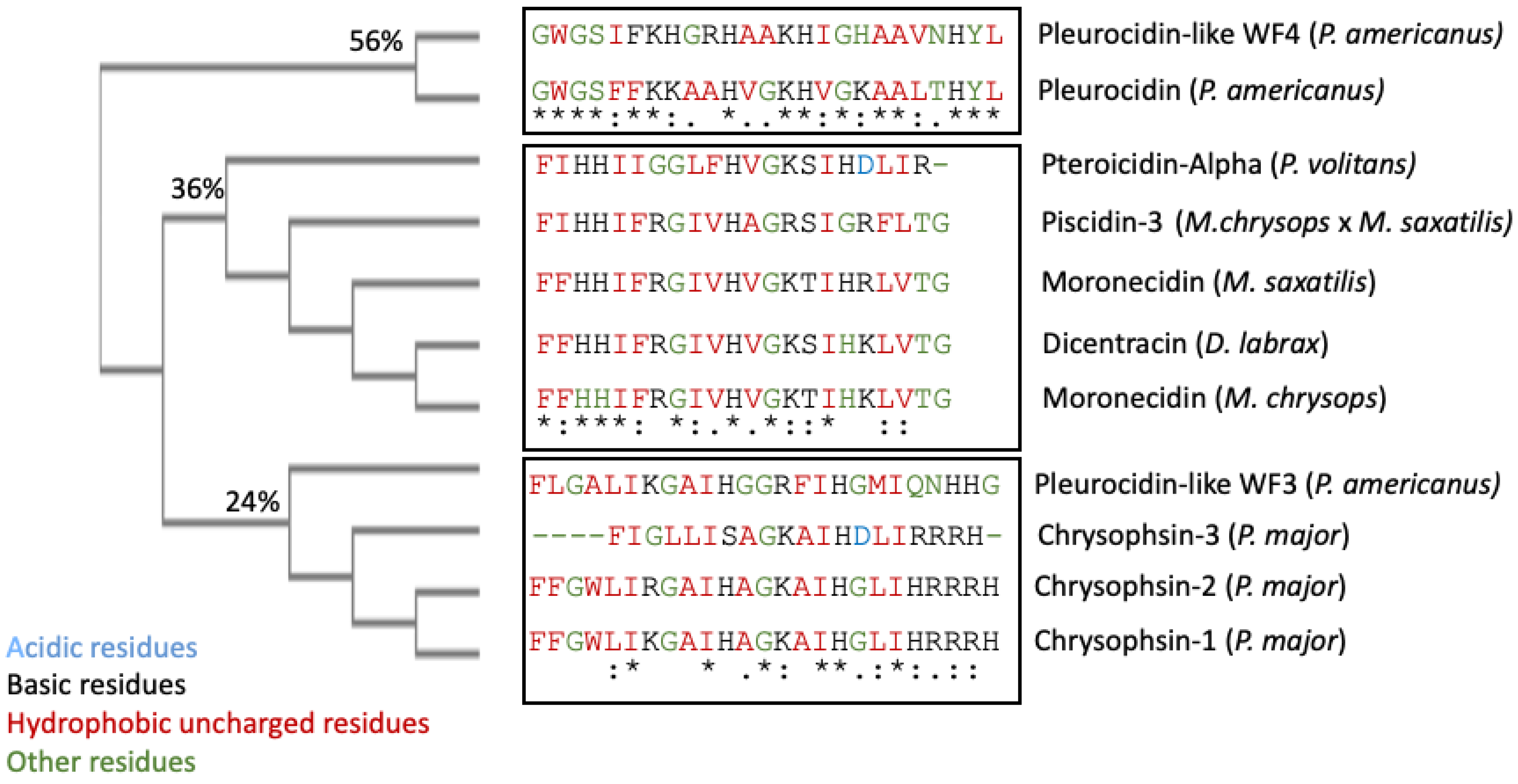
3. Evolutionary Diversity of Piscidins
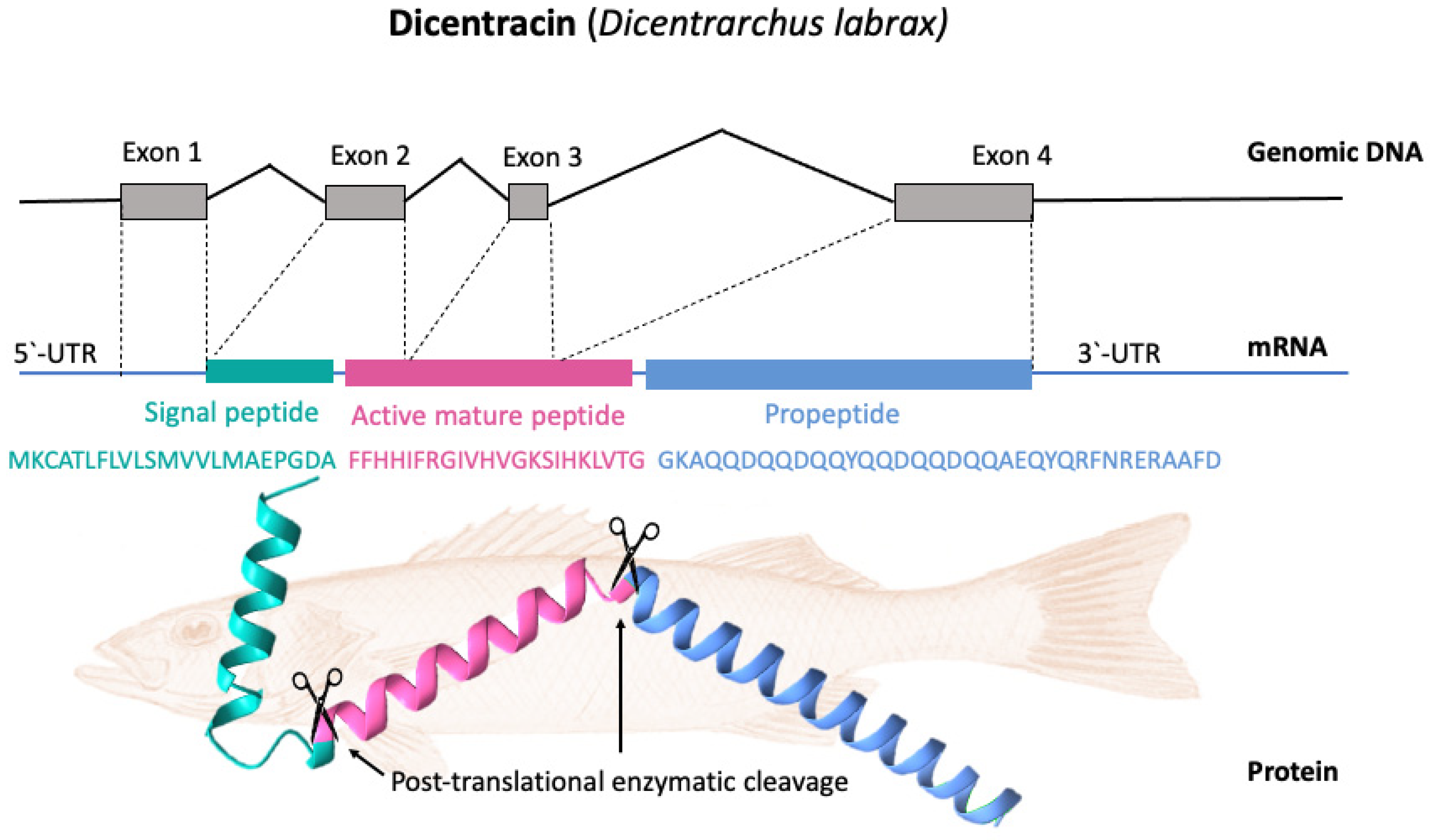
4. Piscidin Gene Arrangement, Processing, and Expression
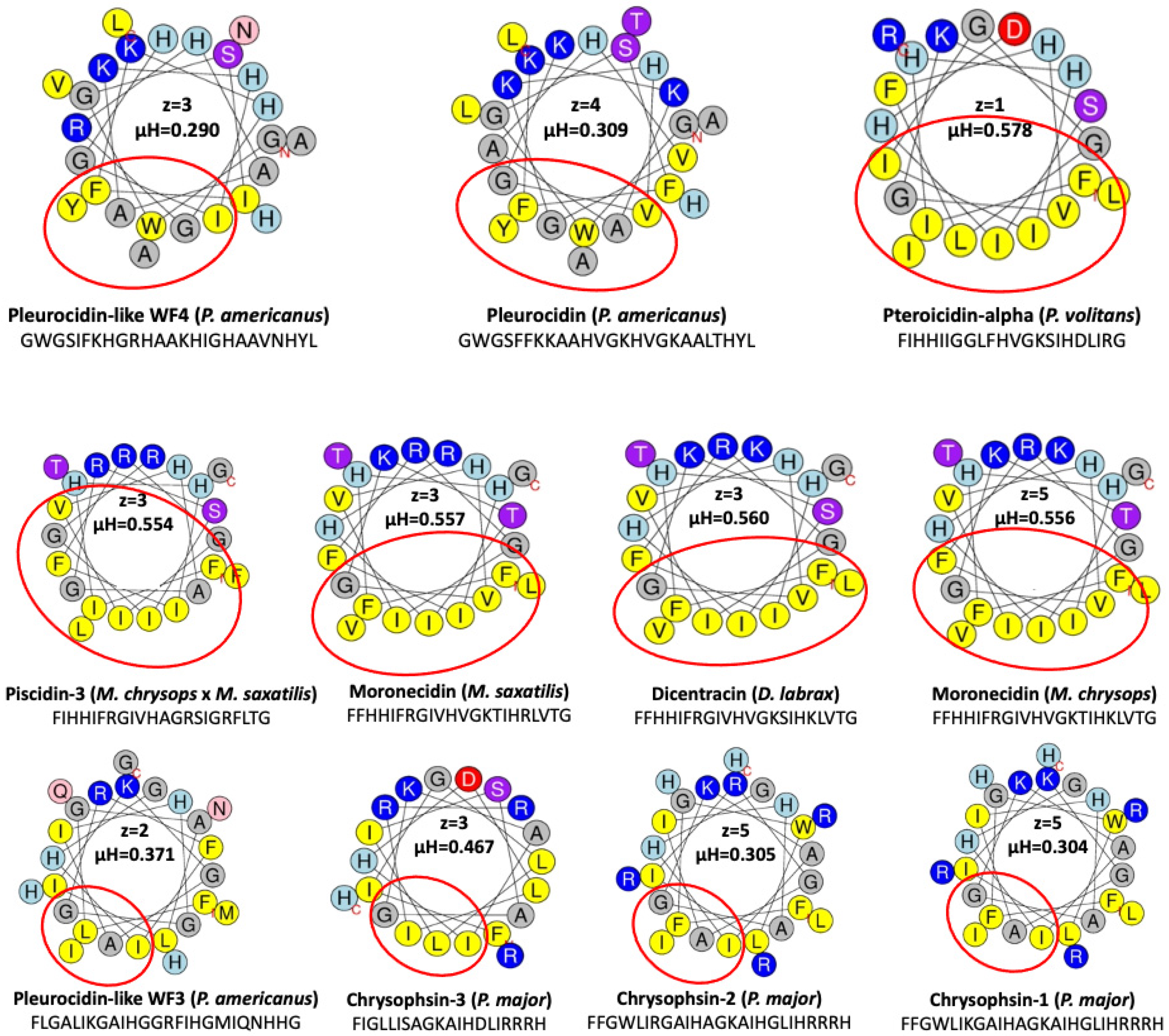
5. Structure
6. Mechanisms of Action
7. Function
8. Conclusions and Future Trends
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, Y.E.; Fischbach, M.A.; Belkaid, Y. Skin microbiota–host interactions. Nature 2018, 553, 427–436. [Google Scholar] [CrossRef]
- Patrzykat, A.; Hancock, R.E. Host defense peptides in fish: From the peculiar to the mainstream. In Fish Defenses; CRC Press: Boca Raton, FL, USA, 2019; pp. 43–61. [Google Scholar]
- Masso-Silva, J.A.; Diamond, G. Antimicrobial Peptides from Fish. Pharmaceuticals 2014, 7, 265–310. [Google Scholar] [CrossRef]
- Ravi, V.; Venkatesh, B. The divergent genomes of teleosts. Annu. Rev. Anim. Biosci. 2018, 6, 47–68. [Google Scholar] [CrossRef]
- Wootton, R.J. Ecology of Teleost Fishes; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012; Volume 1. [Google Scholar]
- Arratia, G. Complexities of early Teleostei and the evolution of particular morphological structures through time. Copeia 2015, 103, 999–1025. [Google Scholar] [CrossRef]
- Witten, P.E.; Hall, B.K. Teleost skeletal plasticity: Modulation, adaptation, and remodelling. Copeia 2015, 103, 727–739. [Google Scholar] [CrossRef]
- Gomez, D.; Sunyer, J.O.; Salinas, I. The mucosal immune system of fish: The evolution of tolerating commensals while fighting pathogens. Fish Shellfish Immunol. 2013, 35, 1729–1739. [Google Scholar] [CrossRef]
- Kelly, C.; Salinas, I. Under pressure: Interactions between commensal microbiota and the teleost immune system. Front. Immunol. 2017, 8, 559. [Google Scholar] [CrossRef]
- Xu, Z.; Parra, D.; Gómez, D.; Salinas, I.; Zhang, Y.-A.; von Gersdorff Jørgensen, L.; Heinecke, R.D.; Buchmann, K.; LaPatra, S.; Sunyer, J.O. Teleost skin, an ancient mucosal surface that elicits gut-like immune responses. Proc. Natl. Acad. Sci. USA 2013, 110, 13097–13102. [Google Scholar] [CrossRef]
- Ángeles Esteban, M. An overview of the immunological defenses in fish skin. Int. Sch. Res. Not. 2012, 2012, 853470. [Google Scholar] [CrossRef]
- Kogame, T.; Kabashima, K.; Egawa, G. Putative immunological functions of inducible skin-associated lymphoid tissue in the context of mucosa-associated lymphoid tissue. Front. Immunol. 2021, 12, 733484. [Google Scholar] [CrossRef]
- Dickerson, H.W. The biology of teleost mucosal immunity. Fish Def. Pathog. Parasites Predat. 2009, 2, 1–42. [Google Scholar]
- Fontenot, D.K.; Neiffer, D.L. Wound management in teleost fish: Biology of the healing process, evaluation, and treatment. Vet. Clin. Exot. Anim. Pract. 2004, 7, 57–86. [Google Scholar] [CrossRef]
- Watts, M.; Munday, B.; Burke, C. Immune responses of teleost fish. Aust. Vet. J. 2001, 79, 570–574. [Google Scholar] [CrossRef]
- Ellis, A. Innate host defense mechanisms of fish against viruses and bacteria. Dev. Comp. Immunol. 2001, 25, 827–839. [Google Scholar] [CrossRef]
- Magnadóttir, B. Innate immunity of fish (overview). Fish Shellfish Immunol. 2006, 20, 137–151. [Google Scholar] [CrossRef]
- Plouffe, D.A.; Hanington, P.C.; Walsh, J.G.; Wilson, E.C.; Belosevic, M. Comparison of select innate immune mechanisms of fish and mammals. Xenotransplantation 2005, 12, 266–277. [Google Scholar] [CrossRef]
- Salger, S.A.; Cassady, K.R.; Reading, B.J.; Noga, E.J. A diverse family of host-defense peptides (piscidins) exhibit specialized anti-bacterial and anti-protozoal activities in fishes. PLoS ONE 2016, 11, e0159423. [Google Scholar] [CrossRef]
- Priyam, M.; Bhat, R.A.; Kumar, N. Recent Advances in Antimicrobial Peptides to Improve Fish Health. In Biotechnological Advances in Aquaculture Health Management; Gupta, S.K., Giri, S.S., Eds.; Springer: Singapore, 2021; pp. 165–187. [Google Scholar]
- Silphaduang, U.; Colorni, A.; Noga, E. Evidence for widespread distribution of piscidin antimicrobial peptides in teleost fish. Dis. Aquat. Org. 2006, 72, 241–252. [Google Scholar] [CrossRef]
- Noga, E.; Fan, Z.; Silphaduang, U. Histone-like proteins from fish are lethal to the parasitic dinoflagellate Amyloodinium ocellatum. Parasitology 2001, 123, 57–65. [Google Scholar] [CrossRef]
- Raju, S.V.; Sarkar, P.; Kumar, P.; Arockiaraj, J. Piscidin, fish antimicrobial peptide: Structure, classification, properties, mechanism, gene regulation and therapeutical importance. Int. J. Pept. Res. Ther. 2021, 27, 91–107. [Google Scholar] [CrossRef]
- Hazam, P.K.; Chen, J.-Y. Therapeutic utility of the antimicrobial peptide Tilapia Piscidin 4 (TP4). Aquac. Rep. 2020, 17, 100409. [Google Scholar] [CrossRef]
- Valero, Y.; Saraiva-Fraga, M.; Costas, B.; Guardiola, F.A. Antimicrobial peptides from fish: Beyond the fight against pathogens. Rev. Aquac. 2020, 12, 224–253. [Google Scholar] [CrossRef]
- Mulero, I.; Noga, E.J.; Meseguer, J.; García-Ayala, A.; Mulero, V. The antimicrobial peptides piscidins are stored in the granules of professional phagocytic granulocytes of fish and are delivered to the bacteria-containing phagosome upon phagocytosis. Dev. Comp. Immunol. 2008, 32, 1531–1538. [Google Scholar] [CrossRef]
- Katzenback, B.A. Antimicrobial peptides as mediators of innate immunity in teleosts. Biology 2015, 4, 607–639. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M. Host-defense peptides of the skin with therapeutic potential: From hagfish to human. Peptides 2015, 67, 29–38. [Google Scholar] [CrossRef]
- Shabir, U.; Ali, S.; Magray, A.R.; Ganai, B.A.; Firdous, P.; Hassan, T.; Nazir, R. Fish antimicrobial peptides (AMP’s) as essential and promising molecular therapeutic agents: A review. Microb. Pathog. 2018, 114, 50–56. [Google Scholar] [CrossRef]
- Mokhtar, D.M.; Zaccone, G.; Alesci, A.; Kuciel, M.; Hussein, M.T.; Sayed, R.K. Main Components of Fish Immunity: An Overview of the Fish Immune System. Fishes 2023, 8, 93. [Google Scholar] [CrossRef]
- Valero, Y.; Chaves-Pozo, E.; Meseguer, J.; Esteban, M.A.; Cuesta, A. Biological role of fish antimicrobial peptides. Antimicrob. Pept. 2013, 2, 31–60. [Google Scholar]
- Paria, A.; Vinay, T.; Gupta, S.K.; Choudhury, T.G.; Sarkar, B. Antimicrobial peptides: A promising future alternative to antibiotics in aquaculture. World Aquac. 2018, 49, 67–69. [Google Scholar]
- Hancock, R.E.; Alford, M.A.; Haney, E.F. Antibiofilm activity of host defence peptides: Complexity provides opportunities. Nat. Rev. Microbiol. 2021, 19, 786–797. [Google Scholar] [CrossRef] [PubMed]
- Pérez de la Lastra, J.M.; Anand, U.; González-Acosta, S.; López, M.R.; Dey, A.; Bontempi, E.; Morales delaNuez, A. Antimicrobial resistance in the COVID-19 landscape: Is there an opportunity for anti-infective antibodies and antimicrobial peptides? Front. Immunol. 2022, 13, 921483. [Google Scholar] [CrossRef]
- Bharadwaj, A.; Rastogi, A.; Pandey, S.; Gupta, S.; Sohal, J.S. Multidrug-Resistant Bacteria: Their mechanism of action and prophylaxis. BioMed Res. Int. 2022, 2022, 5419874. [Google Scholar] [CrossRef]
- Sinha, R.; Shukla, P. Antimicrobial peptides: Recent insights on biotechnological interventions and future perspectives. Protein Pept. Lett. 2019, 26, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, P.; Bhat, R.A.H.; Pande, A. Antimicrobial peptides of fish: Innocuous alternatives to antibiotics. Rev. Aquac. 2020, 12, 85–106. [Google Scholar] [CrossRef]
- Bischetti, M.; Alaimo, N.; Nardelli, F.; Punzi, P.; Amariei, C.; Ingenito, R.; Musco, G.; Gallo, M.; Cicero, D.O. Structural insights on the selective interaction of the histidine-rich piscidin antimicrobial peptide Of-Pis1 with membranes. Biochim. Et Biophys. Acta (BBA) Biomembr. 2023, 1865, 184080. [Google Scholar] [CrossRef]
- Yosri, N.; Khalifa, S.A.M.; Guo, Z.; Xu, B.; Zou, X.; El-Seedi, H.R. Marine organisms: Pioneer natural sources of polysaccharides/proteins for green synthesis of nanoparticles and their potential applications. Int. J. Biol. Macromol. 2021, 193, 1767–1798. [Google Scholar] [CrossRef]
- Wang, J.; Su, B.; Dunham, R.A. Genome-wide identification of catfish antimicrobial peptides: A new perspective to enhance fish disease resistance. Rev. Aquac. 2022, 14, 2002–2022. [Google Scholar] [CrossRef]
- Douglas, S.; Gallant, J.; Gong, Z.; Hew, C. Cloning and developmental expression of a family of pleurocidin-like antimicrobial peptides from winter flounder, Pleuronectes americanus (Walbaum). Dev. Comp. Immunol. 2001, 25, 137–147. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Consortium, T.U. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2020, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Barroso, C.; Carvalho, P.; Carvalho, C.; Santarém, N.; Gonçalves, J.F.M.; Rodrigues, P.N.S.; Neves, J.V. The Diverse Piscidin Repertoire of the European Sea Bass (Dicentrarchus labrax): Molecular Characterization and Antimicrobial Activities. Int. J. Mol. Sci. 2020, 21, 4613. [Google Scholar] [CrossRef]
- Buonocore, F.; Randelli, E.; Casani, D.; Picchietti, S.; Belardinelli, M.C.; de Pascale, D.; De Santi, C.; Scapigliati, G. A piscidin-like antimicrobial peptide from the icefish Chionodraco hamatus (Perciformes: Channichthyidae): Molecular characterization, localization and bactericidal activity. Fish Shellfish Immunol. 2012, 33, 1183–1191. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.K.; Yadav, I.S. Biological databases and their application. In Bioinformatics; Elsevier: Amsterdam, The Netherlands, 2022; pp. 17–31. [Google Scholar]
- Mulder, N.J.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Buillard, V.; Cerutti, L.; Copley, R. New developments in the InterPro database. Nucleic Acids Res. 2007, 35, D224–D228. [Google Scholar] [PubMed]
- Qiao, Y.; Ma, X.; Zhang, M.; Zhong, S. Cerocin, a novel piscidin-like antimicrobial peptide from black seabass, Centropristis striata. Fish Shellfish Immunol. 2021, 110, 86–90. [Google Scholar]
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Das, U.; Daugherty, L.; Duquenne, L. InterPro: The integrative protein signature database. Nucleic Acids Res. 2009, 37, D211–D215. [Google Scholar] [CrossRef]
- Fernandes, J.M.; Ruangsri, J.; Kiron, V. Atlantic cod piscidin and its diversification through positive selection. PLoS ONE 2010, 5, e9501. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Higgins, D.G. The clustal omega multiple alignment package. Mult. Seq. Alignment Methods Protoc. 2021, 2231, 3–16. [Google Scholar]
- Ruangsri, J.; Salger, S.A.; Caipang, C.M.; Kiron, V.; Fernandes, J.M. Differential expression and biological activity of two piscidin paralogues and a novel splice variant in Atlantic cod (Gadus morhua L.). Fish Shellfish Immunol. 2012, 32, 396–406. [Google Scholar] [CrossRef]
- Serna-Duque, J.A.; Cuesta, A.; Sánchez-Ferrer, Á.; Esteban, M.Á. Two duplicated piscidin genes from gilthead seabream (Sparus aurata) with different roles in vitro and in vivo. Fish Shellfish Immunol. 2022, 127, 730–739. [Google Scholar] [CrossRef]
- Zaccone, G.; Capillo, G.; Fernandes, J.M.O.; Kiron, V.; Lauriano, E.R.; Alesci, A.; Lo Cascio, P.; Guerrera, M.C.; Kuciel, M.; Zuwala, K. Expression of the antimicrobial peptide piscidin 1 and neuropeptides in fish gill and skin: A potential participation in neuro-immune interaction. Mar. Drugs 2022, 20, 145. [Google Scholar] [CrossRef] [PubMed]
- Wiederanders, B.; Kaulmann, G.; Schilling, K. Functions of propeptide parts in cysteine proteases. Curr. Protein Pept. Sci. 2003, 4, 309–326. [Google Scholar] [CrossRef] [PubMed]
- Xhindoli, D.; Pacor, S.; Benincasa, M.; Scocchi, M.; Gennaro, R.; Tossi, A. The human cathelicidin LL-37—A pore-forming antibacterial peptide and host-cell modulator. Biochim. Et Biophys. Acta (BBA)-Biomembr. 2016, 1858, 546–566. [Google Scholar] [CrossRef] [PubMed]
- Umasuthan, N.; Mothishri, M.; Thulasitha, W.S.; Nam, B.-H.; Lee, J. Molecular, genomic, and expressional delineation of a piscidin from rock bream (Oplegnathus fasciatus) with evidence for the potent antimicrobial activities of Of-Pis1 peptide. Fish Shellfish Immunol. 2016, 48, 154–168. [Google Scholar] [CrossRef]
- Go, H.-J.; Kim, C.-H.; Park, J.B.; Kim, T.Y.; Lee, T.K.; Oh, H.Y.; Park, N.G. Biochemical and molecular identification of a novel hepcidin type 2-like antimicrobial peptide in the skin mucus of the pufferfish Takifugu pardalis. Fish Shellfish Immunol. 2019, 93, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Diamond, G.; Beckloff, N.; Weinberg, A.; Kisich, K.O. The roles of antimicrobial peptides in innate host defense. Curr. Pharm. Des. 2009, 15, 2377–2392. [Google Scholar] [CrossRef]
- Dorschner, R.A.; Lopez-Garcia, B.; Peschel, A.; Kraus, D.; Morikawa, K.; Nizet, V.; Gallo, R.L. The mammalian ionic environment dictates microbial susceptibility to antimicrobial defense peptides. FASEB J. 2006, 20, 35–42. [Google Scholar] [CrossRef]
- Campagna, S.; Saint, N.; Molle, G.; Aumelas, A. Structure and mechanism of action of the antimicrobial peptide piscidin. Biochemistry 2007, 46, 1771–1778. [Google Scholar] [CrossRef]
- Kim, J.-K.; Lee, S.-A.; Shin, S.; Lee, J.-Y.; Jeong, K.-W.; Nan, Y.H.; Park, Y.S.; Shin, S.Y.; Kim, Y. Structural flexibility and the positive charges are the key factors in bacterial cell selectivity and membrane penetration of peptoid-substituted analog of Piscidin 1. Biochim. Et Biophys. Acta (BBA)-Biomembr. 2010, 1798, 1913–1925. [Google Scholar] [CrossRef]
- Huang, Y.; Huang, J.; Chen, Y. Alpha-helical cationic antimicrobial peptides: Relationships of structure and function. Protein Cell 2010, 1, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Gautier, R.; Douguet, D.; Antonny, B.; Drin, G. HELIQUEST: A web server to screen sequences with specific α-helical properties. Bioinformatics 2008, 24, 2101–2102. [Google Scholar] [CrossRef] [PubMed]
- Sohlenkamp, C.; Geiger, O. Bacterial membrane lipids: Diversity in structures and pathways. FEMS Microbiol. Rev. 2016, 40, 133–159. [Google Scholar] [CrossRef]
- Zhang, S.-K.; Song, J.-w.; Gong, F.; Li, S.-B.; Chang, H.-Y.; Xie, H.-M.; Gao, H.-W.; Tan, Y.-X.; Ji, S.-P. Design of an α-helical antimicrobial peptide with improved cell-selective and potent anti-biofilm activity. Sci. Rep. 2016, 6, 27394. [Google Scholar] [CrossRef]
- Papo, N.; Shai, Y. Can we predict biological activity of antimicrobial peptides from their interactions with model phospholipid membranes? Peptides 2003, 24, 1693–1703. [Google Scholar] [CrossRef]
- Song, C.; de Groot, B.L.; Sansom, M.S.P. Lipid Bilayer Composition Influences the Activity of the Antimicrobial Peptide Dermcidin Channel. Biophys. J. 2019, 116, 1658–1666. [Google Scholar] [CrossRef]
- Cetuk, H.; Maramba, J.; Britt, M.; Scott, A.J.; Ernst, R.K.; Mihailescu, M.; Cotten, M.L.; Sukharev, S. Differential interactions of piscidins with phospholipids and lipopolysaccharides at membrane interfaces. Langmuir 2020, 36, 5065–5077. [Google Scholar] [CrossRef]
- Talandashti, R.; Mehrnejad, F.; Rostamipour, K.; Doustdar, F.; Lavasanifar, A. Molecular insights into pore formation mechanism, membrane perturbation, and water permeation by the antimicrobial peptide Pleurocidin: A combined all-atom and coarse-grained molecular dynamics simulation study. J. Phys. Chem. B 2021, 125, 7163–7176. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, O.R. Antimicrobial Peptides in Jawed and Jawless Vertebrates; University of Waikato: Hamilton, New Zealand, 2016. [Google Scholar]
- Rai, R.K.; De Angelis, A.; Greenwood, A.; Opella, S.J.; Cotten, M.L. Metal-ion Binding to Host Defense Peptide Piscidin 3 Observed in Phospholipid Bilayers by Magic Angle Spinning Solid-state NMR. ChemPhysChem 2019, 20, 295–301. [Google Scholar] [CrossRef]
- Patrzykat, A.; Gallant, J.W.; Seo, J.-K.; Pytyck, J.; Douglas, S.E. Novel antimicrobial peptides derived from flatfish genes. Antimicrob. Agents Chemother. 2003, 47, 2464–2470. [Google Scholar] [CrossRef]
- Douglas, S.E.; Patrzykat, A.; Pytyck, J.; Gallant, J.W. Identification, structure and differential expression of novel pleurocidins clustered on the genome of the winter flounder, Pseudopleuronectes americanus (Walbaum). Eur. J. Biochem. 2003, 270, 3720–3730. [Google Scholar] [CrossRef]
- Houyvet, B.; Bouchon-Navaro, Y.; Bouchon, C.; Goux, D.; Bernay, B.; Corre, E.; Zatylny-Gaudin, C. Identification of a moronecidin-like antimicrobial peptide in the venomous fish Pterois volitans: Functional and structural study of pteroicidin-α. Fish Shellfish Immunol. 2018, 72, 318–324. [Google Scholar] [CrossRef]
- Oludiran, A.; Courson, D.S.; Stuart, M.D.; Radwan, A.R.; Poutsma, J.C.; Cotten, M.L.; Purcell, E.B. How oxygen availability affects the antimicrobial efficacy of host defense peptides: Lessons learned from studying the copper-binding peptides piscidins 1 and 3. Int. J. Mol. Sci. 2019, 20, 5289. [Google Scholar] [CrossRef] [PubMed]
- Sung, W.S.; Lee, J.; Lee, D.G. Fungicidal effect of piscidin on Candida albicans: Pore formation in lipid vesicles and activity in fungal membranes. Biol. Pharm. Bull. 2008, 31, 1906–1910. [Google Scholar] [CrossRef]
- Chen, X.; Wu, X.; Wang, S. An optimized antimicrobial peptide analog acts as an antibiotic adjuvant to reverse methicillin-resistant Staphylococcus aureus. npj Sci. Food 2022, 6, 57. [Google Scholar] [CrossRef]
- Taheri, B.; Mohammadi, M.; Nabipour, I.; Momenzadeh, N.; Roozbehani, M. Identification of novel antimicrobial peptide from Asian sea bass (Lates calcarifer) by in silico and activity characterization. PLoS ONE 2018, 13, e0206578. [Google Scholar] [CrossRef] [PubMed]
- Sung, W.S.; Lee, J.; Lee, D.G. Fungicidal effect and the mode of action of piscidin 2 derived from hybrid striped bass. Biochem. Biophys. Res. Commun. 2008, 371, 551–555. [Google Scholar] [CrossRef] [PubMed]
- Libardo, M.D.J.; Bahar, A.A.; Ma, B.; Fu, R.; McCormick, L.E.; Zhao, J.; McCallum, S.A.; Nussinov, R.; Ren, D.; Angeles-Boza, A.M. Nuclease activity gives an edge to host-defense peptide piscidin 3 over piscidin 1, rendering it more effective against persisters and biofilms. FEBS J. 2017, 284, 3662–3683. [Google Scholar] [CrossRef]
- Wang, K.; Hou, L.; Sun, Z.-A.; Wang, W. Antibacterial activity of chrysophsin-3 against oral pathogens and Streptococcus mutans biofilms. Cell. Mol. Biol. 2022, 68, 21–27. [Google Scholar] [CrossRef]
- Tripathi, A.K.; Kumari, T.; Harioudh, M.K.; Yadav, P.K.; Kathuria, M.; Shukla, P.; Mitra, K.; Ghosh, J.K. Identification of GXXXXG motif in Chrysophsin-1 and its implication in the design of analogs with cell-selective antimicrobial and anti-endotoxin activities. Sci. Rep. 2017, 7, 3384. [Google Scholar] [CrossRef]
- Wang, W.; Tao, R.; Tong, Z.; Ding, Y.; Kuang, R.; Zhai, S.; Liu, J.; Ni, L. Effect of a novel antimicrobial peptide chrysophsin-1 on oral pathogens and Streptococcus mutans biofilms. Peptides 2012, 33, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Lomize, M.A.; Pogozheva, I.D.; Joo, H.; Mosberg, H.I.; Lomize, A.L. OPM database and PPM web server: Resources for positioning of proteins in membranes. Nucleic Acids Res. 2012, 40, D370–D376. [Google Scholar] [CrossRef]
- He, Y.; Lazaridis, T. Activity determinants of helical antimicrobial peptides: A large-scale computational study. PLoS ONE 2013, 8, e66440. [Google Scholar] [CrossRef] [PubMed]
- Pirtskhalava, M.; Vishnepolsky, B.; Grigolava, M.; Managadze, G. Physicochemical Features and Peculiarities of Interaction of AMP with the Membrane. Pharmaceuticals 2021, 14, 471. [Google Scholar] [CrossRef]
- Comert, F.; Heinrich, F.; Chowdhury, A.; Schoeneck, M.; Darling, C.; Anderson, K.W.; Libardo, M.D.J.; Angeles-Boza, A.M.; Silin, V.; Cotten, M.L. Metallated Anticancer Peptides: An Expanded Mechanism that Encompasses Physical and Chemical Bilayer Disruption. FASEB J. 2022, 36. [Google Scholar] [CrossRef]
- Portelinha, J.; Heilemann, K.; Jin, J.; Angeles-Boza, A.M. Unraveling the implications of multiple histidine residues in the potent antimicrobial peptide Gaduscidin-1. J. Inorg. Biochem. 2021, 219, 111391. [Google Scholar] [CrossRef]
- Juliano IV, S.A. From Antimicrobial Activity to Zinc Binding: An In-Depth Analysis of the Tunicate Host Defense Peptide Clavanin A. Ph.D. Thesis, University of Connecticut, Mansfield, CT, USA, 2020. [Google Scholar]
- Fu, R.; Rooney, M.T.; Zhang, R.; Cotten, M.L. Coordination of redox ions within a membrane-binding peptide: A tale of aromatic rings. J. Phys. Chem. Lett. 2021, 12, 4392–4399. [Google Scholar] [CrossRef] [PubMed]
- Paredes, S.D.; Kim, S.; Rooney, M.T.; Greenwood, A.I.; Hristova, K.; Cotten, M.L. Enhancing the membrane activity of Piscidin 1 through peptide metallation and the presence of oxidized lipid species: Implications for the unification of host defense mechanisms at lipid membranes. Biochim. Et Biophys. Acta (BBA) Biomembr. 2020, 1862, 183236. [Google Scholar] [CrossRef]
- Łoboda, D.; Kozłowski, H.; Rowińska-Żyrek, M. Antimicrobial peptide–metal ion interactions—A potential way of activity enhancement. New J. Chem. 2018, 42, 7560–7568. [Google Scholar] [CrossRef]
- Alexander, J.L.; Thompson, Z.; Cowan, J. Antimicrobial metallopeptides. ACS Chem. Biol. 2018, 13, 844–853. [Google Scholar] [CrossRef]
- Portelinha, J.; Duay, S.S.; Yu, S.I.; Heilemann, K.; Libardo, M.D.J.; Juliano, S.A.; Klassen, J.L.; Angeles-Boza, A.M. Antimicrobial peptides and copper (II) ions: Novel therapeutic opportunities. Chem. Rev. 2021, 121, 2648–2712. [Google Scholar] [CrossRef]
- Aptekmann, A.A.; Buongiorno, J.; Giovannelli, D.; Glamoclija, M.; Ferreiro, D.U.; Bromberg, Y. mebipred: Identifying metal-binding potential in protein sequence. Bioinformatics 2022, 38, 3532–3540. [Google Scholar] [CrossRef]
- Terova, G.; Cattaneo, A.G.; Preziosa, E.; Bernardini, G.; Saroglia, M. Impact of acute stress on antimicrobial polypeptides mRNA copy number in several tissues of marine sea bass (Dicentrarchus labrax). BMC Immunol. 2011, 12, 69. [Google Scholar] [CrossRef]
- Erdem Büyükkiraz, M.; Kesmen, Z. Antimicrobial peptides (AMPs): A promising class of antimicrobial compounds. J. Appl. Microbiol. 2022, 132, 1573–1596. [Google Scholar] [CrossRef] [PubMed]
- Duque, H.M.; Rodrigues, G.; Santos, L.S.; Franco, O.L. The biological role of charge distribution in linear antimicrobial peptides. Expert Opin. Drug Discov. 2023, 18, 287–302. [Google Scholar] [CrossRef]
- Singh, T.; Choudhary, P.; Singh, S. Antimicrobial Peptides: Mechanism of Action. Insights Antimicrob. Pept. 2022, 23, 1417. [Google Scholar]
- Pirtskhalava, M.; Amstrong, A.A.; Grigolava, M.; Chubinidze, M.; Alimbarashvili, E.; Vishnepolsky, B.; Gabrielian, A.; Rosenthal, A.; Hurt, D.E.; Tartakovsky, M. DBAASP v3: Database of antimicrobial/cytotoxic activity and structure of peptides as a resource for development of new therapeutics. Nucleic Acids Res. 2021, 49, D288–D297. [Google Scholar] [CrossRef] [PubMed]
- Falco, A.; Ortega-Villaizan, M.; Chico, V.; Brocal, I.; Perez, L.; Coll, J.; Estepa, A. Antimicrobial peptides as model molecules for the development of novel antiviral agents in aquaculture. Mini Rev. Med. Chem. 2009, 9, 1159–1164. [Google Scholar] [CrossRef]
- Chinchar, V.; Bryan, L.; Silphadaung, U.; Noga, E.; Wade, D.; Rollins-Smith, L. Inactivation of viruses infecting ectothermic animals by amphibian and piscine antimicrobial peptides. Virology 2004, 323, 268–275. [Google Scholar] [CrossRef]
- García-Valtanen, P.; Martinez-Lopez, A.; Ortega-Villaizan, M.; Perez, L.; Coll, J.; Estepa, A. In addition to its antiviral and immunomodulatory properties, the zebrafish β-defensin 2 (zfBD2) is a potent viral DNA vaccine molecular adjuvant. Antivir. Res. 2014, 101, 136–147. [Google Scholar] [CrossRef]
- Gui, L.; Zhang, P.; Zhang, Q.; Zhang, J. Two hepcidins from spotted scat (Scatophagus argus) possess antibacterial and antiviral functions in vitro. Fish Shellfish Immunol. 2016, 50, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Kong, R.; Yang, G.; Xue, R.; Liu, M.; Wang, F.; Hu, J.; Guo, X.; Chang, S. COVID-19 Docking Server: A meta server for docking small molecules, peptides and antibodies against potential targets of COVID-19. Bioinformatics 2020, 36, 5109–5111. [Google Scholar] [CrossRef] [PubMed]
- Pérez de la Lastra, J.M.; Asensio-Calavia, P.; González-Acosta, S.; Baca-González, V.; Morales-delaNuez, A. Bioinformatic Analysis of Genome-Predicted Bat Cathelicidins. Molecules 2021, 26, 1811. [Google Scholar] [CrossRef]
- Mustafa, S.; Balkhy, H.; Gabere, M. Peptide-Protein Interaction Studies of Antimicrobial Peptides Targeting Middle East Respiratory Syndrome Coronavirus Spike Protein: An In Silico Approach. Adv Bioinform. 2019, 2019, 6815105. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Hu, B.; Yang, X.; Gong, L.; Tan, J.; Deng, L. The putative mature peptide of piscidin-1 modulates global transcriptional profile and proliferation of splenic lymphocytes in orange-spotted grouper (Epinephelus coioides). Fish Shellfish Immunol. 2019, 86, 1035–1043. [Google Scholar] [CrossRef]
- Kang, H.K.; Lee, H.H.; Seo, C.H.; Park, Y. Antimicrobial and immunomodulatory properties and applications of marine-derived proteins and peptides. Mar. Drugs 2019, 17, 350. [Google Scholar] [CrossRef]
- Pavlicevic, M.; Marmiroli, N.; Maestri, E. Immunomodulatory peptides—A promising source for novel functional food production and drug discovery. Peptides 2022, 148, 170696. [Google Scholar] [CrossRef]
- Amagai, R.; Takahashi, T.; Terui, H.; Fujimura, T.; Yamasaki, K.; Aiba, S.; Asano, Y. The Antimicrobial Peptide Cathelicidin Exerts Immunomodulatory Effects via Scavenger Receptors. Int. J. Mol. Sci. 2023, 24, 875. [Google Scholar] [CrossRef]
- Guryanova, S.V.; Ovchinnikova, T.V. Immunomodulatory and allergenic properties of antimicrobial peptides. Int. J. Mol. Sci. 2022, 23, 2499. [Google Scholar] [CrossRef]
- Afacan, N.J.; Janot, L.M.; Hancock, R.E. Host defense peptides: Immune modulation and antimicrobial activity in vivo. Antimicrob. Pept. Innate Immun. 2013, 2, 321–358. [Google Scholar]
- Bagwe, P.V.; Bagwe, P.V.; Ponugoti, S.S.; Joshi, S.V. Peptide-based vaccines and therapeutics for COVID-19. Int. J. Pept. Res. Ther. 2022, 28, 94. [Google Scholar] [CrossRef] [PubMed]
- Herath, H.; Elvitigala, D.A.S.; Godahewa, G.; Umasuthan, N.; Whang, I.; Noh, J.K.; Lee, J. Molecular characterization and comparative expression analysis of two teleostean pro-inflammatory cytokines, IL-1β and IL-8, from Sebastes schlegeli. Gene 2016, 575, 732–742. [Google Scholar] [CrossRef] [PubMed]
- de Freitas, T.V.; Karmakar, U.; Vasconcelos, A.; Santos, M.; do Vale Lira, B.O.; Costa, S.; Barbosa, E.; Cardozo-Fh, J.; Correa, R.; Ribeiro, D.S. Release of immunomodulatory peptides at bacterial membrane interfaces as a novel strategy to fight microorganisms. J. Biol. Chem. 2023, 299, 103056. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, G.; Chaudhary, K.; Agrawal, P.; Raghava, G.P.S. Computer-aided prediction of antigen presenting cell modulators for designing peptide-based vaccine adjuvants. J. Transl. Med. 2018, 16, 181. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Madhu, M.K.; Sharma, A.K.; Sharma, V.K. ProInflam: A webserver for the prediction of proinflammatory antigenicity of peptides and proteins. J. Transl. Med. 2016, 14, 178. [Google Scholar] [CrossRef]
- Khatun, M.; Hasan, M.; Kurata, H. PreAIP: Computational prediction of anti-inflammatory peptides by integrating multiple complementary features. Front. Genet. 2019, 10, 129. [Google Scholar] [CrossRef]
- Dimitrov, I.; Bangov, I.; Flower, D.R.; Doytchinova, I. AllerTOP v. 2—A server for in silico prediction of allergens. J. Mol. Model. 2014, 20, 2278. [Google Scholar] [CrossRef]
- Chen, W.-F.; Huang, S.-Y.; Liao, C.-Y.; Sung, C.-S.; Chen, J.-Y.; Wen, Z.-H. The use of the antimicrobial peptide piscidin (PCD)-1 as a novel anti-nociceptive agent. Biomaterials 2015, 53, 1–11. [Google Scholar] [CrossRef]
- Lee, E.; Shin, A.; Jeong, K.-W.; Jin, B.; Jnawali, H.N.; Shin, S.; Shin, S.Y.; Kim, Y. Role of phenylalanine and valine10 residues in the antimicrobial activity and cytotoxicity of piscidin-1. PLoS ONE 2014, 9, e114453. [Google Scholar] [CrossRef]
- Gulati, K.; Guhathakurta, S.; Joshi, J.; Rai, N.; Ray, A. Cytokines and their role in health and disease: A brief overview. MOJ Immunol. 2016, 4, 00121. [Google Scholar]
- Klimpel, A.; Lützenburg, T.; Neundorf, I. Recent advances of anti-cancer therapies including the use of cell-penetrating peptides. Curr. Opin. Pharmacol. 2019, 47, 8–13. [Google Scholar] [CrossRef]
- Renz, A.; Berdel, W.E.; Kreuter, M.; Belka, C.; Schulze-Osthoff, K.; Los, M. Rapid extracellular release of cytochrome c is specific for apoptosis and marks cell death in vivo. Blood J. Am. Soc. Hematol. 2001, 98, 1542–1548. [Google Scholar] [CrossRef] [PubMed]
- Kurrikoff, K.; Aphkhazava, D.; Langel, Ü. The future of peptides in cancer treatment. Curr. Opin. Pharmacol. 2019, 47, 27–32. [Google Scholar] [CrossRef]
- Liscano, Y.; Oñate-Garzón, J.; Delgado, J.P. Peptides with dual antimicrobial–anticancer activity: Strategies to overcome peptide limitations and rational design of anticancer peptides. Molecules 2020, 25, 4245. [Google Scholar] [CrossRef]
- Chen, J.; Cheong, H.H.; Siu, S.W.I. xDeep-AcPEP: Deep Learning Method for Anticancer Peptide Activity Prediction Based on Convolutional Neural Network and Multitask Learning. J. Chem. Inf. Model. 2021, 61, 3789–3803. [Google Scholar] [CrossRef]
- Najm, A.A.; Azfaralarriff, A.; Dyari, H.R.E.; Alwi, S.S.S.; Khalili, N.; Othman, B.A.; Law, D.; Shahid, M.; Fazry, S. A Systematic Review of Antimicrobial Peptides from Fish with Anticancer Properties. Pertanika J. Sci. Technol. 2022, 30, 1171–1196. [Google Scholar] [CrossRef]
- Parmar, H.S.S.; Tripathi, V.; Jaiswal, P.; Sahu, K.; Majumder, S.K.; Kashyap, D.; Dixit, A.K.; Jha, H.C. Repurposing of Metabolic Drugs and Mitochondrial Modulators as an Emerging Class of Cancer Therapeutics with a Special Focus on Breast Cancer. Adv. Cancer Biol. Metastasis 2022, 6, 100065. [Google Scholar] [CrossRef]
- Lin, H.-J.; Huang, T.-C.; Muthusamy, S.; Lee, J.-F.; Duann, Y.-F.; Lin, C.-H. Piscidin-1, an antimicrobial peptide from fish (hybrid striped bass morone saxatilis x M. chrysops), induces apoptotic and necrotic activity in HT1080 cells. Zool. Sci. 2012, 29, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, A.; Ghavimi, R.; Momenzadeh, N.; Hajian, S.; Mohammadi, M. Characterization of antitumor activity of a synthetic moronecidin-like peptide computationally predicted from the tiger tail seahorse hippocampus comes in tumor-bearing mice. Int. J. Pept. Res. Ther. 2021, 27, 2391–2401. [Google Scholar] [CrossRef]
- Zheng, L.; Qiu, J.; Liu, H.; Chi, C.; Lin, L. Potential anticancer activity analysis of piscidin 5-like from Larimichthys crocea. Acta Oceanol. Sin. 2022, 41, 53–60. [Google Scholar] [CrossRef]


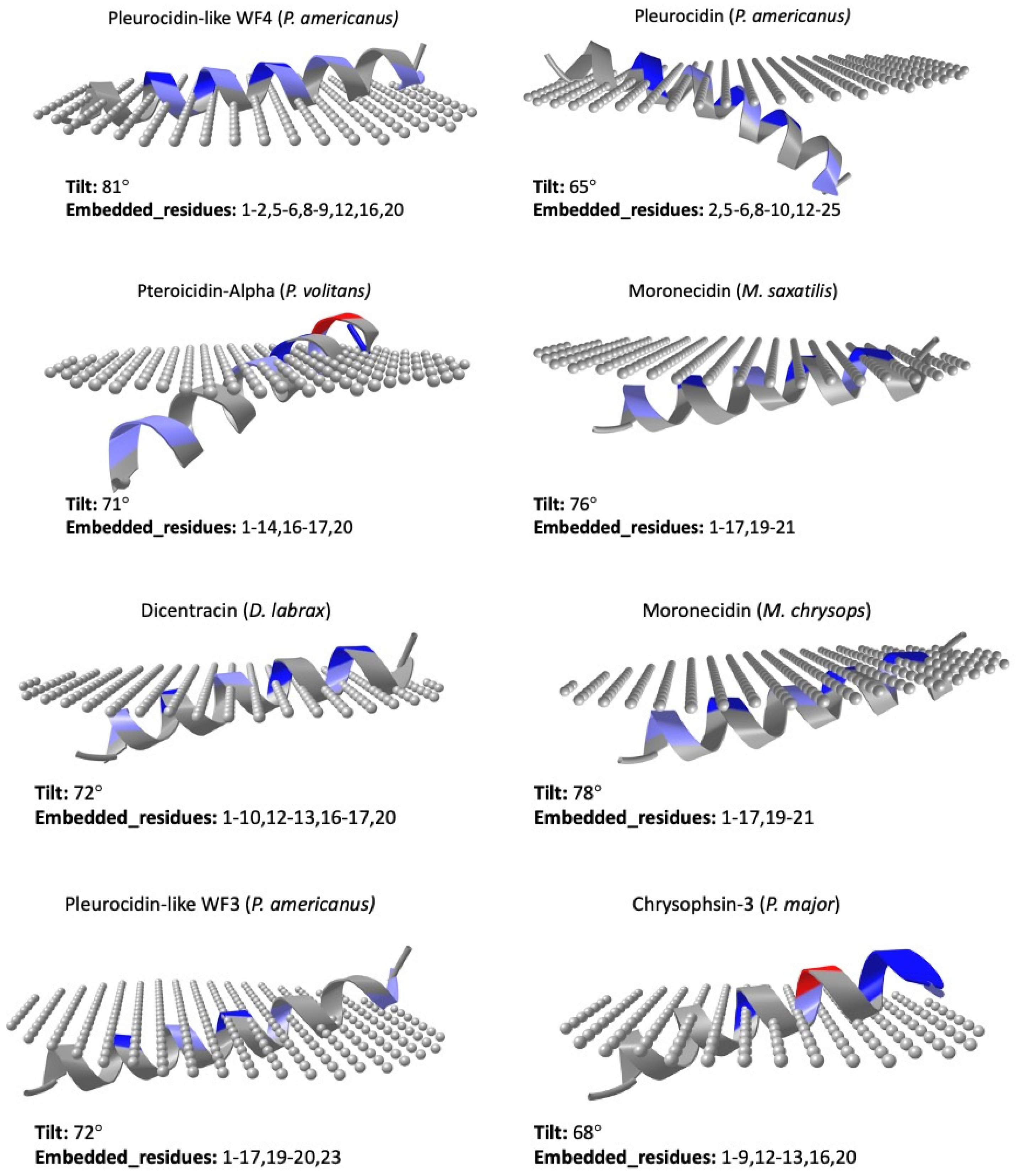
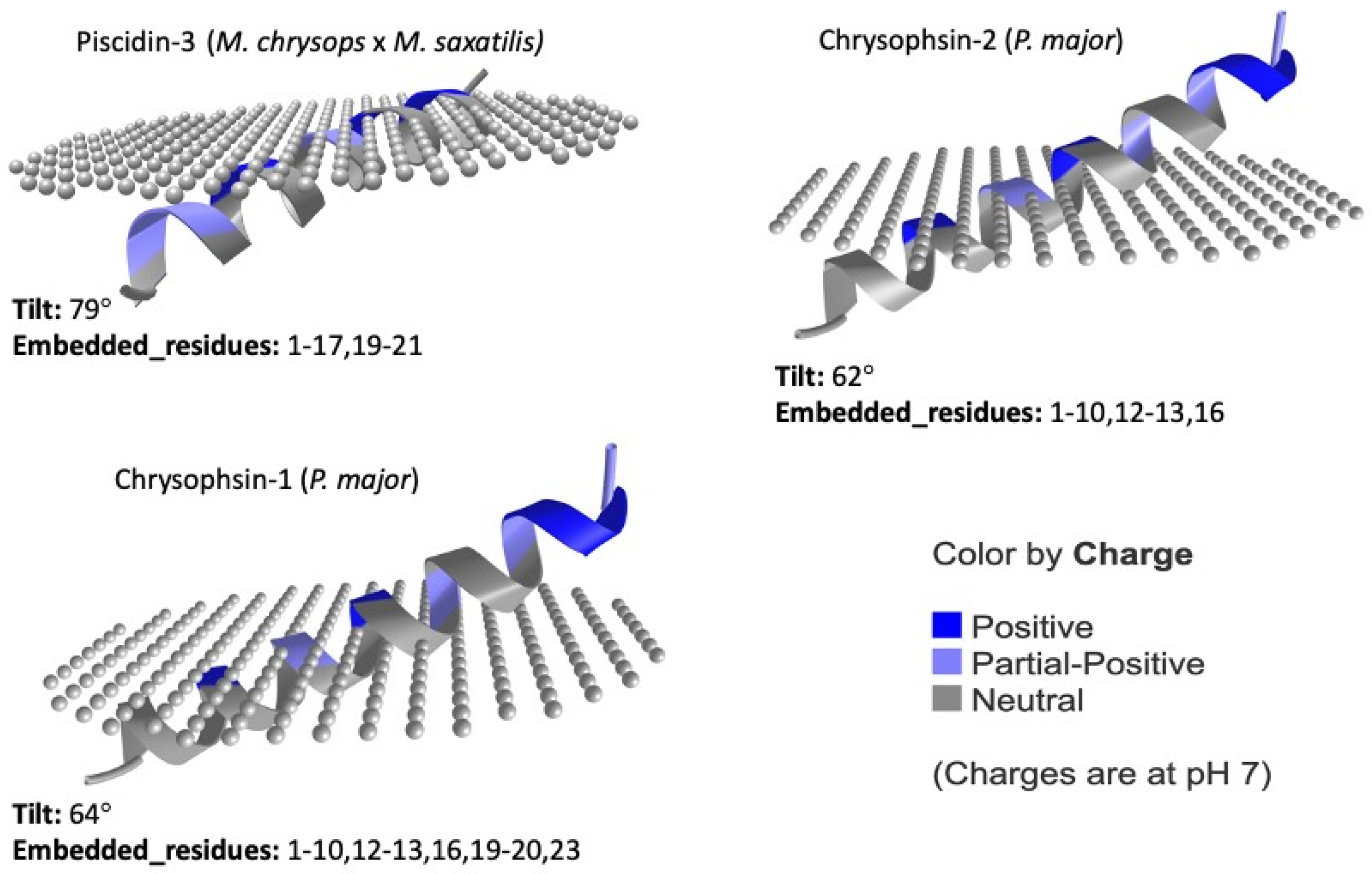
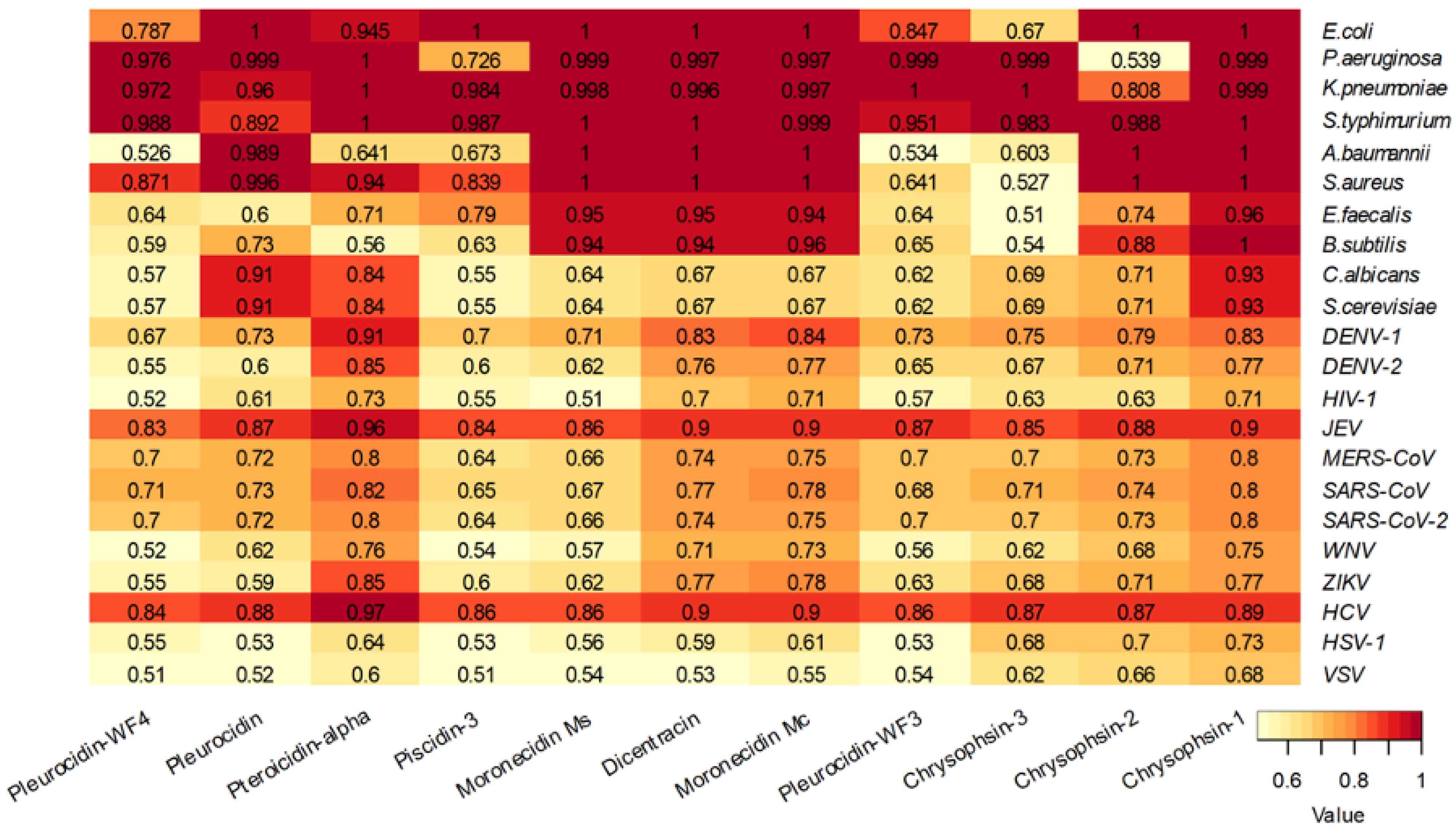
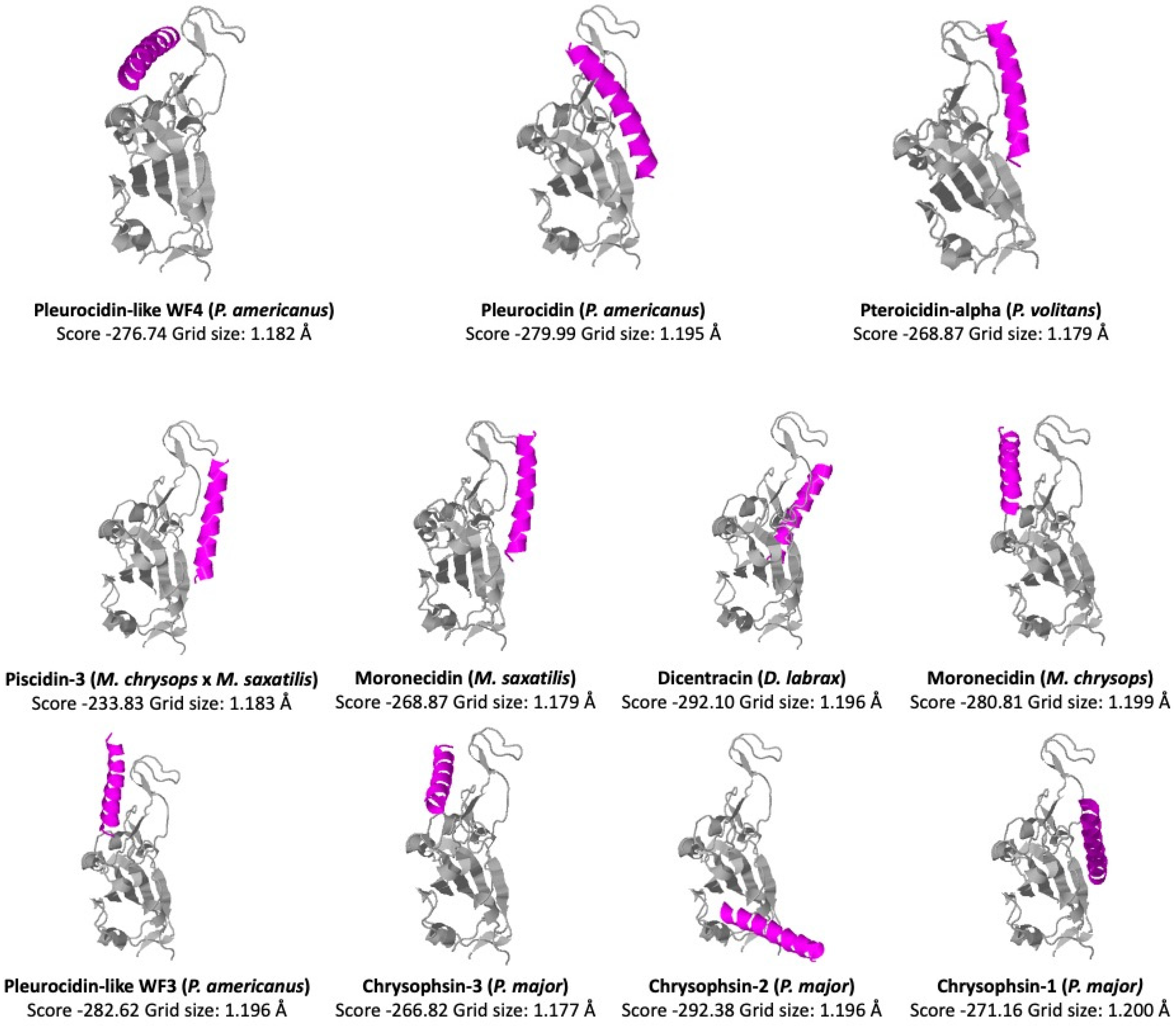
| Accession | Name | Species | Length |
|---|---|---|---|
| Q90ZX8 | Pleurocidin-WF4 | Pseudopleuronectes americanus (Winter flounder) | 25 |
| P81941 | Pleurocidin | Pseudopleuronectes americanus (Winter flounder) | 25 |
| P0DUJ5 | Pteroicidin-alpha | Pterois volitans (Red lionfish) | 21 |
| P0C006 | Piscidin-3 | Morone chrysops × Morone saxatilis (White bass × Striped bass) | 22 |
| Q8UUG0 | Moronecidin Ms | Morone saxatilis (Striped bass) | 22 |
| P59906 | Dicentracin | Dicentrarchus labrax (European seabass) | 22 |
| Q8UUG2 | Moronecidin Mc | Morone chrysops (White bass) | 22 |
| Q90VW7 | Pleurocidin-WF3 | Pseudopleuronectes americanus (Winter flounder) | 25 |
| Q90VW7 | Chrysophsin-3 | Pagrus major (Red seabream) | 20 |
| P83546 | Chrysophsin-3 | Pagrus major (Red seabream) | 25 |
| P83545 | Chrysophsin-1 | Pagrus major (Red seabream) | 25 |
| Piscidin Peptide | Hydrophobic | Hydrophilic | Positive | Negative | Neutral | pI | Mw |
|---|---|---|---|---|---|---|---|
| Pleurocidin-like WF4 | 15 | 10 | 8 | 0 | 17 | 10.29 | 2765.14 |
| Pleurocidin | 16 | 9 | 7 | 0 | 18 | 10.18 | 2711.17 |
| Pteroicidin-Alpha | 13 | 8 | 6 | 1 | 14 | 8.78 | 2409.87 |
| Piscidin-3 | 14 | 8 | 6 | 0 | 16 | 12.30 | 2491.93 |
| Moronecidin M.s | 13 | 9 | 7 | 0 | 15 | 12.01 | 2572.06 |
| Dicentracin | 13 | 9 | 7 | 0 | 15 | 11.17 | 2530.02 |
| Moronecidin M.c | 13 | 9 | 7 | 0 | 15 | 11.17 | 2544.05 |
| Pleurocidin-like WF3 | 16 | 9 | 6 | 0 | 19 | 11.00 | 2682.15 |
| Chrysophsin-3 | 12 | 8 | 6 | 1 | 13 | 11.71 | 2286.75 |
| Chrysophsin-2 | 16 | 9 | 9 | 0 | 16 | 12.48 | 2920.47 |
| Chrysophsin-1 | 16 | 9 | 9 | 0 | 16 | 12.31 | 2892.46 |
| Name | Antimicrobial Activity | References |
|---|---|---|
| Pleurocidin-WF4 | A. salmonicida, S. enterica, P. aeruginosa, E. coli, S. epidermidis | [75] |
| Pleurocidin | P. aeruginosa, E. coli, S. epidermidis, S. aureus, C. albicans | [76] |
| Pteroicidin-alpha | L. monocytogenes, E. faecalis, S. aureus, E. coli, A. salmonicida, V. vulnificus | [77] |
| Piscidin-3 | C. difficile, M. furfur, T. beigelii, C. albicans | [78,79] |
| Moronecidin Ms | C. difficile, S. aureus (MRSA), M. furfur, T. beigelii, C. albicans | [78,79,80] |
| Dicentracin | E. coli, S. aureus, S. epidermidis, C. albicans, C. tropicalis | [81] |
| Moronecidin Mc | M. furfur, T. beigelii, C. albicans | [82] |
| Pleurocidin-WF3 | E. coli, S. aureus, P. aeruginosa | [83] |
| Chrysophsin-3 | S. mutans, E. faecalis | [84] |
| Chrysophsin-2 | E. coli, S. aureus | [85] |
| Chrysophsin-1 | S. mutans, S. sanguinis, S. sobrinus, L. acidophilus, E. faecalis | [86] |
| Piscidin Peptide | Ca2+ | Co2+ | Cu2+ | Fe2+/Fe3+ | K | Mg2+ | Mn2+ | Na | Ni2+ | Zn2+ |
|---|---|---|---|---|---|---|---|---|---|---|
| Pleurocidin-like WF4 | - | - | - | - | - | - | - | - | - | + |
| Pleurocidin | - | - | - | - | - | - | - | - | - | - |
| Pteroicidin-Alpha | - | - | - | - | - | - | - | - | - | - |
| Piscidin-3 | - | - | - | - | - | - | - | - | - | - |
| Moronecidin M.s | - | - | - | - | - | - | - | - | - | - |
| Dicentracin | - | - | - | - | - | - | - | - | - | - |
| Moronecidin M.c | - | - | - | - | - | - | - | - | - | - |
| Pleurocidin-like WF3 | - | - | - | - | - | - | - | - | - | - |
| Chrysophsin-3 | - | - | - | - | - | - | - | - | - | + |
| Chrysophsin-2 | - | - | + | - | - | - | - | - | - | - |
| Chrysophsin-1 | - | - | + | - | - | - | - | - | - | - |
| Piscidin Peptide | Immunomodulatory | Pro-Inflammatory | Anti-Inflammatory | Allergenic |
|---|---|---|---|---|
| Pleurocidin-like WF4 | No | Yes | Yes | No |
| Pleurocidin | No | Yes | Yes | Yes |
| Pteroicidin-Alpha | No | Yes | Yes | No |
| Piscidin-3 | No | Yes | Yes | Yes |
| Moronecidin M.s | Yes | No | Yes | No |
| Dicentracin | Yes | Yes | Yes | No |
| Moronecidin M.c | Yes | Yes | Yes | No |
| Pleurocidin-like WF3 | No | Yes | Yes | No |
| Chrysophsin-3 | No | Yes | Yes | No |
| Chrysophsin-2 | No | No | Yes | No |
| Chrysophsin-1 | Yes | Yes | Yes | Yes |
| Piscidin Peptide | Breast | Cervix | Colon | Lung | Prostate | Skin |
|---|---|---|---|---|---|---|
| Pleurocidin-like WF4 | 34.1 µM | 10.7 µM | 28.2 µM | 71.8 µM | 97.1 µM | 7.0 µM |
| Pleurocidin | OAD | OAD | OAD | OAD | OAD | OAD |
| Pteroicidin-Alpha | 7.8 µM | 14.5 µM | 26.7 µM | 13.5 µM | 28.0 µM | 4.5 µM |
| Piscidin-3 | 33.2 µM | 25.0 µM | 62.4 µM | 58.4 µM | 226.7 µM | 6.5 µM |
| Moronecidin M.s | 15.7 µM | 12.5 µM | 23.9 µM | 25.0 µM | 82.6 µM | 6.0 µM |
| Dicentracin | 15.4 µM | 12.7 µM | 25.5 µM | 26.3 µM | 63.9 µM | 6.8 µM |
| Moronecidin M.c | 15.6 µM | 14.4 µM | 22.5 µM | 25.1 µM | 59.6 µM | 8.0 µM |
| Pleurocidin-like WF3 | 6.9 µM | 63.5 µM | 116.1 µM | 20.4 µM | 100.5 µM | 8.2 µM |
| Chrysophsin-3 | 9.6 µM | 95.0 µM | 62.4 µM | 19.8 µM | 19.3 µM | 7.3 µM |
| Chrysophsin-2 | OAD | OAD | OAD | OAD | OAD | OAD |
| Chrysophsin-1 | OAD | OAD | OAD | OAD | OAD | OAD |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asensio-Calavia, P.; González-Acosta, S.; Otazo-Pérez, A.; López, M.R.; Morales-delaNuez, A.; Pérez de la Lastra, J.M. Teleost Piscidins—In Silico Perspective of Natural Peptide Antibiotics from Marine Sources. Antibiotics 2023, 12, 855. https://doi.org/10.3390/antibiotics12050855
Asensio-Calavia P, González-Acosta S, Otazo-Pérez A, López MR, Morales-delaNuez A, Pérez de la Lastra JM. Teleost Piscidins—In Silico Perspective of Natural Peptide Antibiotics from Marine Sources. Antibiotics. 2023; 12(5):855. https://doi.org/10.3390/antibiotics12050855
Chicago/Turabian StyleAsensio-Calavia, Patricia, Sergio González-Acosta, Andrea Otazo-Pérez, Manuel R. López, Antonio Morales-delaNuez, and José Manuel Pérez de la Lastra. 2023. "Teleost Piscidins—In Silico Perspective of Natural Peptide Antibiotics from Marine Sources" Antibiotics 12, no. 5: 855. https://doi.org/10.3390/antibiotics12050855
APA StyleAsensio-Calavia, P., González-Acosta, S., Otazo-Pérez, A., López, M. R., Morales-delaNuez, A., & Pérez de la Lastra, J. M. (2023). Teleost Piscidins—In Silico Perspective of Natural Peptide Antibiotics from Marine Sources. Antibiotics, 12(5), 855. https://doi.org/10.3390/antibiotics12050855