Progressive Sub-MIC Exposure of Klebsiella pneumoniae 43816 to Cephalothin Induces the Evolution of Beta-Lactam Resistance without Acquisition of Beta-Lactamase Genes
Abstract
:1. Introduction
2. Results
2.1. Experimental Design of Progressive Sub-MIC Exposure to Cephalothin
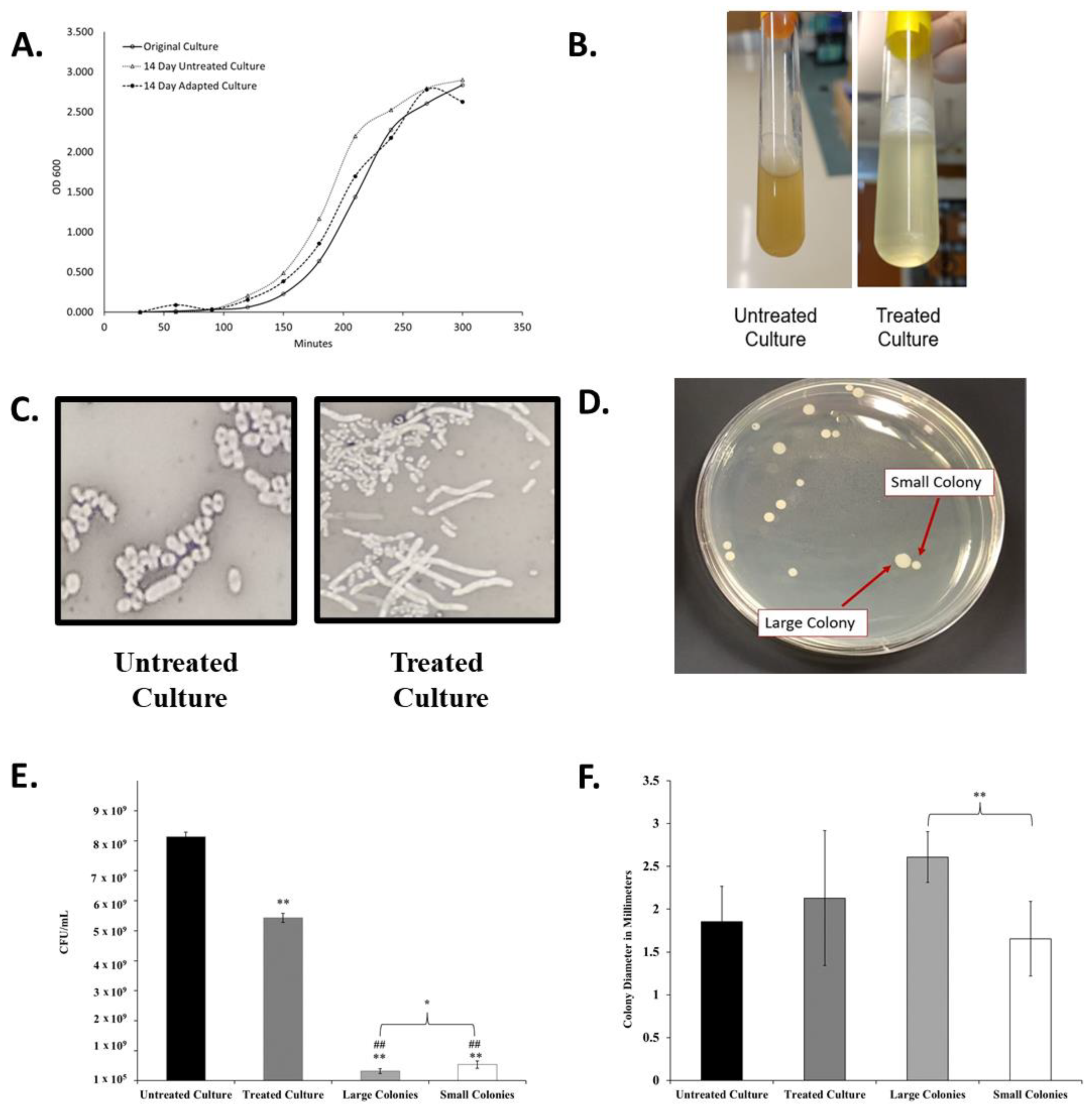
2.2. Progressive Antibiotic Exposure Alters K. pneumoniae Growth, Cellular Morphology, and Colony Morphology
2.3. Progressive Antibiotic Exposure Induces Clinical Resistance to Multiple Classes of Antibiotics
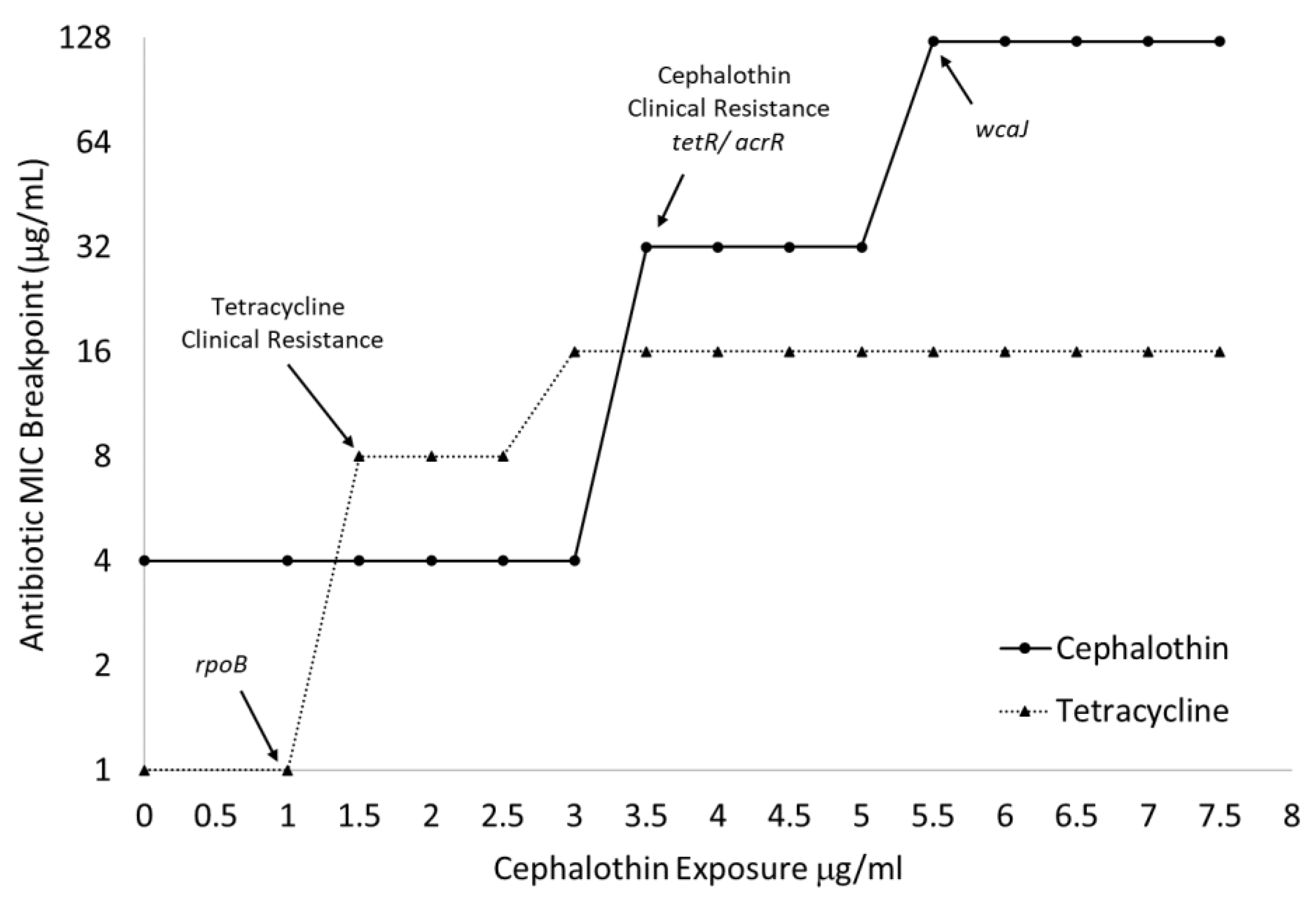
2.4. Whole Genome Sequencing Reveals Distinctive Genetic Changes Because of Progressive Antibiotic Exposure
2.5. Genome Sequencing Allows for Correlation of Genetic Changes with Increased MICs
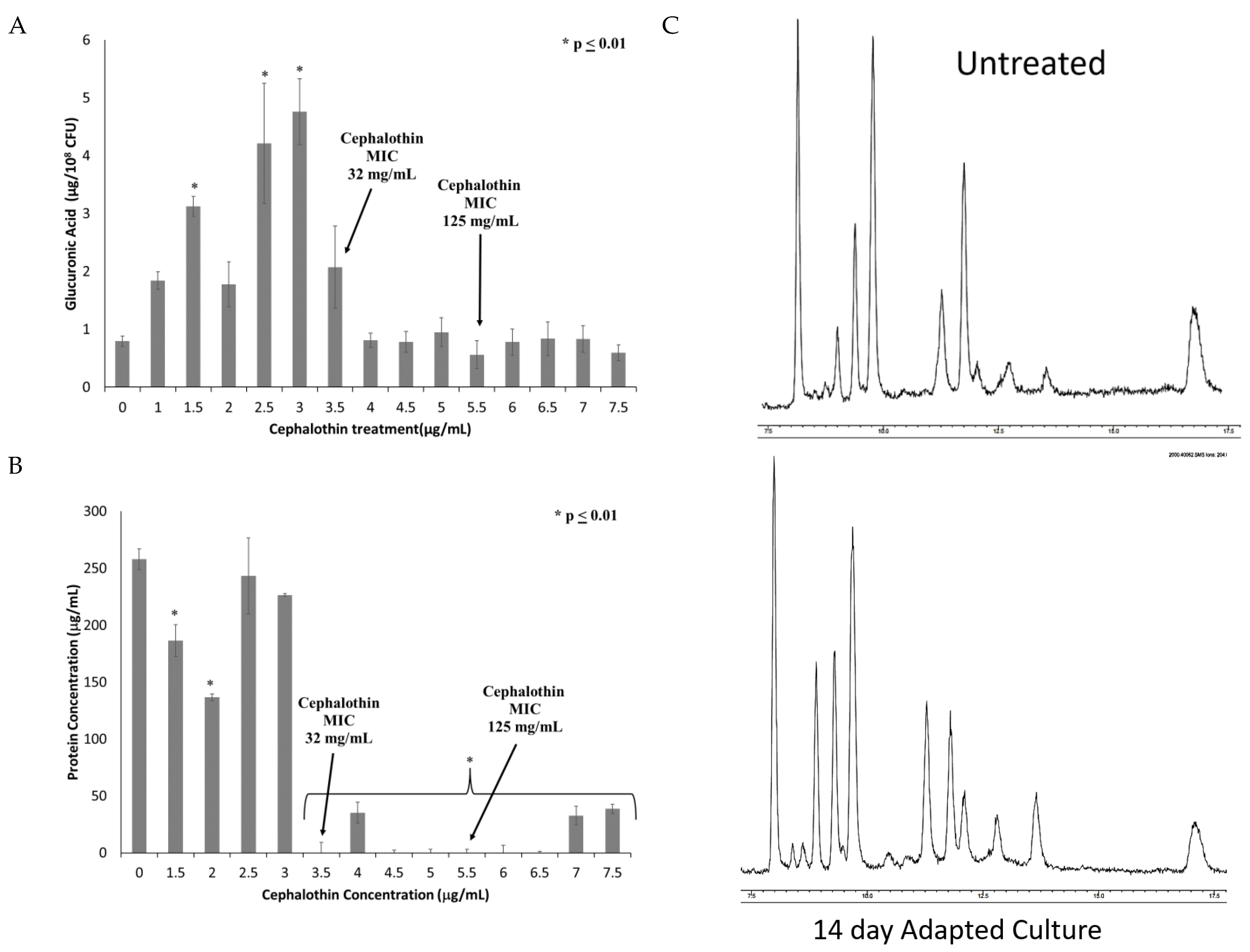
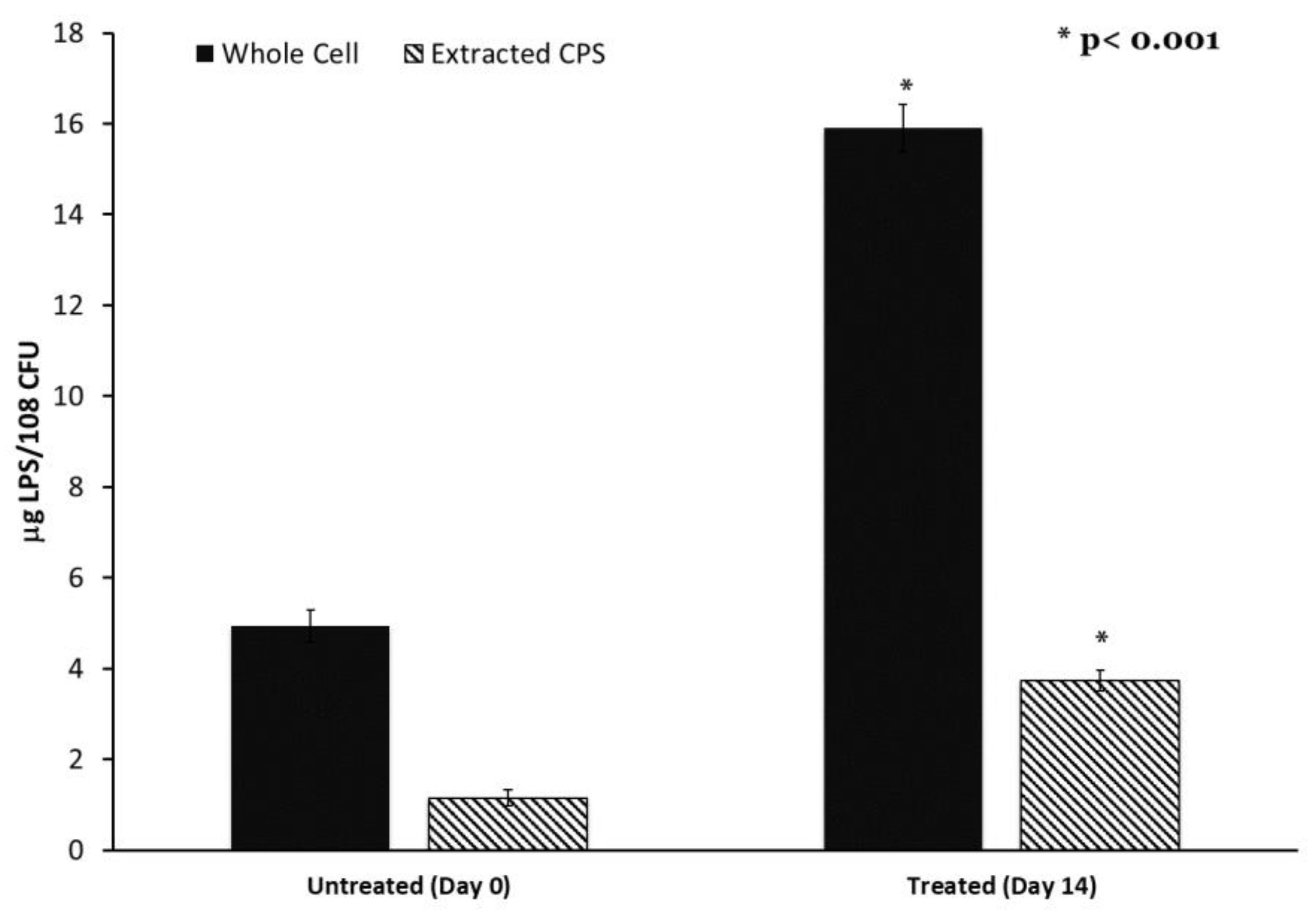
2.6. Genetic Changes in Progressive Antibiotic Adapted Cultures Are Associated with Alterations to Capsule and LPS
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Bacterial Strains and Progressive Antibiotic Exposure
5.2. Bacterial Growth and Morphology
5.3. Determination of Minimum Inhibitory Concentration Breakpoints
5.4. Genomic DNA Isolation
5.5. Genome Sequencing
5.6. Data Availability
5.7. Capsule Extraction and Characterization
5.8. Quantification of Lipopolysaccharide
5.9. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Patel, M.; Kumar, R.; Kishor, K.; Mlsna, T.; Pittman, C.U.; Mohan, D. Pharmaceuticals of Emerging Concern in Aquatic Systems: Chemistry, Occurrence, Effects, and Removal Methods. Chem. Rev. 2019, 119, 3510–3673. [Google Scholar] [CrossRef] [PubMed]
- Polianciuc, S.I.; Gurzău, A.E.; Kiss, B.; Georgia Ștefan, M.; Loghin, F. Antibiotics in the Environment: Causes and Consequences. Med. Pharm. Reports 2020, 93, 231–240. [Google Scholar] [CrossRef] [PubMed]
- National Action Plan to Combat Antibiotic-Resistant Bacteria. 2015. Available online: https://www.cdc.gov/drugresistance/us-activities/national-action-plan.html (accessed on 11 April 2023).
- Kampf, G. Acquired Resistance to Chlorhexidine—Is It Time to Establish an ‘Antiseptic Stewardship’ Initiative? J. Hosp. Infect. 2016, 94, 213–227. [Google Scholar] [CrossRef]
- Nair, C.G.; Chao, C.; Ryall, B.; Williams, H.D. Sub-Lethal Concentrations of Antibiotics Increase Mutation Frequency in the Cystic Fibrosis Pathogen Pseudomonas Aeruginosa. Lett. Appl. Microbiol. 2013, 56, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Bhunia, A.K. Animal-Use Antibiotics Induce Cross-Resistance in Bacterial Pathogens to Human Therapeutic Antibiotics. Curr. Microbiol. 2019, 76, 1112–1117. [Google Scholar] [CrossRef] [PubMed]
- Singer, A.C.; Shaw, H.; Rhodes, V.; Hart, A. Review of Antimicrobial Resistance in the Environment and Its Relevance to Environmental Regulators. Front. Microbiol. 2016, 7, 1728. [Google Scholar] [CrossRef]
- Drlica, K. The Mutant Selection Window and Antimicrobial Resistance. J. Antimicrob. Chemother. 2003, 52, 11–17. [Google Scholar] [CrossRef]
- Drlica, K.; Zhao, X. Mutant Selection Window Hypothesis Updated. Clin. Infect. Dis. 2007, 44, 681–688. [Google Scholar] [CrossRef]
- Gullberg, E.; Cao, S.; Berg, O.G.; Ilbäck, C.; Sandegren, L.; Hughes, D.; Andersson, D.I. Selection of Resistant Bacteria at Very Low Antibiotic Concentrations. PLoS Pathog. 2011, 7, e1002158. [Google Scholar] [CrossRef]
- Low, Y.M.; Chong, C.W.; Yap, I.K.S.; Chai, L.C.; Clarke, S.C.; Ponnampalavanar, S.; Abdul Jabar, K.; Md Yusof, M.Y.; Teh, C.S.J. Elucidating the Survival and Response of Carbapenem Resistant Klebsiella Pneumoniae after Exposure to Imipenem at Sub-Lethal Concentrations. Pathog. Glob. Health 2018, 112, 378–386. [Google Scholar] [CrossRef]
- Migliorini, L.B.; Brüggemann, H.; De Sales, R.O.; Koga, P.C.M.; De Souza, A.V.; Martino, M.D.V.; Galhardo, R.S.; Severino, P. Mutagenesis Induced by Sub-Lethal Doses of Ciprofloxacin: Genotypic and Phenotypic Differences between the Pseudomonas Aeruginosa Strain PA14 and Clinical Isolates. Front. Microbiol. 2019, 10, 1553. [Google Scholar] [CrossRef] [PubMed]
- Paczosa, M.K.; Mecsas, J. Klebsiella Pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Simmonds, A.; Uhlemann, A.C. Clinical Implications of Genomic Adaptation and Evolution of Carbapenem-Resistant Klebsiella Pneumoniae. J. Infect. Dis. 2017, 215, S18–S27. [Google Scholar] [CrossRef] [PubMed]
- Broberg, C.A.; Wu, W.; Cavalcoli, J.D.; Miller, V.L.; Bachman, M.A. Complete Genome Sequence of Klebsiella Pneumoniae Strain ATCC 43816 KPPR1, a Rifampin-Resistant Mutant Commonly Used in Animal, Genetic, and Molecular Biology Studies. Genome Announc. 2014, 2, e00924-14. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI: Wayne, PA, USA, 2020. [Google Scholar]
- Barrick, J.E. Breseq Manual—Breseq 0.35.2rc1 Documentation. Available online: https://barricklab.org/twiki/pub/Lab/ToolsBacterialGenomeResequencing/documentation/index.html (accessed on 11 April 2023).
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Hendriksen, R.S.; Bortolaia, V.; Tate, H.; Tyson, G.H.; Aarestrup, F.M.; McDermott, P.F. Using Genomics to Track Global Antimicrobial Resistance. Front. Public Health 2019, 7, 242. [Google Scholar] [CrossRef]
- Campbell, E.A.; Korzheva, N.; Mustaev, A.; Murakami, K.; Nair, S.; Goldfarb, A.; Darst, S.A. Structural Mechanism for Rifampicin Inhibition of Bacterial RNA Polymerase. Cell 2001, 104, 901–912. [Google Scholar] [CrossRef]
- Cai, R.; Wang, G.; Le, S.; Wu, M.; Cheng, M.; Guo, Z.; Ji, Y.; Xi, H.; Zhao, C.; Wang, X.; et al. Three Capsular Polysaccharide Synthesis-Related Glucosyltransferases, GT-1, GT-2 and WcaJ, Are Associated with Virulence and Phage Sensitivity of Klebsiella Pneumoniae. Front. Microbiol. 2019, 10, 1189. [Google Scholar] [CrossRef]
- Lee, C.H.; Chang, C.C.; Liu, J.W.; Chen, R.F.; Yang, K.D. Sialic Acid Involved in Hypermucoviscosity Phenotype of Klebsiella Pneumoniae and Associated with Resistance to Neutrophil Phagocytosis. Virulence 2014, 5, 673–679. [Google Scholar] [CrossRef]
- Walker, K.A.; Treat, L.P.; Sepúlveda, V.E.; Miller, V.L. The Small Protein RmpD Drives Hypermucoviscosity in Klebsiella Pneumoniae. MBio 2020, 11, e01750-20. [Google Scholar] [CrossRef]
- Lenski, R.E. Experimental Evolution and the Dynamics of Adaptation and Genome Evolution in Microbial Populations. ISME J. 2017, 11, 2181–2194. [Google Scholar] [CrossRef] [PubMed]
- Good, B.H.; McDonald, M.J.; Barrick, J.E.; Lenski, R.E.; Desai, M.M. The Dynamics of Molecular Evolution over 60,000 Generations. Nature 2017, 551, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Kleinman, A. Fixation and Adaptation in the Lenski E. Coli Long Term Evolution Experiment. Biomed. J. Sci. Tech. Res. 2019, 20, 14754–14760. [Google Scholar] [CrossRef]
- Regué, M.; Hita, B.; Piqué, N.; Izquierdo, L.; Merino, S.; Fresno, S.; Benedí, V.J.; Tomás, J.M. A Gene, Uge, Is Essential for Klebsiella Pneumoniae Virulence. Infect. Immun. 2004, 72, 54–61. [Google Scholar] [CrossRef]
- Martinez, J.L.; Baquero, F. Mutation Frequencies and Antibiotic Resistance. Antimicrob. Agents Chemother. 2000, 44, 1771–1777. [Google Scholar] [CrossRef] [PubMed]
- Long, H.; Miller, S.F.; Strauss, C.; Zhao, C.; Cheng, L.; Ye, Z.; Griffin, K.; Te, R.; Lee, H.; Chen, C.C.; et al. Antibiotic Treatment Enhances the Genome-Wide Mutation Rate of Target Cells. Proc. Natl. Acad. Sci. USA 2016, 113, E2498–E2505. [Google Scholar] [CrossRef]
- Sta Ana, K.M.; Madriaga, J.; Espino, M.P. β-Lactam Antibiotics and Antibiotic Resistance in Asian Lakes and Rivers: An Overview of Contamination, Sources and Detection Methods. Environ. Pollut. 2021, 275, 116624. [Google Scholar] [CrossRef]
- Brumfitt, W.; Kosmidis, J.; Hamilton-Miller, J.M.; Gilchrist, J.N. Cefoxitin and Cephalothin: Antimicrobial Activity, Human Pharmacokinetics, and Toxicology. Antimicrob. Agents Chemother. 1974, 6, 290–299. [Google Scholar] [CrossRef]
- Udikovic-Kolic, N.; Wichmann, F.; Broderick, N.A.; Handelsman, J. Bloom of Resident Antibiotic-Resistant Bacteria in Soil Following Manure Fertilization. Proc. Natl. Acad. Sci. USA 2014, 111, 15202–15207. [Google Scholar] [CrossRef]
- De Maio, N.; Shaw, L.P.; Hubbard, A.; George, S.; Sanderson, N.D.; Swann, J.; Wick, R.; Oun, M.A.; Stubberfield, E.; Hoosdally, S.J.; et al. Comparison of Long-Read Sequencing Technologies in the Hybrid Assembly of Complex Bacterial Genomes. Microb. Genom. 2019, 5, e000294. [Google Scholar] [CrossRef]
- Slupska, M.M.; Chiang, J.H.; Luther, W.M.; Stewart, J.L.; Amii, L.; Conrad, A.; Miller, J.H. Genes Involved in the Determination of the Rate of Inversions at Short Inverted Repeats. Genes Cells 2000, 5, 425–437. [Google Scholar] [CrossRef] [PubMed]
- Moskowitz, S.M.; Ernst, R.K.; Miller, S.I. PmrAB, a Two-Component Regulatory System of Pseudomonas Aeruginosa That Modulates Resistance to Cationic Antimicrobial Peptides and Addition of Aminoarabinose to Lipid A. J. Bacteriol. 2004, 186, 575–579. [Google Scholar] [CrossRef] [PubMed]
- Van Dyk, T.K.; DeRose, E.J.; Gonye, G.E. LuxArray, a High-Density, Genomewide Transcription Analysis of Escherichia Coli Using Bioluminescent Reporter Strains. J. Bacteriol. 2001, 183, 5496–5505. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.J.; Gross, C.A. Mapping and Sequencing of Mutations in the Escherichia ColirpoB Gene That Lead to Rifampicin Resistance. J. Mol. Biol. 1988, 202, 45–58. [Google Scholar] [CrossRef]
- Severinov, K.; Soushko, M.; Goldfarb, A.; Nikiforov, V. RifR Mutations in the Beginning of the Escherichia Coli RpoB Gene. MGG Mol. Gen. Genet. 1994, 244, 120–126. [Google Scholar] [CrossRef]
- Alifano, P.; Palumbo, C.; Pasanisi, D.; Talà, A. Rifampicin-Resistance, RpoB Polymorphism and RNA Polymerase Genetic Engineering. J. Biotechnol. 2015, 202, 60–77. [Google Scholar] [CrossRef]
- Colclough, A.L.; Scadden, J.; Blair, J.M.A. TetR-Family Transcription Factors in Gram-Negative Bacteria: Conservation, Variation and Implications for Efflux-Mediated Antimicrobial Resistance. BMC Genom. 2019, 20, 731. [Google Scholar] [CrossRef]
- Du, S.; Lutkenhaus, J. SlmA Antagonism of FtsZ Assembly Employs a Two-Pronged Mechanism like MinCD. PLoS Genet. 2014, 10, e1004460. [Google Scholar] [CrossRef]
- Yoshino, M.; Aihara, M.; Gotoh, Y.; Akimoto, M.; Tatsuhara, W.; Kiyosuke, M.; Matsushima, Y.; Uchiumi, T.; Hayashi, T.; Kang, D. Stepwise Evolution of a Klebsiella Pneumoniae Clone within a Host Leading to Increased Multidrug Resistance. mSphere 2021, 6, e00734-21. [Google Scholar] [CrossRef]
- Lopatkin, A.J.; Bening, S.C.; Manson, A.L.; Stokes, J.M.; Kohanski, M.A.; Badran, A.H.; Earl, A.M.; Cheney, N.J.; Yang, J.H.; Collins, J.J. Clinically Relevant Mutations in Core Metabolic Genes Confer Antibiotic Resistance. Science 2021, 371, eaba0862. [Google Scholar] [CrossRef]
- Pal, S.; Verma, J.; Mallick, S.; Rastogi, S.K.; Kumar, A.; Ghosh, A.S. Absence of the Glycosyltransferase Wcaj in Klebsiella Pneumoniae Atcc13883 Affects Biofilm Formation, Increases Polymyxin Resistance and Reduces Murine Macrophage Activation. Microbiology 2019, 165, 891–904. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Kahne, D.; Kishony, R. Distinct Single-Cell Morphological Dynamics under Beta-Lactam Antibiotics. Mol. Cell 2012, 48, 705–712. [Google Scholar] [CrossRef] [PubMed]
- Inui, T.; Endo, T.; Matsushita, T. Morphological Changes and Lysis Induced by Beta-Lactams Associated with the Characteristic Profiles of Affinities of Penicillin-Binding Proteins in Actinobacillus Pleuropneumoniae. Antimicrob. Agents Chemother. 2000, 44, 1518–1523. [Google Scholar] [CrossRef] [PubMed]
- Kessler, N.G.; Caraballo Delgado, D.M.; Shah, N.K.; Dickinson, J.A.; Moorea, S.D. Exopolysaccharide Anchoring Creates an Extreme Resistance to Sedimentation. J. Bacteriol. 2021, 203, e00023-21. [Google Scholar] [CrossRef] [PubMed]
- Ojkic, N.; Serbanescu, D.; Banerjee, S. Antibiotic Resistance via Bacterial Cell Shape-Shifting. MBio 2022, 13, e00659-22. [Google Scholar] [CrossRef]
- Meredith, T.C.; Mamat, U.; Kaczynski, Z.; Lindner, B.; Holst, O.; Woodard, R.W. Modification of Lipopolysaccharide with Colanic Acid (M-Antigen) Repeats in Escherichia Coli. J. Biol. Chem. 2007, 282, 7790–7798. [Google Scholar] [CrossRef]
- Ren, G.; Wang, Z.; Li, Y.; Hu, X.; Wang, X. Effects of Lipopolysaccharide Core Sugar Deficiency on Colanic Acid Biosynthesis in Escherichia Coli. J. Bacteriol. 2016, 198, 1576–1584. [Google Scholar] [CrossRef]
- Borić, M.; Danevčič, T.; Stopar, D. Viscosity Dictates Metabolic Activity of Vibrio Ruber. Front. Microbiol. 2012, 3, 255. [Google Scholar] [CrossRef]
- Cephalosporins—Infectious Diseases—Merck Manuals Professional Edition. Available online: https://www.merckmanuals.com/professional/infectious-diseases/bacteria-and-antibacterial-drugs/cephalosporins (accessed on 26 November 2022).
- Palace, S.G.; Wang, Y.; Rubin, D.H.F.; Welsh, M.A.; Mortimer, T.D.; Cole, K.; Eyre, D.W.; Walker, S.; Grad, Y.H. Rna Polymerase Mutations Cause Cephalosporin Resistance in Clinical Neisseria Gonorrhoeae Isolates. Elife 2020, 9, e51407. [Google Scholar] [CrossRef]
- Nelson, K.; Selander, R.K. Intergeneric Transfer and Recombination of the 6-Phosphogluconate Dehydrogenase Gene (Gnd) in Enteric Bacteria. Proc. Natl. Acad. Sci. USA 1994, 91, 10227–10231. [Google Scholar] [CrossRef]
- Arakawa, Y.; Wacharotayankun, R.; Nagatsuka, T.; Ito, H.; Kato, N.; Ohta, M. Genomic Organization of the Klebsiella Pneumoniae Cps Region Responsible for Serotype K2 Capsular Polysaccharide Synthesis in the Virulent Strain Chedid. J. Bacteriol. 1995, 177, 1788–1796. [Google Scholar] [CrossRef] [PubMed]
- Jousset, A.B.; Bonnin, R.A.; Rosinski-Chupin, I.; Girlich, D.; Cuzon, G.; Cabanel, N.; Frech, H.; Farfour, E.; Dortet, L.; Glaser, P.; et al. A 4.5-Year Within-Patient Evolution of a Colistin-Resistant Klebsiella Pneumoniae Carbapenemase-Producing K. Pneumoniae Sequence Type 258. Clin. Infect. Dis. 2018, 67, 1388–1394. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Wang, Q.; Perlaza-Jiménez, L.; Zheng, X.; Zhao, Y.; Dhanasekaran, V.; Fang, R.; Li, J.; Wang, C.; Liu, H.; et al. First Description of Antimicrobial Resistance in Carbapenem-Susceptible Klebsiella Pneumoniae after Imipenem Treatment, Driven by Outer Membrane Remodeling. BMC Microbiol. 2020, 20, 218. [Google Scholar] [CrossRef] [PubMed]
- Aihara, M.; Nishida, R.; Akimoto, M.; Gotoh, Y.; Kiyosuke, M.; Uchiumi, T.; Nishioka, M.; Matsushima, Y.; Hayashi, T.; Kang, D. Within-Host Evolution of a Klebsiella Pneumoniae Clone: Selected Mutations Associated with the Alteration of Outer Membrane Protein Expression Conferred Multidrug Resistance. J. Antimicrob. Chemother. 2021, 76, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Chebotar, I.V.; Emelyanova, M.A.; Bocharova, J.A.; Mayansky, N.A.; Kopantseva, E.E.; Mikhailovich, V.M. The Classification of Bacterial Survival Strategies in the Presence of Antimicrobials. Microb. Pathog. 2021, 155, 104901. [Google Scholar] [CrossRef] [PubMed]
- Wright, M.H.; Adelskov, J.; Greene, A.C. Bacterial DNA Extraction Using Individual Enzymes and Phenol/Chloroform Separation. J. Microbiol. Biol. Educ. 2017, 18, 18–22. [Google Scholar] [CrossRef]
- Baym, M.; Kryazhimskiy, S.; Lieberman, T.D.; Chung, H.; Desai, M.M.; Kishony, R.K. Inexpensive Multiplexed Library Preparation for Megabase-Sized Genomes. PLoS ONE 2015, 10, e0128036. [Google Scholar] [CrossRef]
- Deatherage, D.E.; Barrick, J.E. Identification of Mutations in Laboratory-Evolved Microbes from next-Generation Sequencing Data Using Breseq. Methods Mol. Biol. 2014, 1151, 165–188. [Google Scholar] [CrossRef]
- Martin, A.P. Phylogenetic Approaches for Describing and Comparing the Diversity of Microbial Communities. Appl. Environ. Microbiol. 2002, 68, 3673–3682. [Google Scholar] [CrossRef]
- Domenico, P.; Schwartz, S.; Cunha, B.A. Reduction of Capsular Polysaccharide Production in Klebsiella Pneumoniae by Sodium Salicylate. Infect. Immun. 1989, 57, 3778–3782. [Google Scholar] [CrossRef]
- Lin, T.L.; Yang, F.L.; Yang, A.S.; Peng, H.P.; Li, T.L.; Tsai, M.D.; Wu, S.H.; Wang, J.T. Amino Acid Substitutions of MagA in Klebsiella Pneumoniae Affect the Biosynthesis of the Capsular Polysaccharide. PLoS ONE 2012, 7, e46783. [Google Scholar] [CrossRef] [PubMed]
- Brunson, D.N.; Maldosevic, E.; Velez, A.; Figgins, E.; Ellis, T.N. Porin Loss in Klebsiella Pneumoniae Clinical Isolates Impacts Production of Virulence Factors and Survival within Macrophages. Int. J. Med. Microbiol. 2019, 309, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Kachlany, S.C.; Levery, S.B.; Kim, J.S.; Reuhs, B.L.; Lion, L.W.; Ghiorse, W.C. Structure and Carbohydrate Analysis of the Exopolysaccharide Capsule of Pseudomonas Putida G7. Environ. Microbiol. 2001, 3, 774–784. [Google Scholar] [CrossRef] [PubMed]
- York, W.S.; Darvill, A.G.; McNeil, M.; Stevenson, T.T.; Albersheim, P. Isolation and Characterization of Plant Cell Walls and Cell Wall Components. Methods Enzymol. 1986, 118, 3–40. [Google Scholar] [CrossRef]
- Turner, K.L.; Cahill, B.K.B.K.; Dilello, S.K.S.K.; Gutel, D.; Brunson, D.N.D.N.; Albertí, S.; Ellis, T.N.T.N. Porin Loss Impacts the Host Inflammatory Response to Outer Membrane Vesicles of Klebsiella Pneumoniae. Antimicrob. Agents Chemother. 2015, 60, 1360–1369. [Google Scholar] [CrossRef]
- Velkov, T.; Soon, R.L.; Chong, P.L.; Huang, J.X.; Cooper, M.A.; Azad, M.A.K.; Baker, M.A.; Thompson, P.E.; Roberts, K.; Nation, R.L.; et al. Molecular Basis for the Increased Polymyxin Susceptibility of Klebsiella Pneumoniae Strains with Under-Acylated Lipid A. Innate Immun. 2013, 19, 265–277. [Google Scholar] [CrossRef]

| Time (Minutes) from Subculture to OD600 1.0 | ||||
|---|---|---|---|---|
| Elapsed Time of Culture (Hours) | Cephalothin Dose for Antibiotic Treated Culture (μg/mL) | Untreated Culture | Antibiotic Treated Culture | Increase in Growth Time (Minutes) of Treated Culture |
| 36 | 1 | 120 | 195 | 53 |
| 60 | 1.5 | 180 | 195 | 53 |
| 84 | 2 | 135 | 177 | 35 |
| 108 | 2.5 | 130 | 165 | 23 |
| 132 | 3 | 140 | 160 | 18 |
| 156 | 3.5 | 130 | 155 | 13 |
| 180 | 4 | 132.5 | 180 | 38 |
| 204 | 4.5 | 135 | 165 | 23 |
| 228 | 5 | 137 | 165 | 23 |
| 252 | 5.5 | 150 | 240 | 98 |
| 276 | 6 | 145 | 375 | 233 |
| 300 | 6.5 | 152 | 390 | 248 |
| 324 | 7 | 148 | 245 | 103 |
| 348 | 7.5 | 155 | 350 | 208 |
| 372 | 7.5 (final culture) | 155 | 180 | 38 |
| Antibiotic | K. pneumoniae 43816 | Day 14 Untreated | Day 14 Adapted | K. pneumoniae 43816 ΔwcaJ | Adapted Large Colony | Adapted Small Colony |
|---|---|---|---|---|---|---|
| Tetracycline | 0.5 | 1 | 16 * | 4 | 8 * | 8 * |
| Cephalothin | 4 | 4 | 125 * | 16 ** | 500 * | 125 * |
| Cefoxitin | 2 | 8 | 64 * | 16 ** | 64 * | 128 * |
| Cefotaxime | 1 | 8 | 8 | 1 | 1 | 8 |
| Cefepime | 1 | 8 | 8 | 1 | 1 | 8 |
| Imipenem | 1 | 1 | 1 | 1 | 2 | 1 |
| Amikacin | 0.5 | 1 | 2 | 1 | 0.5 | 0.5 |
| Position | Change in Sequence | Depth of Coverage | Gene/Promoter | Gene Family |
|---|---|---|---|---|
| 10,279 | G→C | 43 | Gene | SGNH/GDSL hydrolase |
| 10,285 | G→C | 43 | Gene | SGNH/GDSL hydrolase |
| 10,412 | T Deletion | 56 | Promoter | SGNH/GDSL hydrolase |
| 412,527 | G Deletion | 100 | Gene | N-acetyltransferase |
| 1,125,537 | Insertion T | 100 | Gene | Globin |
| 2,008,934 | Insertion TTTCGCTA | 100 | Gene | TetR/AcrR Transcription regulator |
| 2,063,994 | C Deletion | 100 | Gene | ABC Transporter |
| 3,225,592 | C Deletion | 100 | Gene | RmuC DNA Recombination |
| 5,188,116 | T→G | 100 | Gene | UDP Phosphate Glucose Phosphotransferse |
| 2,657,272 | G Deletion | 100 | Promoter | Peptide Chain Release Factor 3 |
| 1,078,904 | C Deletion | 100 | Promoter | LysR Transcriptional Regulator |
| 1,294,495 | G Deletion | 100 | Promoter | Type II Aspariginase |
| 1,823,692 | A Insertion | 100 | Promoter | Pyrimidine Photo-lyase |
| 5,050,494 | A Insertion | 100 | Promoter | Cyclic Di-GMP Phosphodiesterase |
| Colony Variant | Position | Original | New | Coverage (Small/Large) | Gene |
|---|---|---|---|---|---|
| Small and Large | 2,008,948 | Insert TATTTCGC | 100/52 | TetR/AcrR family transcriptional regulator | |
| Small and Large | 3,191,984 | A | T | 100/100 | rpoB/DA-directed RNA polymerase subunit beta |
| Large | 1,548,640 | Unspecified | Unspecified | 15 | ComEC family protein |
| Small | 4,676,685 | T | G | 100 | YfiR family protein |
| Small | 5,187,773 | Deletion 2103 bp. | 100 | wcaJ and gndA | |
| Final adapted mixture | 5,187,772 | T | G | 64 | wcaJ Undecaprenyl phosphate-gluocse phosphotransferase |
| Final adapted mixture | 5,189,876 | unspecified | unspecified | 64 | gndA NADP-dependent phosphogluconate dehydrogenase |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anderson, J.R.; Lam, N.B.; Jackson, J.L.; Dorenkott, S.M.; Ticer, T.; Maldosevic, E.; Velez, A.; Camden, M.R.; Ellis, T.N. Progressive Sub-MIC Exposure of Klebsiella pneumoniae 43816 to Cephalothin Induces the Evolution of Beta-Lactam Resistance without Acquisition of Beta-Lactamase Genes. Antibiotics 2023, 12, 887. https://doi.org/10.3390/antibiotics12050887
Anderson JR, Lam NB, Jackson JL, Dorenkott SM, Ticer T, Maldosevic E, Velez A, Camden MR, Ellis TN. Progressive Sub-MIC Exposure of Klebsiella pneumoniae 43816 to Cephalothin Induces the Evolution of Beta-Lactam Resistance without Acquisition of Beta-Lactamase Genes. Antibiotics. 2023; 12(5):887. https://doi.org/10.3390/antibiotics12050887
Chicago/Turabian StyleAnderson, Jasmine R., Nghi B. Lam, Jazmyne L. Jackson, Sean M. Dorenkott, Taylor Ticer, Emir Maldosevic, Amanda Velez, Megan R. Camden, and Terri N. Ellis. 2023. "Progressive Sub-MIC Exposure of Klebsiella pneumoniae 43816 to Cephalothin Induces the Evolution of Beta-Lactam Resistance without Acquisition of Beta-Lactamase Genes" Antibiotics 12, no. 5: 887. https://doi.org/10.3390/antibiotics12050887
APA StyleAnderson, J. R., Lam, N. B., Jackson, J. L., Dorenkott, S. M., Ticer, T., Maldosevic, E., Velez, A., Camden, M. R., & Ellis, T. N. (2023). Progressive Sub-MIC Exposure of Klebsiella pneumoniae 43816 to Cephalothin Induces the Evolution of Beta-Lactam Resistance without Acquisition of Beta-Lactamase Genes. Antibiotics, 12(5), 887. https://doi.org/10.3390/antibiotics12050887






