Molecular Mechanisms of Biofilm Formation in Helicobacter pylori
Abstract
1. Introduction
2. Search Method
3. Evidence on How H. pylori Forms Biofilm
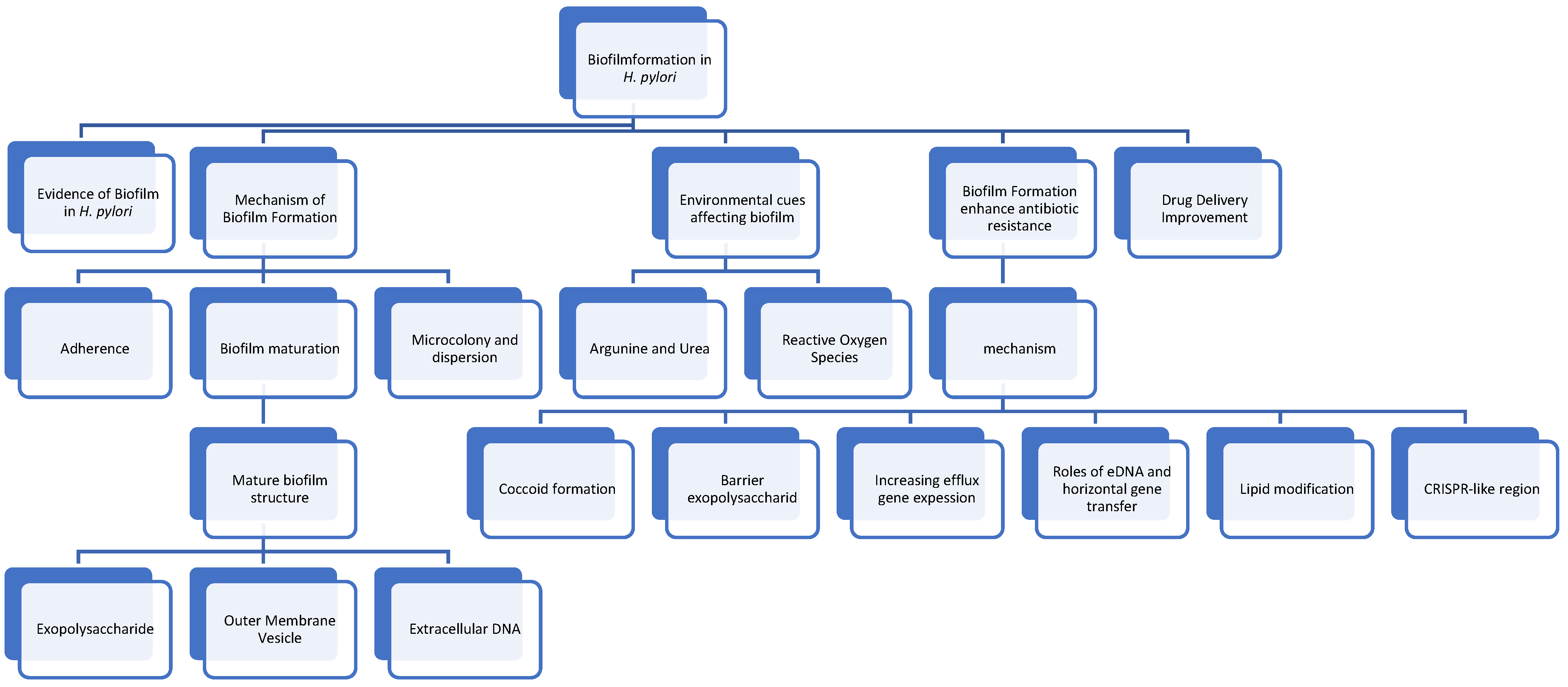
4. Mechanism of Biofilm Formation
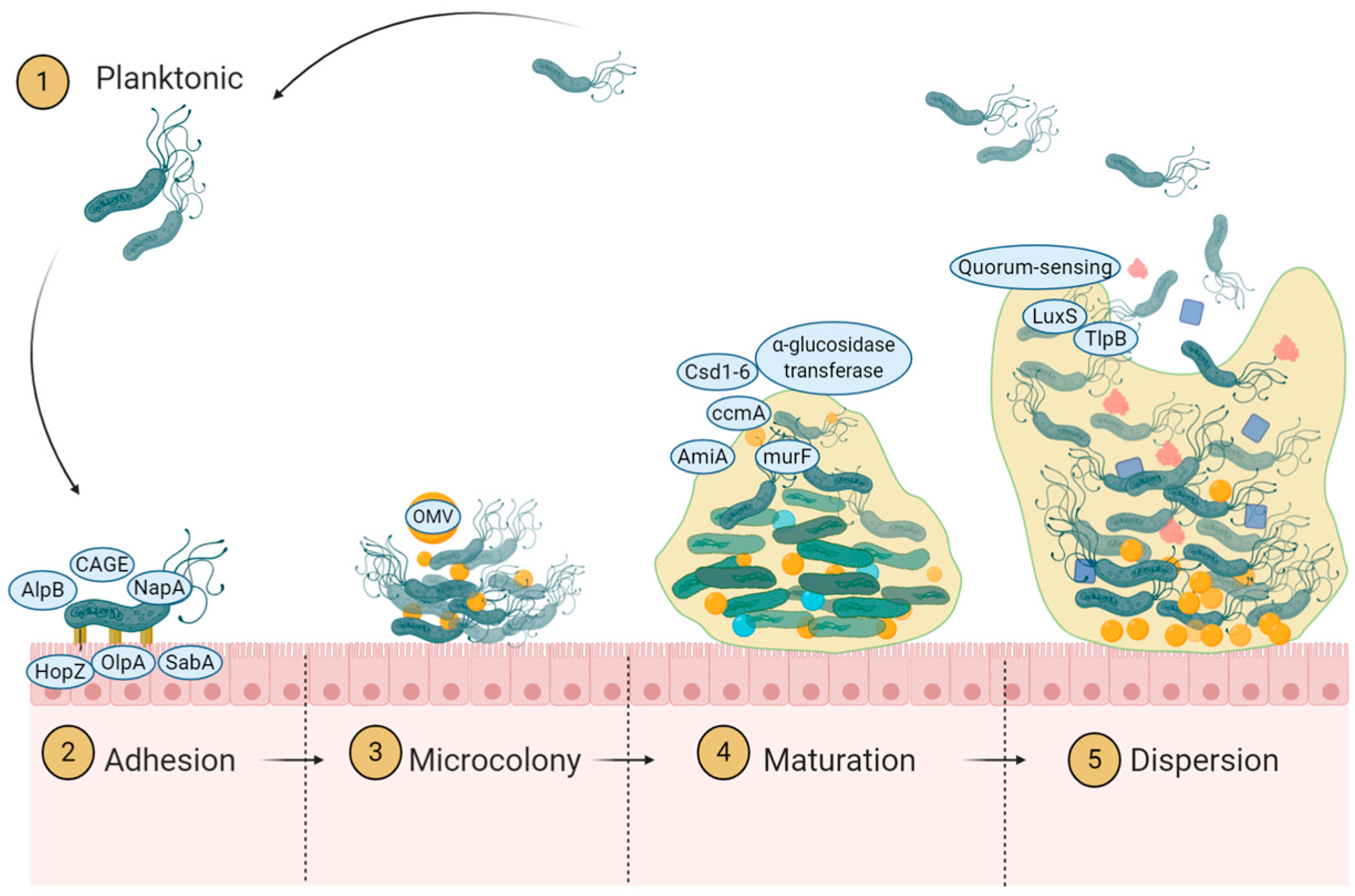
4.1. Initiation of Adherence
4.1.1. Role of Adhesion Protein
4.1.2. Role of Mucin Binding Protein
4.1.3. Role of Chemotaxis
4.2. Microcolony Formation and Dispersion
4.2.1. Definition of Microcolony Formation
4.2.2. Role of Quorum Sensing
4.3. Biofilm Maturation
4.3.1. Extracellular Polysaccharide Deposit
4.3.2. Outer Membrane Vesicle as Extracellular Substance
4.3.3. Extracellular DNA
5. Environmental Cues Affecting Biofilm Formation
5.1. Arginine and Urea
5.2. Reactive Oxygen Species
6. Biofilm Formation Enhances Antibiotic Resistance
6.1. Coccoid Formation
6.2. The Barrier of Extracellular Polysaccharide
6.3. The Increasing Expression of Efflux Gene
6.4. The Roles of eDNA and Horizontal Gene Transfer
6.5. Lipid Modification
6.6. CRISPR-like Region
7. Discussion: Alternative Solution Against the Biofilm and Resistance
8. Future Research
9. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Correa, P.; Piazuelo, M.B. Natural history of Helicobacter pylori infection. Dig. Liver Dis. 2008, 40, 490–496. [Google Scholar] [CrossRef] [PubMed]
- Correa, P.; Houghton, J. Carcinogenesis of Helicobacter pylori. Gastroenterology 2007, 133, 659–672. [Google Scholar] [CrossRef] [PubMed]
- Vogiatzi, P.; Cassone, M.; Luzzi, I.; Lucchetti, C.; Otvos, L., Jr.; Giordano, A. Helicobacter pylori as a class I carcinogen: Physiopathology and management strategies. J. Cell. Biochem. 2007, 102, 264–273. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, Y.; Graham, D.Y. Helicobacter pylori virulence and cancer pathogenesis. Future Oncol. 2014, 10, 1487–1500. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.T.; Liou, J.M.; El-Omar, E.M.; Wu, J.Y.; Leow, A.H.R.; Goh, K.L.; Das, R.; Lu, H.; Lin, J.T.; Tu, Y.K.; et al. Primary antibiotic resistance in Helicobacter pylori in the Asia-Pacific region: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2017, 2, 707–715. [Google Scholar] [CrossRef]
- Blair, J.M.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Yonezawa, H.; Osaki, T.; Kamiya, S. Biofilm Formation by Helicobacter pylori and Its Involvement for Antibiotic Resistance. BioMed Res. Int. 2015, 2015, 914791. [Google Scholar] [CrossRef]
- Malfertheiner, P.; Megraud, F.; Rokkas, T.; Gisbert, J.P.; Liou, J.M.; Schulz, C.; Gasbarrini, A.; Hunt, R.H.; Leja, M.; O’Morain, C.; et al. Management of Helicobacter pylori infection: The Maastricht VI/Florence consensus report. Gut 2022, 71, 1724–1762. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Monds, R.D.; O’Toole, G.A. The developmental model of microbial biofilms: Ten years of a paradigm up for review. Trends Microbiol. 2009, 17, 73–87. [Google Scholar] [CrossRef]
- Jefferson, K.K. What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 2004, 236, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Coticchia, J.; Zuliani, G.; Coleman, C.; Carron, M.; Gurrola, J., 2nd; Haupert, M.; Berk, R. Biofilm surface area in the pediatric nasopharynx: Chronic rhinosinusitis vs obstructive sleep apnea. Arch. Otolaryngol.-Head Neck Surg. 2007, 133, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Carron, M.A.; Tran, V.R.; Sugawa, C.; Coticchia, J.M. Identification of Helicobacter pylori biofilms in human gastric mucosa. J. Gastrointest. Surg. 2006, 10, 712–717. [Google Scholar] [CrossRef] [PubMed]
- Cole, S.P.; Harwood, J.; Lee, R.; She, R.; Guiney, D.G. Characterization of monospecies biofilm formation by Helicobacter pylori. J. Bacteriol. 2004, 186, 3124–3132. [Google Scholar] [CrossRef]
- Attaran, B.; Falsafi, T.; Moghaddam, A.N. Study of biofilm formation in C57Bl/6J mice by clinical isolates of Helicobacter pylori. Saudi J. Gastroenterol. 2016, 22, 161–168. [Google Scholar] [CrossRef]
- Park, S.; Mackay, W.; Reid, D. Helicobacter sp. recovered from drinking water biofilm sampled from a water distribution system. Water Res. 2001, 35, 1624–1626. [Google Scholar] [CrossRef]
- Giao, M.S.; Azevedo, N.; Wilks, S.A.; Vieira, M.; Keevil, C.W. Persistence of Helicobacter pylori in heterotrophic drinking-water biofilms. Appl. Environ. Microbiol. 2008, 74, 5898–5904. [Google Scholar] [CrossRef]
- Azevedo, N.; Pinto, A.; Reis, N.; Vieira, M.; Keevil, C. Shear stress, temperature, and inoculation concentration influence the adhesion of water-stressed Helicobacter pylori to stainless steel 304 and polypropylene. Appl. Environ. Microbiol. 2006, 72, 2936–2941. [Google Scholar] [CrossRef][Green Version]
- Bragança, S.M.; Azevedo, N.F.; Simões, L.C.; Keevil, C.; Vieira, M. Use of fluorescent in situ hybridisation for the visualisation of Helicobacter pylori in real drinking water biofilms. Water Sci. Technol. 2007, 55, 387–393. [Google Scholar] [CrossRef]
- Percival, S.L.; Thomas, J.G. Transmission of Helicobacter pylori and the role of water and biofilms. J. Water Health 2009, 7, 469–477. [Google Scholar] [CrossRef]
- Zhao, Y.; Cai, Y.; Chen, Z.; Li, H.; Xu, Z.; Li, W.; Jia, J.; Sun, Y. SpoT-mediated NapA upregulation promotes oxidative stress-induced Helicobacter pylori biofilm formation and confers multidrug resistance. Antimicrob. Agents Chemother. 2023, 65, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Yonezawa, H.; Osaki, T.; Kurata, S.; Zaman, C.; Hanawa, T.; Kamiya, S. Assessment of in vitro biofilm formation by Helicobacter pylori. J. Gastroenterol. Hepatol. 2010, 25 (Suppl. S1), S90–S94. [Google Scholar] [CrossRef] [PubMed]
- Hathroubi, S.; Servetas, S.L.; Windham, I.; Merrell, D.S. Helicobacter pylori Biofilm Formation and Its Potential Role in Pathogenesis. Microbiol. Mol. Biol. Rev. 2018, 82, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Mah, T.F. Biofilm-specific antibiotic resistance. Future Microbiol. 2012, 7, 1061–1072. [Google Scholar] [CrossRef]
- Yonezawa, H.; Osaki, T.; Fukutomi, T.; Hanawa, T.; Kurata, S.; Zaman, C.; Hojo, F.; Kamiya, S. Diversification of the AlpB outer membrane protein of Helicobacter pylori affects biofilm formation and cellular adhesion. J. Bacteriol. 2017, 199, 10–1128. [Google Scholar] [CrossRef]
- Wong, E.H.; Ng, C.G.; Chua, E.G.; Tay, A.C.; Peters, F.; Marshall, B.J.; Ho, B.; Goh, K.L.; Vadivelu, J.; Loke, M.F. Comparative Genomics Revealed Multiple Helicobacter pylori Genes Associated with Biofilm Formation In Vitro. PLoS ONE 2016, 11, e0166835. [Google Scholar] [CrossRef]
- Backert, S.; Tegtmeyer, N.; Fischer, W. Composition, structure and function of the Helicobacter pylori cag pathogenicity island encoded type IV secretion system. Future Microbiol. 2015, 10, 955–965. [Google Scholar] [CrossRef]
- Zhang, Z.-W.; Dorrell, N.; Wren, B.W.; Farthing, M.J. Helicobacter pylori adherence to gastric epithelial cells: A role for non-adhesin virulence genes. J. Med. Microbiol. 2002, 51, 495–502. [Google Scholar] [CrossRef][Green Version]
- Namavar, F.; Sparrius, M.; Veerman, E.C.; Appelmelk, B.J.; Vandenbroucke-Grauls, C.M. Neutrophil-activating protein mediates adhesion of Helicobacter pylori to sulfated carbohydrates on high-molecular-weight salivary mucin. Infect. Immun. 1998, 66, 444–447. [Google Scholar] [CrossRef]
- Skoog, E. Helicobacter spp. Interactions with Mucins: Adhesion and Mucin Regulation of Pathogen Proliferation and Gene Expression. Ph.D. Thesis, University of Gothenburg, Göteborg, Sweden, 2014. [Google Scholar]
- Yang, F.L.; Hassanbhai, A.M.; Chen, H.Y.; Huang, Z.Y.; Lin, T.L.; Wu, S.H.; Ho, B. Proteomannans in biofilm of Helicobacter pylori ATCC 43504. Helicobacter 2011, 16, 89–98. [Google Scholar] [CrossRef]
- Rizzato, C.; Torres, J.; Kasamatsu, E.; Camorlinga-Ponce, M.; Bravo, M.M.; Canzian, F.; Kato, I. Potential Role of Biofilm Formation in the Development of Digestive Tract Cancer With Special Reference to Helicobacter pylori Infection. Front. Microbiol. 2019, 10, 846. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lertsethtakarn, P.; Mariscal, V.T.; Yildiz, F.; Ottemann, K.M. Counterclockwise rotation of the flagellum promotes biofilm initiation in Helicobacter pylori. Mbio 2024, e00440-24. [Google Scholar] [CrossRef] [PubMed]
- Rani, S.A.; Pitts, B.; Beyenal, H.; Veluchamy, R.A.; Lewandowski, Z.; Davison, W.M.; Buckingham-Meyer, K.; Stewart, P.S. Spatial patterns of DNA replication, protein synthesis, and oxygen concentration within bacterial biofilms reveal diverse physiological states. J. Bacteriol. 2007, 189, 4223–4233. [Google Scholar] [CrossRef]
- Anderson, J.K.; Huang, J.Y.; Wreden, C.; Sweeney, E.G.; Goers, J.; Remington, S.J.; Guillemin, K. Chemorepulsion from the Quorum Signal Autoinducer-2 Promotes Helicobacter pylori Biofilm Dispersal. mBio 2015, 6, e00379. [Google Scholar] [CrossRef]
- De la Cruz, M.A.; Ares, M.A.; von Bargen, K.; Panunzi, L.G.; Martínez-Cruz, J.; Valdez-Salazar, H.A.; Jiménez-Galicia, C.; Torres, J. Gene expression profiling of transcription factors of Helicobacter pylori under different environmental conditions. Front. Microbiol. 2017, 8, 615. [Google Scholar] [CrossRef]
- Lee, A.Y.; Kao, C.Y.; Wang, Y.K.; Lin, S.Y.; Lai, T.Y.; Sheu, B.S.; Lo, C.J.; Wu, J.J. Inactivation of ferric uptake regulator (Fur) attenuates Helicobacter pylori J99 motility by disturbing the flagellar motor switch and autoinducer-2 production. Helicobacter 2017, 22, e12388. [Google Scholar] [CrossRef]
- Joyce, E.A.; Bassler, B.L.; Wright, A. Evidence for a signaling system in Helicobacter pylori: Detection of a luxS-encoded autoinducer. J. Bacteriol. 2000, 182, 3638–3643. [Google Scholar] [CrossRef]
- Shen, F.; Hobley, L.; Doherty, N.; Loh, J.T.; Cover, T.L.; Sockett, R.E.; Hardie, K.R.; Atherton, J.C. In Helicobacter pylori auto-inducer-2, but not LuxS/MccAB catalysed reverse transsulphuration, regulates motility through modulation of flagellar gene transcription. BMC Microbiol. 2010, 10, 210. [Google Scholar] [CrossRef]
- Rader, B.A.; Campagna, S.R.; Semmelhack, M.F.; Bassler, B.L.; Guillemin, K. The quorum-sensing molecule autoinducer 2 regulates motility and flagellar morphogenesis in Helicobacter pylori. J. Bacteriol. 2007, 189, 6109–6117. [Google Scholar] [CrossRef]
- Osaki, T.; Hanawa, T.; Manzoku, T.; Fukuda, M.; Kawakami, H.; Suzuki, H.; Yamaguchi, H.; Yan, X.; Taguchi, H.; Kurata, S.; et al. Mutation of luxS affects motility and infectivity of Helicobacter pylori in gastric mucosa of a Mongolian gerbil model. J. Med. Microbiol. 2006, 55, 1477–1485. [Google Scholar] [CrossRef]
- Davies, D. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2003, 2, 114. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Igarashi, M.; Hayashi, C.; Shitara, T.; Nomoto, A.; Mizote, T.; Shibasaki, M. Identification of self-growth-inhibiting compounds lauric acid and 7-(Z)-tetradecenoic acid from Helicobacter pylori. Microbiology 2015, 161, 1231–1239. [Google Scholar] [CrossRef]
- Krzyzek, P.; Gosciniak, G. A proposed role for diffusible signal factors in the biofilm formation and morphological transformation of Helicobacter pylori. Turk. J. Gastroenterol. 2017, 29, 7–13. [Google Scholar] [CrossRef]
- Krzyżek, P.; Migdał, P.; Grande, R.; Gościniak, G. Biofilm Formation of Helicobacter pylori in Both Static and Microfluidic Conditions Is Associated with Resistance to Clarithromycin. Front. Cell. Infect. Microbiol. 2022, 12, 868905. [Google Scholar] [CrossRef]
- Hathroubi, S.; Zerebinski, J.; Ottemann, K.M. Helicobacter pylori Biofilm Involves a Multigene Stress-Biased Response, Including a Structural Role for Flagella. mBio 2018, 9, e01973-18. [Google Scholar] [CrossRef]
- Gu, H. Role of Flagella in the Pathogenesis of Helicobacter pylori. Curr. Microbiol. 2017, 74, 863–869. [Google Scholar] [CrossRef]
- Hathroubi, S.; Hu, S.; Ottemann, K.M. Genetic requirements and transcriptomics of Helicobacter pylori biofilm formation on abiotic and biotic surfaces. NPJ Biofilms Microbiomes 2020, 6, 56. [Google Scholar] [CrossRef]
- Baker, P.; Hill, P.J.; Snarr, B.D.; Alnabelseya, N.; Pestrak, M.J.; Lee, M.J.; Jennings, L.K.; Tam, J.; Melnyk, R.A.; Parsek, M.R.; et al. Exopolysaccharide Biosynthetic Glycoside Hydrolases Can Be Utilized to Disrupt and Prevent Pseudomonas aeruginosa biofilms. Sci. Adv. 2015, 2, e1501632. [Google Scholar] [CrossRef]
- Stark, R.M.; Gerwig, G.J.; Pitman, R.S.; Potts, L.F.; Williams, N.A.; Greenman, J.; Weinzweig, I.P.; Hirst, T.R.; Millar, M.R. Biofilm formation by Helicobacter pylori. Lett. Appl. Microbiol. 1999, 28, 121–126. [Google Scholar] [CrossRef]
- Hassanbhai, A.M.; Phoon, M.C.; Chow, V.T.; Ho, B. The Association of Helicobacter pylori Biofilm with Enterovirus 71 Prolongs Viral Viability and Survival. Int. J. Mol. Sci. 2023, 24, 14500. [Google Scholar] [CrossRef]
- Kulp, A.; Kuehn, M.J. Biological Functions and Biogenesis of Secreted Bacterial Outer Membrane Vesicles. Annu. Rev. Microbiol. 2010, 64, 163–184. [Google Scholar] [CrossRef] [PubMed]
- Yonezawa, H.; Osaki, T.; Woo, T.; Kurata, S.; Zaman, C.; Hojo, F.; Hanawa, T.; Kato, S.; Kamiya, S. Analysis of outer membrane vesicle protein involved in biofilm formation of Helicobacter pylori. Anaerobe 2011, 17, 388–390. [Google Scholar] [CrossRef] [PubMed]
- Altındiş, E.; Fu, Y.; Mekalanos, J.J. Proteomic Analysis of Vibrio cholerae Outer Membrane Vesicles. Proc. Natl. Acad. Sci. USA 2014, 111, E1548–E1556. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.-Y.; Choi, D.-S.; Kim, K.P.; Gho, Y.S. Proteomics in Gram-negative Bacterial Outer Membrane Vesicles. Mass. Spectrom. Rev. 2008, 27, 535–555. [Google Scholar] [CrossRef]
- Lee, J.W.; Kim, O.Y.; Gho, Y.S. Proteomic Profiling of Gram-negative Bacterial Outer Membrane Vesicles: Current Perspectives. Proteom.-Clin. Appl. 2016, 10, 897–909. [Google Scholar] [CrossRef]
- Cooke, A.C.; Nello, A.V.; Ernst, R.K.; Schertzer, J.W. Analysis of Pseudomonas aeruginosa Biofilm Membrane Vesicles Supports Multiple Mechanisms of Biogenesis. PLoS ONE 2019, 14, e0212275. [Google Scholar] [CrossRef]
- Wang, Y.; Hoffmann, J.P.; Baker, S.M.; Bentrup, K.H.z.; Wimley, W.C.; Fuselier, J.A.; Bitoun, J.P.; Morici, L.A. Inhibition of Streptococcus mutans Biofilms with Bacterial-Derived Outer Membrane Vesicles. BMC Microbiol. 2021, 21, 1–12. [Google Scholar] [CrossRef]
- Toyofuku, M.; Roschitzki, B.; Riedel, K.; Eberl, L. Identification of Proteins Associated with the Pseudomonas aeruginosa Biofilm Extracellular Matrix. J. Proteome Res. 2012, 11, 4906–4915. [Google Scholar] [CrossRef]
- Cooke, A.C.; Florez, C.; Dunshee, E.B.; Lieber, A.D.; Terry, M.; Light, C.J.; Schertzer, J.W. Pseudomonas Quinolone Signal-Induced Outer Membrane Vesicles Enhance Biofilm Dispersion in Pseudomonas aeruginosa. Msphere 2020, 5, 10–1128. [Google Scholar] [CrossRef]
- An, S.J.; Ha, K.-W.; Jun, H.K.; Kim, H.; Choi, B.K. Reduced Proinflammatory Activity of Outer Membrane Vesicles of Tannerella forsythia Treated with Quorum Sensing Inhibitors. Mol. Oral. Microbiol. 2022, 38, 71–81. [Google Scholar] [CrossRef]
- Potapova, A.; Garvey, W.; Dahl, P.; Guo, S.; Chang, Y.; Schwechheimer, C.; Trebino, M.A.; Floyd, K.A.; Phinney, B.S.; Liu, J.; et al. Outer Membrane Vesicles and the Outer Membrane Protein OmpU Govern Vibrio cholerae Biofilm Matrix Assembly. Mbio 2024, 15, e0330423. [Google Scholar] [CrossRef] [PubMed]
- Grande, R.; Di Marcantonio, M.C.; Robuffo, I.; Pompilio, A.; Celia, C.; Di Marzio, L.; Paolino, D.; Codagnone, M.; Muraro, R.; Stoodley, P.; et al. Helicobacter pylori ATCC 43629/NCTC 11639 Outer Membrane Vesicles (OMVs) from Biofilm and Planktonic Phase Associated with Extracellular DNA (eDNA). Front. Microbiol. 2015, 6, 1369. [Google Scholar] [CrossRef] [PubMed]
- Grande, R.; Di Giulio, M.; Bessa, L.J.; Di Campli, E.; Baffoni, M.; Guarnieri, S.; Cellini, L. Extracellular DNA in Helicobacter pylori biofilm: A backstairs rumour. J. Appl. Microbiol. 2011, 110, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Marks, L.R.; Reddinger, R.M.; Hakansson, A.P. High Levels of Genetic Recombination during Nasopharyngeal Carriage and Biofilm Formation in Streptococcus pneumoniae. mBio 2012, 3, e00200-12. [Google Scholar] [CrossRef]
- Yonezawa, H.; Osaki, T.; Hanawa, T.; Kurata, S.; Ochiai, K.; Kamiya, S. Impact of Helicobacter pylori biofilm formation on clarithromycin susceptibility and generation of resistance mutations. PLoS ONE 2013, 8, e73301. [Google Scholar] [CrossRef]
- Ansari, S.; Yamaoka, Y. Survival of Helicobacter pylori in gastric acidic territory. Helicobacter 2017, 22, e12386. [Google Scholar] [CrossRef]
- Ranieri, M.R.; Whitchurch, C.B.; Burrows, L.L. Mechanisms of biofilm stimulation by subinhibitory concentrations of antimicrobials. Curr. Opin. Microbiol. 2018, 45, 164–169. [Google Scholar] [CrossRef]
- Bessa, L.J.; Grande, R.; Iorio, D.D.I.; Giulio, M.D.I.; Campli, E.D.I.; Cellini, L. Helicobacter pylori free-living and biofilm modes of growth: Behavior in response to different culture media. Apmis 2013, 121, 549–560. [Google Scholar] [CrossRef]
- McGee, D.J.; Kumar, S.; Viator, R.J.; Bolland, J.R.; Ruiz, J.; Spadafora, D.; Testerman, T.L.; Kelly, D.J.; Pannell, L.K.; Windle, H.J. Helicobacter pylori thioredoxin is an arginase chaperone and guardian against oxidative and nitrosative stresses. J. Biol. Chem. 2006, 281, 3290–3296. [Google Scholar] [CrossRef]
- Wang, G.; Yang, H.; Olczak, A.A.; Maier, S.E.; Maier, R.J. Dual Roles of Helicobacter pylori NapA in Inducing and Combating Oxidative Stress. Infect. Immun. 2006, 74, 6839–6846. [Google Scholar] [CrossRef]
- Hong, T.C.; El-Omar, E.M.; Kuo, Y.T.; Wu, J.Y.; Chen, M.J.; Chen, C.C.; Fang, Y.J.; Leow, A.H.R.; Lu, H.; Lin, J.T.; et al. Primary antibiotic resistance of Helicobacter pylori in the Asia-Pacific region between 1990 and 2022: An updated systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2024, 9, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Xue, J.; Lin, F.; Liu, D.; Zhang, W.; Ru, S.; Jiang, F. Global Primary Antibiotic Resistance Rate of Helicobacter pylori in Recent 10 years: A Systematic Review and Meta-Analysis. Helicobacter 2024, 29, e13103. [Google Scholar] [CrossRef] [PubMed]
- Azzaya, D.; Gantuya, B.; Oyuntsetseg, K.; Davaadorj, D.; Matsumoto, T.; Akada, J.; Yamaoka, Y. High Antibiotic Resistance of Helicobacter pylori and Its Associated Novel Gene Mutations among the Mongolian Population. Microorganisms 2020, 8, 1062. [Google Scholar] [CrossRef] [PubMed]
- Cimuanga-Mukanya, A.; Tshibangu-Kabamba, E.; Kisoko Pd, J.N.; Fauzia, K.A.; Tshibangu, F.M.; Wola, A.T.; Kashala, P.T.; Ngoyi, D.M.; Ahuka-Mundeke, S.; Revathi, G.; et al. Synergistic effects of novel penicillin-binding protein 1A amino acid substitutions contribute to high-level amoxicillin resistance of Helicobacter pylori. mSphere 2024, 9, e0008924. [Google Scholar] [CrossRef]
- Khangai, A.; Saruuljavkhlan, B.; Azzaya, D.; Gantuya, B.; Oyuntsetseg, K.; Akada, J.; Matsumoto, T.; Yamaoka, Y. Exploring Alternative Treatment Choices for Multidrug-Resistant Clinical Strains of Helicobacter pylori in Mongolia. Microorganisms 2023, 11, 2852. [Google Scholar] [CrossRef]
- Fauzia, K.A.; Miftahussurur, M.; Syam, A.F.; Waskito, L.A.; Doohan, D.; Rezkitha, Y.A.A.; Matsumoto, T.; Tuan, V.P.; Akada, J.; Yonezawa, H.; et al. Biofilm Formation and Antibiotic Resistance Phenotype of Helicobacter pylori Clinical Isolates. Toxins 2020, 12, 473. [Google Scholar] [CrossRef]
- Lewis, K. Multidrug tolerance of biofilms and persister cells. Curr. Top. Microbiol. Immunol. 2008, 322, 107–131. [Google Scholar]
- Reshetnyak, V.I.; Reshetnyak, T.M. Significance of dormant forms of Helicobacter pylori in ulcerogenesis. World J. Gastroenterol. 2017, 23, 4867–4878. [Google Scholar] [CrossRef]
- Faghri, J.; Poursina, F.; Moghim, S.; Esfahani, H.; Esfahani, B.N.; Fazeli, H.; Mirzaei, N.; Jamshidian, A.; Safaei, H.G. Morphological and Bactericidal Effects of Different Antibiotics on Helicobacter pylori. Jundishapur J. Microbiol. 2014, 7, e8704. [Google Scholar] [CrossRef]
- Cole, S.P.; Cirillo, D.M.; Kagnoff, M.F.; Guiney, D.G.; Eckmann, L. Coccoid and Spiral Helicobacter pylori Differ in Their Abilities to Adhere to Gastric Epithelial Cells and Induce Interleukin-8 Secretion. Infect. Immun. 1997, 65, 843–846. [Google Scholar] [CrossRef]
- Kabir, F.; Hasan, M.I.; Aggarwal, S. Beyond Microbial Inactivation: Unveiling the Potential of Detachment-Promoting Agents in Water Distribution System Biofilm Control. Acs Est. Water 2024, 4, 2088–2100. [Google Scholar] [CrossRef]
- Chaput, C.; Ecobichon, C.; Pouradier, N.; Rousselle, J.-C.; Namane, A.; Boneca, I.G. Role of the N-Acetylmuramoyl-l-Alanyl Amidase, AmiA, of Helicobacter pylori in Peptidoglycan Metabolism, Daughter Cell Separation, and Virulence. Microb. Drug Resist. 2016, 22, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Bahrami, A.R.; Rahimi, E.; Safaei, H.G. Detection of Helicobacter pylori in City Water, Dental Units′ Water, and Bottled Mineral Water in Isfahan, Iran. Sci. World J. 2013, 2013, 280510. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Han, L.; Chen, J.; Lin, X.; Chen, H.; She, F. Proliferative and Apoptotic Effects of Gastric Epithelial Cells Induced by Coccoid Helicobacter pylori. J. Basic. Microbiol. 2012, 53, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Kadkhodaei, S.; Siavoshi, F. Mucoid and coccoid Helicobacter pylori with fast growth and antibiotic resistance. Helicobacter 2020, 25, e12678. [Google Scholar] [CrossRef]
- Tamer, Y.T.; Toprak, E. On the Race to Starvation: How Do Bacteria Survive High Doses of Antibiotics? Mol. Cell 2017, 68, 1019–1021. [Google Scholar] [CrossRef]
- Hall, C.W.; Mah, T.-F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef]
- Singh, S.; Datta, S.; Narayanan, K.B.; Rajnish, K.N. Bacterial Exo-Polysaccharides in Biofilms: Role in Antimicrobial Resistance and Treatments. J. Genet. Eng. Biotechnol. 2021, 19, 140. [Google Scholar] [CrossRef]
- Balducci, E.; Papi, F.; Capialbi, D.E.; Bino, L.D. Polysaccharides’ Structures and Functions in Biofilm Architecture of Antimicrobial-Resistant (AMR) Pathogens. Int. J. Mol. Sci. 2023, 24, 4030. [Google Scholar] [CrossRef]
- Reighard, K.P. Antibacterial Action of Nitric Oxide-Releasing Chitosan Oligosaccharides Against Pseudomonas aeruginosa Under Aerobic and Anaerobic Conditions. Antimicrob. Agents Chemother. 2015, 59, 6506–6513. [Google Scholar] [CrossRef][Green Version]
- Tseng, Y.S.; Wu, D.C.; Chang, C.Y.; Kuo, C.H.; Yang, Y.C.; Jan, C.M.; Su, Y.C.; Kuo, F.C.; Chang, L.L. Amoxicillin resistance with beta-lactamase production in Helicobacter pylori. Eur. J. Clin. Investig. 2009, 39, 807–812. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-Q.; Zheng, P.-Y.; Yang, P.-C. Efflux pump gene hefA of Helicobacter pylori plays an important role in multidrug resistance. World J. Gastroenterol. WJG 2008, 14, 5217–5222. [Google Scholar] [CrossRef] [PubMed]
- Attaran, B.; Falsafi, T.; Ghorbanmehr, N. Effect of biofilm formation by clinical isolates of Helicobacter pylori on the efflux-mediated resistance to commonly used antibiotics. World J. Gastroenterol. 2017, 23, 1163–1170. [Google Scholar] [CrossRef] [PubMed]
- Yonezawa, H.; Osaki, T.; Hojo, F.; Kamiya, S. Effect of Helicobacter pylori biofilm formation on susceptibility to amoxicillin, metronidazole and clarithromycin. Microb. Pathog. 2019, 132, 100–108. [Google Scholar] [CrossRef]
- Cai, Y.; Wang, C.; Chen, Z. Transporters HP0939, HP0497, and HP0471 participate in intrinsic multidrug resistance and biofilm formation in Helicobacter pylori by enhancing drug efflux. Helicobacter 2020, 25, e12715. [Google Scholar] [CrossRef]
- Grande, R.; Campli, E.D.; Bartolomeo, S.D.; Verginelli, F.; Giulio, M.D.; Baffoni, M.; Bessa, L.J.; Cellini, L. Helicobacter pylori Biofilm: A Protective Environment for Bacterial Recombination. J. Appl. Microbiol. 2012, 113, 669–676. [Google Scholar] [CrossRef]
- Krause, S.; Pansegrau, W.; Lurz, R.; de la Cruz, F.; Lanka, E. Enzymology of type IV macromolecule secretion systems: The conjugative transfer regions of plasmids RP4 and R388 and the cag pathogenicity island of Helicobacter pylori encode structurally and functionally related nucleoside triphosphate hydrolases. J. Bacteriol. 2000, 182, 2761–2770. [Google Scholar] [CrossRef][Green Version]
- Odenbreit, S.; Haas, R. Helicobacter pylori: Impact of gene transfer and the role of the cag pathogenicity island for host adaptation and virulence. Curr. Top. Microbiol. Immunol. 2002, 264, 1–22. [Google Scholar]
- Gaddy, J.A.; Radin, J.N.; Cullen, T.W.; Chazin, W.J.; Skaar, E.P.; Trent, M.S.; Algood, H.M. Helicobacter pylori Resists the Antimicrobial Activity of Calprotectin via Lipid A Modification and Associated Biofilm Formation. mBio 2015, 6, e01349-15. [Google Scholar] [CrossRef]
- Hsu, C.Y.; Yeh, J.Y.; Chen, C.Y.; Wu, H.; Chiang, M.H.; Wu, C.L.; Lin, H.J.; Chiu, C.H.; Lai, C.H. Helicobacter pylori Cholesterol-A-Glucosyltransferase Manipulates Cholesterol for Bacterial Adherence to Gastric Epithelial Cells. Virulence 2021, 12, 2341–2351. [Google Scholar] [CrossRef]
- Cui, L.; Wang, X.; Huang, D.; Zhao, Y.; Feng, J.; Lu, Q.; Pu, Q.; Wang, Y.; Cheng, G.; Wu, M.; et al. CRISPR-cas3 of Salmonella Upregulates Bacterial Biofilm Formation and Virulence to Host Cells by Targeting Quorum-Sensing Systems. Pathogens 2020, 9, 53. [Google Scholar] [CrossRef] [PubMed]
- Hałasa, R.; Turecka, K.; Mizerska, U.; Krauze-Baranowska, M. Anti-Helicobacter pylori Biofilm Extracts from Rubus idaeus and Rubus occidentalis. Pharmaceutics 2024, 16, 501. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Yang, H. In Vitro Effects of Lactobacillus plantarum LN66 and Antibiotics Used Alone or in Combination on Helicobacter pylori Mature Biofilm. Microorganisms 2021, 9, 424. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.; Yang, H. Effects of Lactobacillus salivarius LN12 in Combination with Amoxicillin and Clarithromycin on Helicobacter pylori Biofilm In Vitro. Microorganisms 2021, 9, 1611. [Google Scholar] [CrossRef]
- Boyanova, L.; Medeiros, J.; Yordanov, D.; Gergova, R.; Markovska, R. Turmeric and curcumin as adjuncts in controlling Helicobacter pylori-associated diseases: A narrative review. Lett. Appl. Microbiol. 2024, 77, ovae049. [Google Scholar] [CrossRef]
- Li, R.-J.; Xu, J.-Y.; Wang, X.; Liao, L.-J.; Wei, X.; Xie, P.; Xu, W.-Y.; Xu, Z.-Y.; Xie, S.-H.; Jiang, Y.-Y.; et al. Therapeutic effect of demethylated hydroxylated phillygenin derivative on Helicobacter pylori infection. Front. Microbiol. 2023, 14, 1071603. [Google Scholar] [CrossRef]
- Fan, J.; Dong, Y.; Sun, Y.; Ji, Y.; Feng, J.; Yan, P.; Zhu, Y. Mucus and Biofilm Penetrating Nanoplatform as an Ultrasound-Induced Free Radical Initiator for Targeted Treatment of Helicobacter pylori Infection. Adv. Healthc. Mater. 2024, 13, 2400363. [Google Scholar] [CrossRef]
- Zhao, L.; Liao, W.; Lin, G.; Yang, J.; Shi, X.; Zheng, Y. Rubropunctatin-silver composite nanoliposomes for eradicating Helicobacter pylori in vitro and in vivo. Int. J. Pharm. 2024, 649, 123655. [Google Scholar] [CrossRef]
- Spósito, L.; Fonseca, D.; Carvalho, S.G.; Sábio, R.M.; Marena, G.D.; Bauab, T.M.; Meneguin, A.B.; Parreira, P.; Martins, M.C.L.; Chorilli, M. Engineering resveratrol-loaded chitosan nanoparticles for potential use against Helicobacter pylori infection. Eur. J. Pharm. Biopharm. 2024, 199, 114280. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, Z.; Kong, J.; Liang, Y.; Chen, K.; Chang, Y.; Yuan, H.; Wang, Y.; Liang, H.; Li, J. Fullerenol nanoparticles eradicate Helicobacter pylori via pH-responsive peroxidase activity. ACS Appl. Mater. Interfaces 2020, 12. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Masanam, E.; Ramkumar, V.S.; Baskaraligam, V.; Selvaraj, G. Influence of N-acylhomoserine lactonase silver nanoparticles on the quorum sensing system of Helicobacter pylori: A potential strategy to combat biofilm formation. J. Basic. Microbiol. 2020, 60, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-J.; Huang, C.-H.; Yang, J.-C.; Wang, C.-H.; Shieh, M.-J. Residence time-extended nanoparticles by magnetic field improve the eradication efficiency of Helicobacter pylori. ACS Appl. Mater. Interfaces 2020, 12, 54316–54327. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Guo, Z.; Yan, J.; Bu, C.; Peng, C.; Li, C.; Mao, R.; Zhang, J.; Wang, Z.; Chen, S. Gastric Acid-Responsive ROS Nanogenerators for Effective Treatment of Helicobacter pylori Infection without Disrupting Homeostasis of Intestinal Flora. Adv. Sci. 2023, 10, 2206957. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Han, M.; Fu, H.; Xu, Y.; Bai, Y.; Wang, S.; Yu, J.; Men, C.; Yin, Y.; Zhao, X. Mucus-Penetrating Nanoassembly as Potential Oral Phototherapeutic Formulation against Multi-Drug Resistant Helicobacter pylori Infection. Small 2023, 20, 2306909. [Google Scholar] [CrossRef]
- Darmani, H.; Smadi, E.A.; Bataineh, S.M. Blue light emitting diodes enhance the antivirulence effects of Curcumin against Helicobacter pylori. J. Med. Microbiol. 2020, 69, 617–624. [Google Scholar] [CrossRef]
- Jia, J.; Zhang, C.; Liu, Y.; Huang, Y.; Bai, Y.; Hang, X.; Zeng, L.; Zhu, D.; Bi, H. Armeniaspirol A: A novel anti-Helicobacter pylori agent. Microb. Biotechnol. 2022, 15, 442–454. [Google Scholar] [CrossRef]
- Krzyżek, P.; Gościniak, G.; Fijałkowski, K.; Migdał, P.; Dziadas, M.; Owczarek, A.; Czajkowska, J.; Aniołek, O.; Junka, A. Potential of bacterial cellulose chemisorbed with anti-metabolites, 3-bromopyruvate or sertraline, to fight against Helicobacter pylori lawn biofilm. Int. J. Mol. Sci. 2020, 21, 9507. [Google Scholar] [CrossRef]
- Battisti, A.; Morici, P.; Sgarbossa, A. Fluorescence Lifetime Imaging Microscopy of Porphyrins in Helicobacter pylori Biofilms. Pharmaceutics 2021, 13, 1674. [Google Scholar] [CrossRef]
- Yan, J.; Bassler, B.L. Surviving as a Community: Antibiotic Tolerance and Persistence in Bacterial Biofilms. Cell Host Microbe 2019, 26, 15–21. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fauzia, K.A.; Effendi, W.I.; Alfaray, R.I.; Malaty, H.M.; Yamaoka, Y.; Mifthussurur, M. Molecular Mechanisms of Biofilm Formation in Helicobacter pylori. Antibiotics 2024, 13, 976. https://doi.org/10.3390/antibiotics13100976
Fauzia KA, Effendi WI, Alfaray RI, Malaty HM, Yamaoka Y, Mifthussurur M. Molecular Mechanisms of Biofilm Formation in Helicobacter pylori. Antibiotics. 2024; 13(10):976. https://doi.org/10.3390/antibiotics13100976
Chicago/Turabian StyleFauzia, Kartika Afrida, Wiwin Is Effendi, Ricky Indra Alfaray, Hoda M. Malaty, Yoshio Yamaoka, and Muhammad Mifthussurur. 2024. "Molecular Mechanisms of Biofilm Formation in Helicobacter pylori" Antibiotics 13, no. 10: 976. https://doi.org/10.3390/antibiotics13100976
APA StyleFauzia, K. A., Effendi, W. I., Alfaray, R. I., Malaty, H. M., Yamaoka, Y., & Mifthussurur, M. (2024). Molecular Mechanisms of Biofilm Formation in Helicobacter pylori. Antibiotics, 13(10), 976. https://doi.org/10.3390/antibiotics13100976








