The Impact of Heterologous Regulatory Genes from Lipodepsipeptide Biosynthetic Gene Clusters on the Production of Teicoplanin and A40926
Abstract
:1. Introduction
2. Results
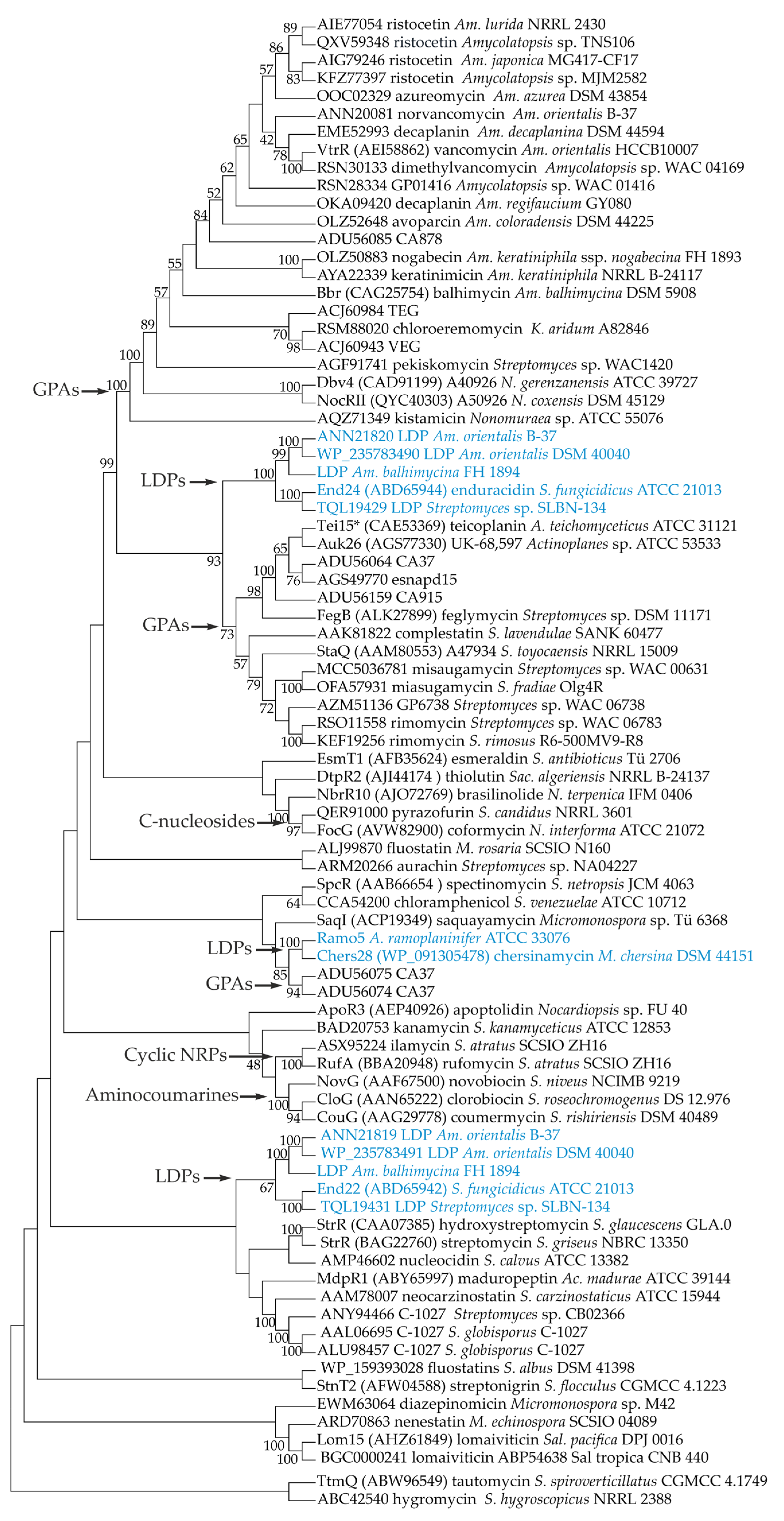
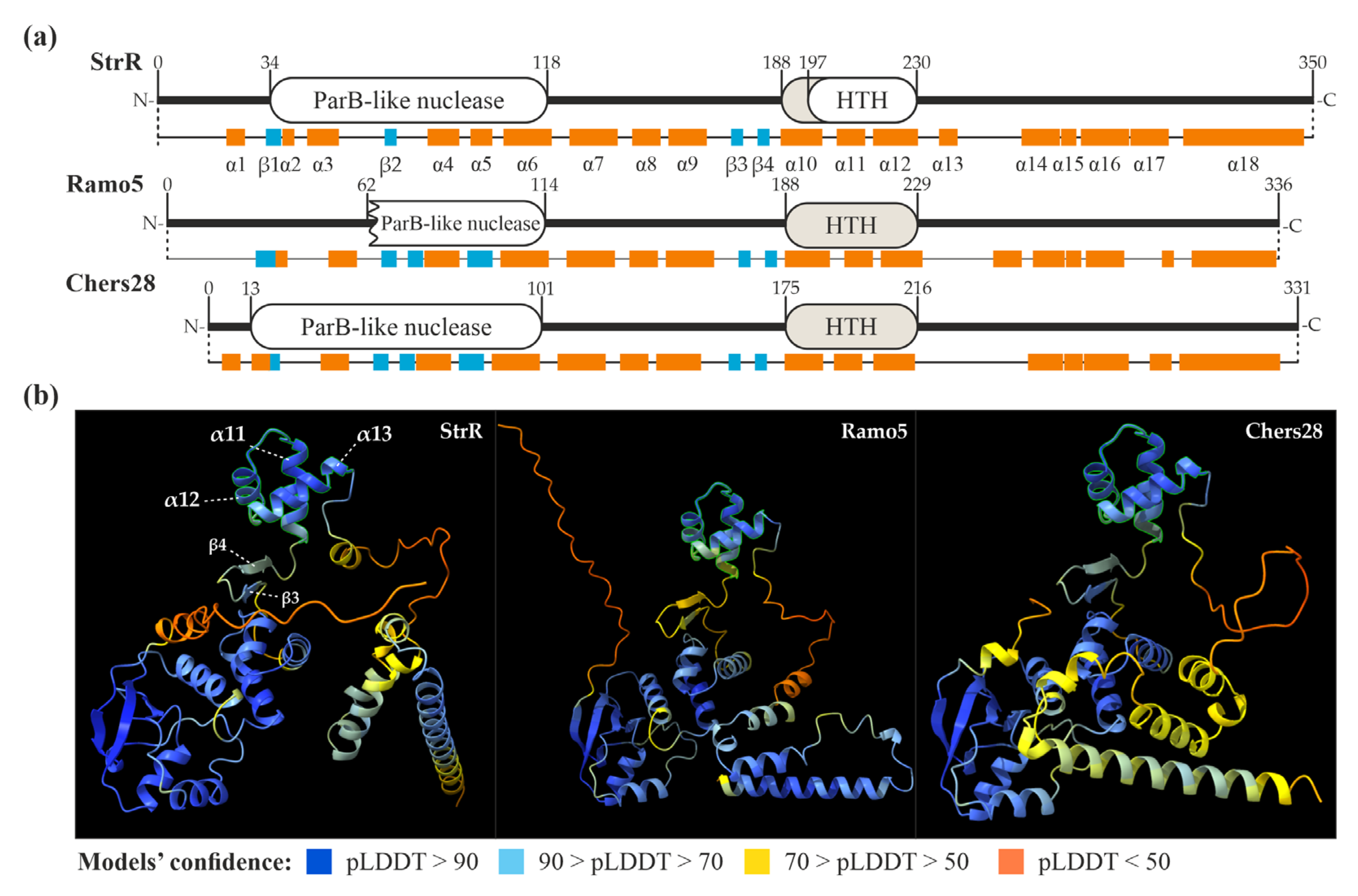
2.1. Phylogenetic Relationships between StrR-like Regulators Coded within Antibiotic BGCs
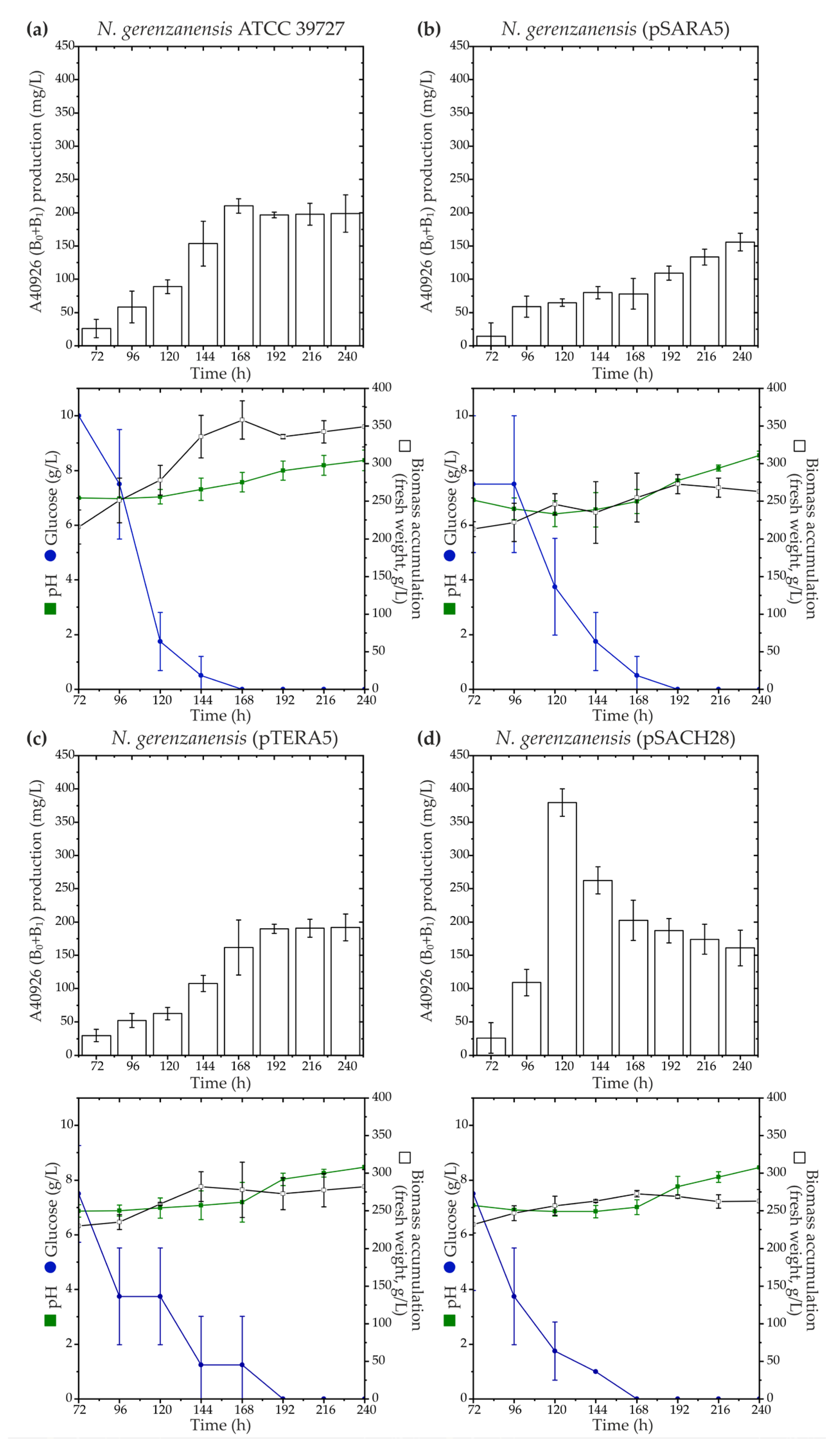
2.2. Expression of Cluster-Situated Regulatory Genes ramo5 and chers28 in N. gerenzanensis ATCC 39727
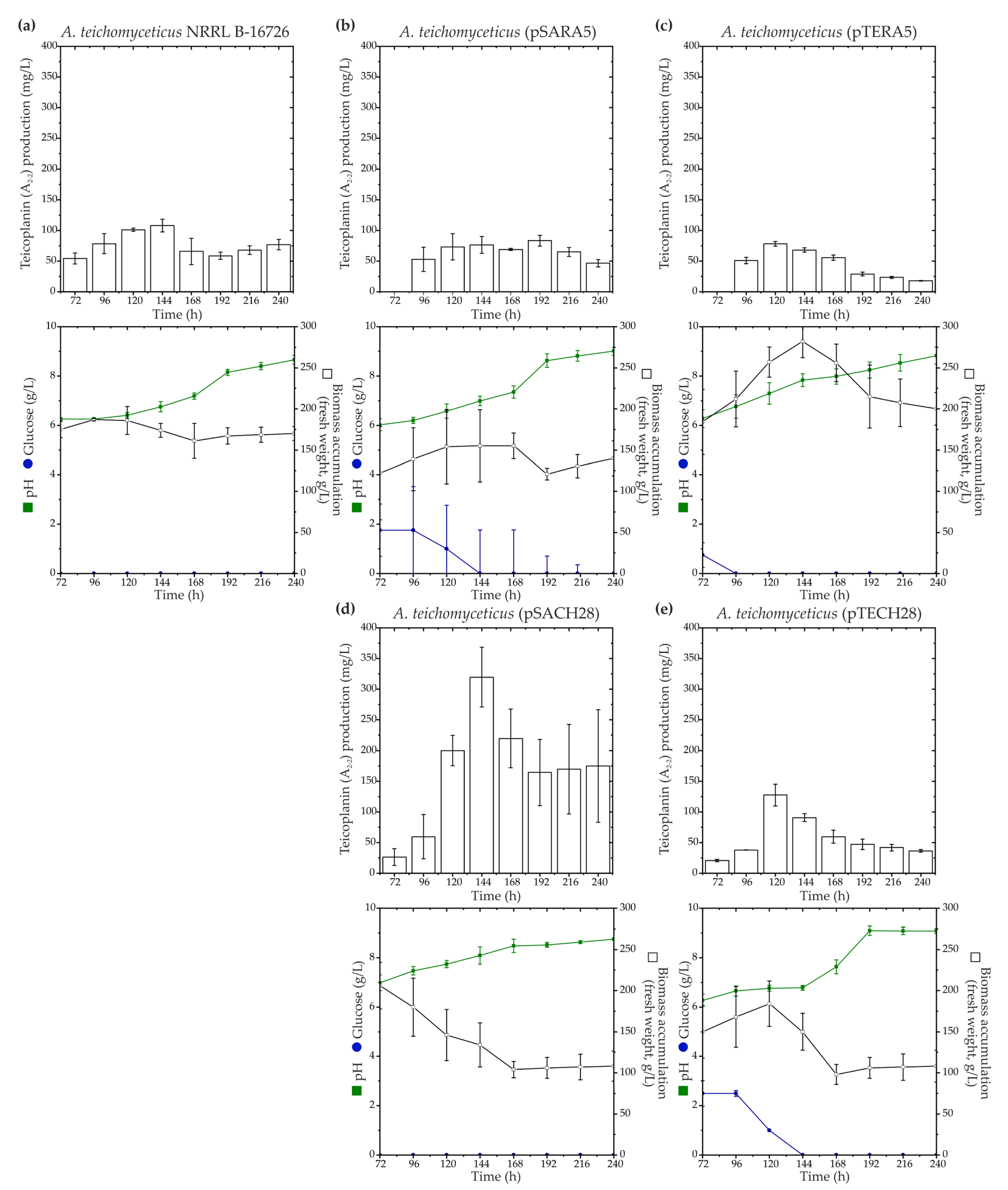
2.3. Expression of ramo5 and chers28 in A. teichomyceticus NRRL B-16726
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Cultivation Conditions
4.2. Extraction of the Genomic DNA
4.3. Generation of the Recombinant Plasmids
4.4. Conjugal Transfer of Plasmids and Strain Verification
4.5. Analysis and Quantification of A40926 and Teicoplanin Production
4.6. Tools for In Silico Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Nazari, B.; Forneris, C.C.; Gibson, M.I.; Moon, K.; Schramma, K.R.; Seyedsayamdost, M.R. Nonomuraea sp. ATCC 55076 harbours the largest actinomycete chromosome to date and the kistamicin biosynthetic gene cluster. MedChemComm 2017, 8, 780–788. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; de Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C.; et al. Minimum Information about a Biosynthetic Gene cluster. Nat. Chem. Biol. 2015, 11, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Hoskisson, P.A.; Seipke, R.F. Cryptic or silent? The known unknowns, unknown knowns, and unknown unknowns of secondary metabolism. mBio 2020, 11, e02642-20. [Google Scholar] [CrossRef] [PubMed]
- Romero-Rodríguez, A.; Robledo-Casados, I.; Sánchez, S. An overview on transcriptional regulators in Streptomyces. Biochim. Biophys. Acta 2015, 1849, 1017–1039. [Google Scholar] [CrossRef] [PubMed]
- Arias, P.; Fernández-Moreno, M.A.; Malpartida, F. Characterization of the pathway-specific positive transcriptional regulator for actinorhodin biosynthesis in Streptomyces coelicolor A3(2) as a DNA-binding protein. J. Bacteriol. 1999, 181, 6958–6968. [Google Scholar] [CrossRef] [PubMed]
- Mingyar, E.; Mühling, L.; Kulik, A.; Winkler, A.; Wibberg, D.; Kalinowski, J.; Blin, K.; Weber, T.; Wohlleben, W.; Stegmann, E. A regulator based “semi-targeted” approach to activate silent biosynthetic gene clusters. Int. J. Mol. Sci. 2021, 22, 7567. [Google Scholar] [CrossRef] [PubMed]
- Krause, J.; Handayani, I.; Blin, K.; Kulik, A.; Mast, Y. Disclosing the potential of the SARP-type regulator PapR2 for the activation of antibiotic gene clusters in streptomycetes. Front. Microbiol. 2020, 11, 225. [Google Scholar] [CrossRef]
- Ye, S.; Molloy, B.; Pérez-Victoria, I.; Montero, I.; Braña, A.F.; Olano, C.; Arca, S.; Martín, J.; Reyes, F.; Salas, J.A.; et al. Uncovering the cryptic gene cluster ahb for 3-amino-4-hydroxybenzoate derived ahbamycins, by searching SARP regulator encoding genes in the Streptomyces argillaceus genome. Int. J. Mol. Sci. 2023, 24, 8197. [Google Scholar] [CrossRef]
- Santos, C.L.; Correia-Neves, M.; Moradas-Ferreira, P.; Mendes, M.V. A walk into the LuxR regulators of Actinobacteria: Phylogenomic distribution and functional diversity. PLoS ONE 2012, 7, e46758. [Google Scholar] [CrossRef]
- Aparicio, J.F.; Caffrey, P.; Gil, J.A.; Zotchev, S.B. Polyene antibiotic biosynthesis gene clusters. Appl. Microbiol. Biotechnol. 2003, 61, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Sekurova, O.N.; Brautaset, T.; Sletta, H.; Borgos, S.E.F.; Jakobsen, Ø.M.; Ellingsen, T.E.; Strøm, A.R.; Valla, S.; Zotchev, S.B. In vivo analysis of the regulatory genes in the nystatin biosynthetic gene cluster of Streptomyces noursei ATCC 11455 reveals their differential control over antibiotic biosynthesis. J. Bacteriol. 2004, 186, 1345–1354. [Google Scholar] [CrossRef] [PubMed]
- Yushchuk, O.; Ostash, I.; Mösker, E.; Vlasiuk, I.; Deneka, M.; Rückert, C.; Busche, T.; Fedorenko, V.; Kalinowski, J.; Süssmuth, R.D.; et al. Eliciting the silent lucensomycin biosynthetic pathway in Streptomyces cyanogenus S136 via manipulation of the global regulatory gene adpA. Sci. Rep. 2021, 11, 3507. [Google Scholar] [CrossRef] [PubMed]
- Beyer, S.; Distler, J.; Piepersberg, W. The str gene cluster for the biosynthesis of 5′-hydroxystreptomycin in Streptomyces glaucescens GLA.0 (ETH 22794): New operons and evidence for pathway-specific regulation by StrR. Mol. Genet. Genom. 1996, 250, 775–784. [Google Scholar] [CrossRef]
- Retzlaff, L.; Distler, J. The regulator of streptomycin gene expression, StrR, of Streptomyces griseus is a DNA binding activator protein with multiple recognition sites. Mol. Microbiol. 1995, 18, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Eustáquio, A.S.; Li, S.-M.; Heide, L. NovG, a DNA-binding protein acting as a positive regulator of novobiocin biosynthesis. Microbiology 2005, 151, 1949–1961. [Google Scholar] [CrossRef]
- van der Heul, H.U.; Bilyk, B.L.; McDowall, K.J.; Seipke, R.F.; van Wezel, G.P. Regulation of antibiotic production in Actinobacteria: New perspectives from the post-genomic era. Nat. Prod. Rep. 2018, 35, 575–604. [Google Scholar] [CrossRef]
- Autret, S.; Nair, R.; Errington, J. Genetic analysis of the chromosome segregation protein Spo0J of Bacillus subtilis: Evidence for separate domains involved in DNA binding and interactions with Soj protein. Mol. Microbiol. 2001, 41, 743–755. [Google Scholar] [CrossRef]
- Liu, K.; Hu, X.-R.; Zhao, L.-X.; Wang, Y.; Deng, Z.; Tao, M. Enhancing ristomycin A production by overexpression of ParB-like StrR family regulators controlling the biosynthesis genes. Appl. Environ. Microbiol. 2021, 87, e0106621Z. [Google Scholar] [CrossRef]
- Nicolaou, K.C.; Boddy, C.N.C.; Bräse, S.; Winssinger, N. Chemistry, biology, and medicine of the glycopeptide antibiotics. Angew. Chem.—Int. Ed. 1999, 38, 2096–2152. [Google Scholar] [CrossRef]
- Butler, M.S.; A Hansford, K.; Blaskovich, M.A.T.; Halai, R.; Cooper, M.A. Glycopeptide antibiotics: Back to the future. J. Antibiot. 2014, 67, 631–644. [Google Scholar] [CrossRef] [PubMed]
- Borghi, A.; Coronelli, C.; Faniuolo, L.; Allievi, G.; Pallanza, R.; Gallo, G.G. Teichomycins, new antibiotics from Actinoplanes teichomyceticus nov. sp. IV. Separation and characterization of the components of teichomycin (teicoplanin). J. Antibiot. 1984, 37, 615–620. [Google Scholar] [CrossRef] [PubMed]
- McCormick, M.H.; McGuire, J.M.; Pittenger, G.E.; Pittenger, R.C.; Stark, W.M. Vancomycin, a new antibiotic. I. Chemical and biologic properties. Antibiot. Annu. 1955, 3, 606–611. [Google Scholar]
- van Wageningen, A.; Kirkpatrick, P.N.; Williams, D.H.; Harris, B.R.; Kershaw, J.K.; Lennard, N.J.; Jones, M.; Jones, S.J.; Solenberg, P.J. Sequencing and analysis of genes involved in the biosynthesis of a vancomycin group antibiotic. Chem. Biol. 1998, 5, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Marcone, G.L.; Binda, E.; Berini, F.; Marinelli, F. Old and new glycopeptide antibiotics: From product to gene and back in the post-genomic era. Biotechnol. Adv. 2018, 36, 534–554. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.H.; Stegmann, E.; Cryle, M.J. Beyond vancomycin: Recent advances in the modification, reengineering, production and discovery of improved glycopeptide antibiotics to tackle multidrug-resistant bacteria. Curr. Opin. Biotechnol. 2022, 77, 102767. [Google Scholar] [CrossRef] [PubMed]
- Yushchuk, O.; Ostash, B. Glycopeptide antibiotics: Genetics, chemistry, and new screening approaches, In Natural Products from Actinomycetes, Diversity, Ecology and Drug Discovery; Springer: Singapore, 2022; pp. 411–444. [Google Scholar] [CrossRef]
- Shawky, R.M.; Puk, O.; Wietzorrek, A.; Pelzer, S.; Takano, E.; Wohlleben, W.; Stegmann, E. The border sequence of the balhimycin biosynthesis gene cluster from Amycolatopsis balhimycina contains bbr, encoding a StrR-like pathway-specific regulator. J. Mol. Microbiol. Biotechnol. 2007, 13, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Spohn, M.; Kirchner, N.; Kulik, A.; Jochim, A.; Wolf, F.; Muenzer, P.; Borst, O.; Gross, H.; Wohlleben, W.; Stegmann, E. Overproduction of ristomycin a by activation of a silent gene cluster in Amycolatopsis japonicum MG417-CF17. Antimicrob. Agents Chemother. 2014, 58, 6185–6196. [Google Scholar] [CrossRef]
- Alduina, R.; Piccolo, L.L.; D’Alia, D.; Ferraro, C.; Gunnarsson, N.; Donadio, S.; Puglia, A.M. Phosphate-controlled regulator for the biosynthesis of the dalbavancin precursor A40926. J. Bacteriol. 2007, 189, 8120–8129. [Google Scholar] [CrossRef]
- Yushchuk, O.; Horbal, L.; Ostash, B.; Marinelli, F.; Wohlleben, W.; Stegmann, E.; Fedorenko, V. Regulation of teicoplanin biosynthesis: Refining the roles of tei cluster-situated regulatory genes. Appl. Microbiol. Biotechnol. 2019, 103, 4089–4102. [Google Scholar] [CrossRef]
- Horbal, L.; Kobylyanskyy, A.; Truman, A.W.; Zaburranyi, N.; Ostash, B.; Luzhetskyy, A.; Marinelli, F.; Fedorenko, V. The pathway-specific regulatory genes, tei15* and tei16*, are the master switches of teicoplanin production in Actinoplanes teichomyceticus. Appl. Microbiol. Biotechnol. 2014, 98, 9295–9309. [Google Scholar] [CrossRef] [PubMed]
- Horbal, L.; Kobylyanskyy, A.; Yushchuk, O.; Zaburannyi, N.; Luzhetskyy, A.; Ostash, B.; Marinelli, F.; Fedorenko, V. Evaluation of heterologous promoters for genetic analysis of Actinoplanes teichomyceticus—Producer of teicoplanin, drug of last defense. J. Biotechnol. 2013, 168, 367–372. [Google Scholar] [CrossRef] [PubMed]
- Alduina, R.; Sosio, M.; Donadio, S. Complex regulatory networks governing production of the glycopeptide A40926. Antibiotics 2018, 7, 30. [Google Scholar] [CrossRef] [PubMed]
- Grasso, L.L.; Maffioli, S.; Sosio, M.; Bibb, M.; Puglia, A.M.; Alduina, R. Two master switch regulators trigger A40926 biosynthesis in Nonomuraea sp. strain ATCC 39727. J. Bacteriol. 2015, 197, 2536–2544. [Google Scholar] [CrossRef] [PubMed]
- Yushchuk, O.; Andreo-Vidal, A.; Marcone, G.L.; Bibb, M.; Marinelli, F.; Binda, E. New molecular tools for regulation and improvement of A40926 glycopeptide antibiotic production in Nonomuraea gerenzanensis ATCC 39727. Front. Microbiol. 2020, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Waglechner, N.; McArthur, A.G.; Wright, G.D. Phylogenetic reconciliation reveals the natural history of glycopeptide antibiotic biosynthesis and resistance. Nat. Microbiol. 2019, 4, 1862–1871. [Google Scholar] [CrossRef] [PubMed]
- Kettenring, J.K.; Ciabatti, R.; Winters, G.; Tamborini, G.; Cavalleri, B. Ramoplanin (A-16686), a new glycolipodepsipeptide antibiotic. IV. Complete sequence determination by homonuclear 2D NMR spectroscopy. J. Antibiot. 1989, 42, 268–275. [Google Scholar] [CrossRef]
- Farnet, C.M.; Zazopoulos, E.; Staffa, A. Gene Cluster for Ramoplanin Biosynthesis (EP1326983B1). European Patent Office. 2001. Available online: https://patents.google.com/patent/EP1326983B1/en (accessed on 15 December 2023).
- Yourassowsky, E.; Monsieur, R. In vitro and in vivo activity of enduracidin on Staphylococcus aureus. Chemotherapy 1972, 17, 182–187. [Google Scholar] [CrossRef]
- Yin, X.; Zabriskie, T.M. The enduracidin biosynthetic gene cluster from Streptomyces fungicidicus. Microbiology 2006, 152, 2969–2983. [Google Scholar] [CrossRef]
- Morgan, K.T.; Zheng, J.; McCafferty, D.G. Discovery of six ramoplanin family gene clusters and the lipoglycodepsipeptide chersinamycin. ChemBioChem 2021, 22, 176–185. [Google Scholar] [CrossRef]
- Bassères, E.; Endres, B.T.; Dotson, K.M.; Alam, M.J.; Garey, K.W. Novel antibiotics in development to treat Clostridium difficile infection. Curr. Opin. Gastroenterol. 2017, 33, 1–7. [Google Scholar] [CrossRef]
- Montecalvo, M.A. Ramoplanin: A novel antimicrobial agent with the potential to prevent vancomycin-resistant enterococcal infection in high-risk patients. J. Antimicrob. Chemother. 2003, 51, 31–35. [Google Scholar] [CrossRef]
- Petrosillo, N.; Granata, G.; Cataldo, M.A. Novel antimicrobials for the treatment of Clostridium difficile infection. Front. Med. 2018, 5, 96. [Google Scholar] [CrossRef] [PubMed]
- Yushchuk, O.; Kseniia, Z.; Fedorenko, V. Insights into the phylogeny of transporters coded within biosynthetic gene clusters for glycopeptides and related antibiotics. Visnyk Lviv Univ. Biol. Ser. 2022, 86, 33–46. [Google Scholar] [CrossRef]
- McCafferty, D.G.; Cudic, P.; Frankel, B.A.; Barkallah, S.; Kruger, R.G.; Li, W. Chemistry and biology of the ramoplanin family of peptide antibiotics. Biopolym.-Pept. Sci. Sect. 2002, 66, 261–284. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, X.; Lv, F.; Li, P. Characterization of three regulatory genes involved in enduracidin biosynthesis and improvement of enduracidin production in Streptomyces fungicidicus. J. Appl. Microbiol. 2019, 127, 1698–1705. [Google Scholar] [CrossRef]
- Terlouw, B.R.; Blin, K.; Navarro-Muñoz, J.C.; Avalon, N.E.; Chevrette, M.G.; Egbert, S.; Lee, S.; Meijer, D.; Recchia, M.J.J.; Reitz, Z.L.; et al. MIBiG 3.0: A community-driven effort to annotate experimentally validated biosynthetic gene clusters. Nucleic Acids Res. 2023, 51, D603–D610. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Thamm, S.; Distler, J. Properties of C-terminal truncated derivatives of the activator, StrR, of the streptomycin biosynthesis in Streptomyces griseus. FEMS Microbiol. Lett. 1997, 149, 265–272. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, W327–W331. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018, 27, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Aravind, L.; Anantharaman, V.; Balaji, S.; Babu, M.; Iyer, L. The many faces of the helix-turn-helix domain: Transcription regulation and beyond. FEMS Microbiol. Rev. 2005, 29, 231–262. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, S.; Siegl, T.; Luzhetska, M.; Petzke, L.; Jilg, C.; Welle, E.; Erb, A.; Leadlay, P.F.; Bechthold, A.; Luzhetskyy, A. Site-specific recombination strategies for engineering actinomycete. Appl. Environ. Microbiol. 2012, 78, 1804–1812. [Google Scholar] [CrossRef] [PubMed]
- Marcone, G.L.; Beltrametti, F.; Binda, E.; Carrano, L.; Foulston, L.; Hesketh, A.; Bibb, M.; Marinelli, F. Novel mechanism of glycopeptide resistance in the A40926 producer Nonomuraea sp. ATCC 39727. Antimicrob. Agents Chemother. 2010, 54, 2465–2472. [Google Scholar] [CrossRef] [PubMed]
- Taurino, C.; Frattini, L.; Marcone, G.L.; Gastaldo, L.; Marinelli, F. Actinoplanes teichomyceticus ATCC 31121 as a cell factory for producing teicoplanin. Microb. Cell Factories 2011, 10, 82. [Google Scholar] [CrossRef] [PubMed]
- Melnyk, S.; Stepanyshyn, A.; Yushchuk, O.; Mandler, M.; Ostash, I.; Koshla, O.; Fedorenko, V.; Kahne, D.; Ostash, B. Genetic approaches to improve clorobiocin production in Streptomyces roseochromogenes NRRL 3504. Appl. Microbiol. Biotechnol. 2022, 106, 1543–1556. [Google Scholar] [CrossRef]
- Shirling, E.B.; Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Evol. Microbiol. 1966, 16, 313–340. [Google Scholar] [CrossRef]
- Paranthaman, S.; Dharmalingam, K. Intergeneric conjugation in Streptomyces peucetius and Streptomyces sp. strain C5: Chromosomal integration and expression of recombinant plasmids carrying the chiC gene. Appl. Environ. Microbiol. 2003, 69, 84–91. [Google Scholar] [CrossRef]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics; John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Ha, H.-S.; Hwang, Y.-I.; Choi, S.-U. Application of conjugation using ϕC31 att/int system for Actinoplanes teichomyceticus, a producer of teicoplanin. Biotechnol. Lett. 2008, 30, 1233–1238. [Google Scholar] [CrossRef] [PubMed]
- Andreo-Vidal, A.; Yushchuk, O.; Marinelli, F.; Binda, E. Cross-talking of pathway-specific regulators in glycopeptide antibiotics (teicoplanin and A40926) production. Antibiotics 2023, 12, 641. [Google Scholar] [CrossRef] [PubMed]
- Yushchuk, O.; Vior, N.M.; Andreo-Vidal, A.; Berini, F.; Rückert, C.; Busche, T.; Binda, E.; Kalinowski, J.; Truman, A.W.; Marinelli, F. Genomic-led discovery of a novel glycopeptide antibiotic by Nonomuraea coxensis DSM 45129. ACS Chem. Biol. 2021, 16, 915–928. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Higgins, D.G. Clustal Omega. Curr. Protoc. Bioinform. 2014, 48, 3–13. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. Mega X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Du, Y.; Derewacz, D.K.; Deguire, S.M.; Teske, J.; Ravel, J.; Sulikowski, G.A.; Bachmann, B.O. Biosynthesis of the apoptolidins in Nocardiopsis sp. FU 40. Tetrahedron 2011, 67, 6568–6575. [Google Scholar] [CrossRef]
- Chiu, H.-T.; Weng, C.-P.; Lin, Y.-C.; Chen, K.-H. Target-specific identification and characterization of the putative gene cluster for brasilinolide biosynthesis revealing the mechanistic insights and combinatorial synthetic utility of 2-deoxy-l-fucose biosynthetic enzymes. Org. Biomol. Chem. 2016, 14, 1988–2006. [Google Scholar] [CrossRef]
- Li, W.; Ju, J.; Rajski, S.R.; Osada, H.; Shen, B. Characterization of the tautomycin biosynthetic gene cluster from Streptomyces spiroverticillatus unveiling new insights into dialkylmaleic anhydride and polyketide biosynthesis. J. Biol. Chem. 2008, 283, 28607–28617. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Nonaka, K.; Nie, L.; Zhang, J.; Christenson, S.D.; Bae, J.; Van Lanen, S.G.; Zazopoulos, E.; Farnet, C.M.; Yang, C.F.; et al. The neocarzinostatin biosynthetic gene cluster from Streptomyces carzinostaticus ATCC 15944 involving two iterative type I polyketide synthases. Chem. Biol. 2005, 12, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Christenson, S.D.; Standage, S.; Shen, B. Biosynthesis of the enediyne antitumor antibiotic C-1027. Science 2002, 297, 1170–1173. [Google Scholar] [CrossRef]
- Van Lanen, S.G.; Oh, T.-J.; Liu, W.; Wendt-Pienkowski, E.; Shen, B. Characterization of the maduropeptin biosynthetic gene cluster from Actinomadura madurae ATCC 39144 supporting a unifying paradigm for enediyne biosynthesis. J. Am. Chem. Soc. 2007, 129, 13082–13094. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lei, X.; Zhang, C.; Jiang, Z.; Shi, Y.; Wang, S.; Wang, L.; Hong, B. Complete genome sequence of Streptomyces globisporus C-1027, the producer of an enediyne antibiotic lidamycin. J. Biotechnol. 2016, 222, 9–10. [Google Scholar] [CrossRef]
- Yan, X.; Ge, H.; Huang, T.; Hindra; Yang, D.; Teng, Q.; Crnovčić, I.; Li, X.; Rudolf, J.D.; Lohman, J.R.; et al. Strain prioritization and genome mining for enediyne natural products. mBio 2016, 7, e02104-16. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Huang, C.; Zhang, W.; Zhu, Y.; Zhang, C. Heterologous expression of fluostatin gene cluster leads to a bioactive heterodimer. Org. Lett. 2015, 17, 5324–5327. [Google Scholar] [CrossRef]
- Erb, A.; Luzhetskyy, A.; Hardter, U.; Bechthold, A. Cloning and sequencing of the biosynthetic gene cluster for saquayamycin Z and galtamycin B and the elucidation of the assembly of their saccharide chains. ChemBioChem 2009, 10, 1392–1401. [Google Scholar] [CrossRef]
- Jin, J.; Yang, X.; Liu, T.; Xiao, H.; Wang, G.; Zhou, M.; Liu, F.; Zhang, Y.; Liu, D.; Chen, M.; et al. Fluostatins M–Q featuring a 6-5-6-6 ring skeleton and high oxidized A-rings from marine Streptomyces sp. PKU-MA00045. Mar. Drugs 2018, 16, 87. [Google Scholar] [CrossRef]
- Jiang, X.; Zhang, Q.; Zhu, Y.; Nie, F.; Wu, Z.; Yang, C.; Zhang, L.; Tian, X.; Zhang, C. Isolation, structure elucidation and biosynthesis of benzo[b]fluorene nenestatin A from deep-sea derived Micromonospora echinospora SCSIO 04089. Tetrahedron 2017, 73, 3585–3590. [Google Scholar] [CrossRef]
- Palaniappan, N.; Ayers, S.; Gupta, S.; Habib, E.-S.; Reynolds, K.A. Production of hygromycin A analogs in Streptomyces hygroscopicus NRRL 2388 through identification and manipulation of the biosynthetic gene cluster. Chem. Biol. 2006, 13, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Yanai, K.; Murakami, T. The kanamycin biosynthetic gene cluster from Streptomyces kanamyceticus. J. Antibiot. 2004, 57, 351–354. [Google Scholar] [CrossRef] [PubMed]
- Lyutzkanova, D.; Distler, J.; Altenbuchner, J. A spectinomyciin resistance determinant from the spectinomycin producer Streptomyces flavopersicus. Microbiology 1997, 143, 2135–2143. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, Y.; Ishikawa, J.; Hara, H.; Suzuki, H.; Ikenoya, M.; Ikeda, H.; Yamashita, A.; Hattori, M.; Horinouchi, S. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 2008, 190, 4050–4060. [Google Scholar] [CrossRef] [PubMed]
- Pojer, F.; Li, S.-M.; Heide, L. Molecular cloning and sequence analysis of the clorobiocin biosynthetic gene cluster: New insights into the biosynthesis of aminocoumarin antibiotics. Microbiology 2002, 148, 3901–3911. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-X.; Li, S.-M.; Heide, L. Identification of the coumermycin A1 biosynthetic gene cluster of Streptomyces rishiriensis DSM 40489. Antimicrob. Agents Chemother. 2000, 44, 3040–3048. [Google Scholar] [CrossRef]
- Steffensky, M.; Mühlenweg, A.; Wang, Z.-X.; Li, S.-M.; Heide, L. Identification of the novobiocin biosynthetic gene cluster of Streptomyces spheroides NCIB 11891. Antimicrob. Agents Chemother. 2000, 44, 1214–1222. [Google Scholar] [CrossRef]
- Xu, F.; Kong, D.; He, X.; Zhang, Z.; Han, M.; Xie, X.; Wang, P.; Cheng, H.; Tao, M.; Zhang, L.; et al. Characterization of streptonigrin biosynthesis reveals a cryptic carboxyl methylation and an unusual oxidative cleavage of a N-C bond. J. Am. Chem. Soc. 2013, 135, 1739–1748. [Google Scholar] [CrossRef]
- Rui, Z.; Ye, M.; Wang, S.; Fujikawa, K.; Akerele, B.; Aung, M.; Floss, H.G.; Zhang, W.; Yu, T.-W. Insights into a divergent phenazine biosynthetic pathway governed by a plasmid-born esmeraldin gene cluster. Chem. Biol. 2012, 19, 1116–1125. [Google Scholar] [CrossRef]
- McAlpine, J.B.; Banskota, A.H.; Charan, R.D.; Schlingmann, G.; Zazopoulos, E.; Piraee, M.; Janso, J.; Bernan, V.S.; Aouidate, M.; Farnet, C.M.; et al. Biosynthesis of diazepinomicin/ECO-4601, a Micromonospora secondary metabolite with a novel ring system. J. Nat. Prod. 2008, 71, 1585–1590. [Google Scholar] [CrossRef]
- Zhang, M.; Yang, C.L.; Xiao, Y.S.; Zhang, B.; Deng, X.Z.; Yang, L.; Shi, J.; Wang, Y.S.; Li, W.; Jiao, R.H.; et al. Aurachin SS, a new antibiotic from Streptomyces sp. NA04227. J. Antibiot. 2017, 70, 853–855. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.M.; Hackl, S.; Thaker, M.N.; Kalan, L.; Weber, C.; Urgast, D.S.; Krupp, E.M.; Brewer, A.; Vanner, S.; Szawiola, A.; et al. Biosynthesis of the fluorinated natural product nucleocidin in Streptomyces calvus is dependent on the bldA-specified Leu-tRNAUUA molecule. ChemBioChem 2015, 16, 2498–2506. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Yao, S.; Rothchild, K.W.; Liu, T.; Liu, Y.; Lian, J.; He, H.; Ryan, K.S.; Du, Y. The biosynthetic gene cluster of pyrazomycin—A C-nucleoside antibiotic with a rare pyrazole moiety. ChemBioChem 2020, 21, 644–649. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, P.; Xu, G.; Zhou, W.; Gao, Y.; Gong, R.; Cai, Y.-S.; Cong, H.; Deng, Z.; Price, N.P.J.; et al. Comparative investigation into formycin A and pyrazofurin A biosynthesis reveals branch pathways for the construction of C-nucleoside scaffolds. Appl. Environ. Microbiol. 2020, 86, e01971-19. [Google Scholar] [CrossRef]
- Huang, S.; Tong, M.H.; Qin, Z.; Deng, Z.; Deng, H.; Yu, Y. Identification and characterization of the biosynthetic gene cluster of thiolutin, a tumor angiogenesis inhibitor, in Saccharothrix algeriensis NRRL B-24137. Anti-Cancer Agents Med. Chem. 2015, 15, 277–284. [Google Scholar] [CrossRef]
- He, J.; Magarvey, N.; Piraee, M.; Vining, L.C. The gene cluster for chloramphenicol biosynthesis in Streptomyces venezuelae ISP5230 includes novel shikimate pathway homologues and a monomodular non-ribosomal peptide synthetase gene. Microbiology 2001, 147, 2817–2829. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Huang, H.; Xie, Y.; Liu, Z.; Zhao, J.; Zhang, C.; Jia, Y.; Zhang, Y.; Zhang, H.; Zhang, T.; et al. Biosynthesis of ilamycins featuring unusual building blocks and engineered production of enhanced anti-tuberculosis agents. Nat. Commun. 2017, 8, 391. [Google Scholar] [CrossRef]
- Tomita, H.; Katsuyama, Y.; Minami, H.; Ohnishi, Y. Identification and characterization of a bacterial cytochrome P450 monooxygenase catalyzing the 3-nitration of tyrosine in rufomycin biosynthesis. J. Biol. Chem. 2017, 292, 15859–15869. [Google Scholar] [CrossRef]
- Gonsior, M.; Mühlenweg, A.; Tietzmann, M.; Rausch, S.; Poch, A.; Süssmuth, R.D. Biosynthesis of the peptide antibiotic feglymycin by a linear nonribosomal peptide synthetase mechanism. ChemBioChem 2015, 16, 2610–2614. [Google Scholar] [CrossRef]
- Pelzer, S.; Süßmuth, R.; Heckmann, D.; Recktenwald, J.; Huber, P.; Jung, G.; Wohlleben, W. Identification and analysis of the balhimycin biosynthetic gene cluster and its use for manipulating glycopeptide biosynthesis in Amycolatopsis mediterranei DSM5908. Antimicrob. Agents Chemother. 1999, 43, 1565–1573. [Google Scholar] [CrossRef]
- Xu, L.; Huang, H.; Wei, W.; Zhong, Y.; Tang, B.; Yuan, H.; Zhu, L.; Huang, W.; Ge, M.; Yang, S.; et al. Complete genome sequence and comparative genomic analyses of the vancomycin-producing Amycolatopsis orientalis. BMC Genom. 2014, 15, 363. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.Y.A.; Robinson, S.; Lacey, E.; Brown, R.; Kim, W.; Goodfellow, M. Amycolatopsis regifaucium sp. nov., a novel actinomycete that produces kigamicins. Int. J. Syst. Evol. Microbiol. 2007, 57, 2562–2567. [Google Scholar] [CrossRef] [PubMed]
- Kaur, N.; Kumar, S.; Bala, M.; Raghava, G.P.S.; Mayilraj, S. Draft genome sequence of Amycolatopsis decaplanina strain DSM 44594T. Genome Announc. 2013, 1, e00138-13. [Google Scholar] [CrossRef] [PubMed]
- Wink, J.M.; Kroppenstedt, R.M.; Ganguli, B.N.; Nadkarni, S.R.; Schumann, P.; Seibert, G.; Stackebrandt, E. Three new antibiotic producing species of the genus Amycolatopsis, Amycolatopsis balhimycina sp. nov., A. tolypomycina sp. nov., A. vancoresmycina sp. nov., and description of Amycolatopsis keratiniphila subsp. keratiniphila subsp. nov. and A. keratiniphiphila subsp. nogabecina subsp. nov. Syst. Appl. Microbiol. 2003, 26, 38–46. [Google Scholar] [CrossRef]
- Labeda, D.P. Amycolatopsis coloradensis sp. nov., the avoparcin (LL-AV290)-producing strain. Int. J. Syst. Bacteriol. 1995, 45, 124–127. [Google Scholar] [CrossRef]
- Xu, F.; Wu, Y.; Zhang, C.; Davis, K.M.; Moon, K.; Bushin, L.B.; Seyedsayamdost, M.R. A genetics-free method for high-throughput discovery of cryptic microbial metabolites. Nat. Chem. Biol. 2019, 15, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Yim, G.; Kalan, L.; Koteva, K.; Thaker, M.N.; Waglechner, N.; Tang, I.; Wright, G.D. Harnessing the synthetic capabilities of glycopeptide antibiotic tailoring enzymes: Characterization of the UK-68, 597 biosynthetic cluster. ChemBioChem 2014, 15, 2613–2623. [Google Scholar] [CrossRef] [PubMed]
- Pootoolal, J.; Thomas, M.G.; Marshall, C.G.; Neu, J.M.; Hubbard, B.K.; Walsh, C.T.; Wright, G.D. Assembling the glycopeptide antibiotic scaffold: The biosynthesis of A47934 from Streptomyces toyocaensis NRRL15009. Proc. Natl. Acad. Sci. USA 2002, 99, 8962–8967. [Google Scholar] [CrossRef]
- Truman, A.W.; Kwun, M.J.; Cheng, J.; Yang, S.H.; Suh, J.-W.; Hong, H.-J. Antibiotic resistance mechanisms inform discovery: Identification and characterization of a novel Amycolatopsis strain producing ristocetin. Antimicrob. Agents Chemother. 2014, 58, 5687–5695. [Google Scholar] [CrossRef]
- Sosio, M.; Kloosterman, H.; Bianchi, A.; de Vreugd, P.; Dijkhuizen, L.; Donadio, S. Organization of the teicoplanin gene cluster in Actinoplanes teichomyceticus. Microbiology 2004, 150, 95–102. [Google Scholar] [CrossRef]
- Sosio, M.; Stinchi, S.; Beltrametti, F.; Lazzarini, A.; Donadio, S. The gene cluster for the biosynthesis of the glycopeptide antibiotic A40926 by Nonomuraea species. Chem. Biol. 2003, 10, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Wang, W.; Waglechner, N.; Culp, E.J.; Guitor, A.K.; Wright, G.D. Phylogeny-informed synthetic biology reveals unprecedented structural novelty in type V glycopeptide antibiotics. ACS Cent. Sci. 2022, 8, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Chiu, H.-T.; Hubbard, B.K.; Shah, A.N.; Eide, J.; Fredenburg, R.A.; Walsh, C.T.; Khosla, C. Molecular cloning and sequence analysis of the complestatin biosynthetic gene cluster. Proc. Natl. Acad. Sci. USA 2001, 98, 8548–8553. [Google Scholar] [CrossRef] [PubMed]
- Banik, J.J.; Craig, J.W.; Calle, P.Y.; Brady, S.F. Tailoring enzyme-rich environmental DNA clones: A source of enzymes for generating libraries of unnatural natural products. J. Am. Chem. Soc. 2010, 132, 15661–15670. [Google Scholar] [CrossRef] [PubMed]
- Owen, J.G.; Reddy, B.V.B.; Ternei, M.A.; Charlop-Powers, Z.; Calle, P.Y.; Kim, J.H.; Brady, S.F. Mapping gene clusters within arrayed metagenomic libraries to expand the structural diversity of biomedically relevant natural products. Proc. Natl. Acad. Sci. USA 2013, 110, 11797–11802. [Google Scholar] [CrossRef] [PubMed]
- Banik, J.J.; Brady, S.F. Cloning and characterization of new glycopeptide gene clusters found in an environmental DNA megalibrary. Proc. Natl. Acad. Sci. USA 2008, 105, 17273–17277. [Google Scholar] [CrossRef]
- Thaker, M.N.; Wang, W.; Spanogiannopoulos, P.; Waglechner, N.; King, A.M.; Medina, R.; Wright, G.D. Identifying producers of antibacterial compounds by screening for antibiotic resistance. Nat. Biotechnol. 2013, 31, 922–927. [Google Scholar] [CrossRef]
- Lei, X.; Yuan, F.; Shi, Y.; Li, X.; Wang, L.; Hong, B. Draft genome sequence of norvancomycin-producing strain Amycolatopsis orientalis CPCC200066. Genome Announc. 2015, 3, e00296-15. [Google Scholar] [CrossRef]
- Khatri, I.; Subramanian, S.; Mayilraj, S. Genome sequencing and annotation of Amycolatopsis azurea DSM 43854T. Genom. Data 2014, 2, 44–45. [Google Scholar] [CrossRef]
| Plasmid | Description | Source or Reference |
|---|---|---|
| pSET152A | φC31-based integrative plasmid, pSET152 derivative carrying aac(3)IVp from pIJ773, AmR | [33] |
| pTES | φC31-based integrative plasmid, pSET152 derivative carrying ermEp flanked by tfd terminator sequences, AmR | [57] |
| pSARA5 | pSET152A derivative carrying ramo5 from ramoplanin BGC under the control of aac(3)IVp, AmR | This work |
| pSACH28 | pSET152A derivative carrying chers28 from chersinamycin BGC under the control of aac(3)IVp, AmR | This work |
| pTERA5 | pTES derivative carrying ramo5 under the control of ermEp, AmR | This work |
| pTEHC28 | pTES derivative carrying chers28 under the control of ermEp, AmR | This work |
| Bacterial Strain | ||
| A. ramoplaninifer | Wild type, ramoplanin producer | ATCC 33076 |
| M. chersina | Wild type, dynemicin and chersinamycin producer | NRRL B-24756 |
| A. teichomyceticus | Wild type, teicoplanin producer | NRRL B-16726 |
| A. teichomyceticus (pSET152A) | Wild type derivative carrying pSET152A | [32] |
| A. teichomyceticus (pSARA5) | Wild type derivative carrying pSARA5 | This work |
| A. teichomyceticus (pSACH28) | Wild type derivative carrying pSACH28 | This work |
| A. teichomyceticus (pTES) | Wild type derivative carrying pTES | This work |
| A. teichomyceticus (pTERA5) | Wild type derivative carrying pTERA5 | This work |
| A. teichomyceticus (pTECH28) | Wild type derivative carrying pTECH28 | This work |
| N. gerenzanensis ATCC 39727 | Wild type, A40926 producer | ATCC 39727 |
| N. gerenzanensis (pSET152A) | Wild type derivative carrying pSET152A | [36] |
| N. gerenzanensis (pSARA5) | Wild type derivative carrying pSARA5 | This work |
| N. gerenzanensis (pSACH28) | Wild type derivative carrying pSACH28 | This work |
| N. gerenzanensis (pTES) | Wild type derivative carrying pTES | This work |
| N. gerenzanensis (pTERA5) | Wild type derivative carrying pTERA5 | This work |
| N. gerenzanensis (pTECH28) | Wild type derivative carrying pTECH28 | This work |
| E. coli DH5α | General cloning host | MBI Fermentas, USA |
| E. coli ET12567 (pUZ8002) | (dam-13::Tn9 dcm-6), pUZ8002 (ΔoriT), used for conjugative transfer of DNA into actinomycetes | [62] |
| Name | Nucleotide Sequence (5′-3′) | Purpose |
|---|---|---|
| ramo5_F ramo5_R | TTTGATATCGGAGGGTTGGTATGGAGTCATTGCACATCG TTTGATATCGCCGCATTCGCTGTTCA | Amplification of ramo5 |
| Mche_StrR_F Mche_StrR_R | TTTGATATCGGAGGGATCGAATGAAGGCGGAGC TTTGAATTCTGTCCGGCTCAGGCGCTGC | Amplification of orf28 |
| pAm_seq_F pAm_seq_R | GATGTCATCAGCGGTGGAG TGAGCGGATAACAATTTCA | Verification of genes cloned into pSET152A |
| pTES_ver_F pTES_ver_R | CGCGTGTTCGTCGGGCTCTT GACCGAGCGCAGCGAGTCAG | Verification of genes cloned into pTES |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhukrovska, K.; Binda, E.; Fedorenko, V.; Marinelli, F.; Yushchuk, O. The Impact of Heterologous Regulatory Genes from Lipodepsipeptide Biosynthetic Gene Clusters on the Production of Teicoplanin and A40926. Antibiotics 2024, 13, 115. https://doi.org/10.3390/antibiotics13020115
Zhukrovska K, Binda E, Fedorenko V, Marinelli F, Yushchuk O. The Impact of Heterologous Regulatory Genes from Lipodepsipeptide Biosynthetic Gene Clusters on the Production of Teicoplanin and A40926. Antibiotics. 2024; 13(2):115. https://doi.org/10.3390/antibiotics13020115
Chicago/Turabian StyleZhukrovska, Kseniia, Elisa Binda, Victor Fedorenko, Flavia Marinelli, and Oleksandr Yushchuk. 2024. "The Impact of Heterologous Regulatory Genes from Lipodepsipeptide Biosynthetic Gene Clusters on the Production of Teicoplanin and A40926" Antibiotics 13, no. 2: 115. https://doi.org/10.3390/antibiotics13020115
APA StyleZhukrovska, K., Binda, E., Fedorenko, V., Marinelli, F., & Yushchuk, O. (2024). The Impact of Heterologous Regulatory Genes from Lipodepsipeptide Biosynthetic Gene Clusters on the Production of Teicoplanin and A40926. Antibiotics, 13(2), 115. https://doi.org/10.3390/antibiotics13020115











