Repurposing Mitomycin C in Combination with Pentamidine or Gentamicin to Treat Infections with Multi-Drug-Resistant (MDR) Pseudomonas aeruginosa
Abstract
1. Introduction
2. Results
2.1. MDR Strains of P. aeruginosa Are Susceptible to the Anticancer Drug, Mitomycin C
2.2. Treatment of G. mellonella Larvae Infected with Different P. aeruginosa Strains with Mitomycin C Results in Significant Therapeutic Benefit
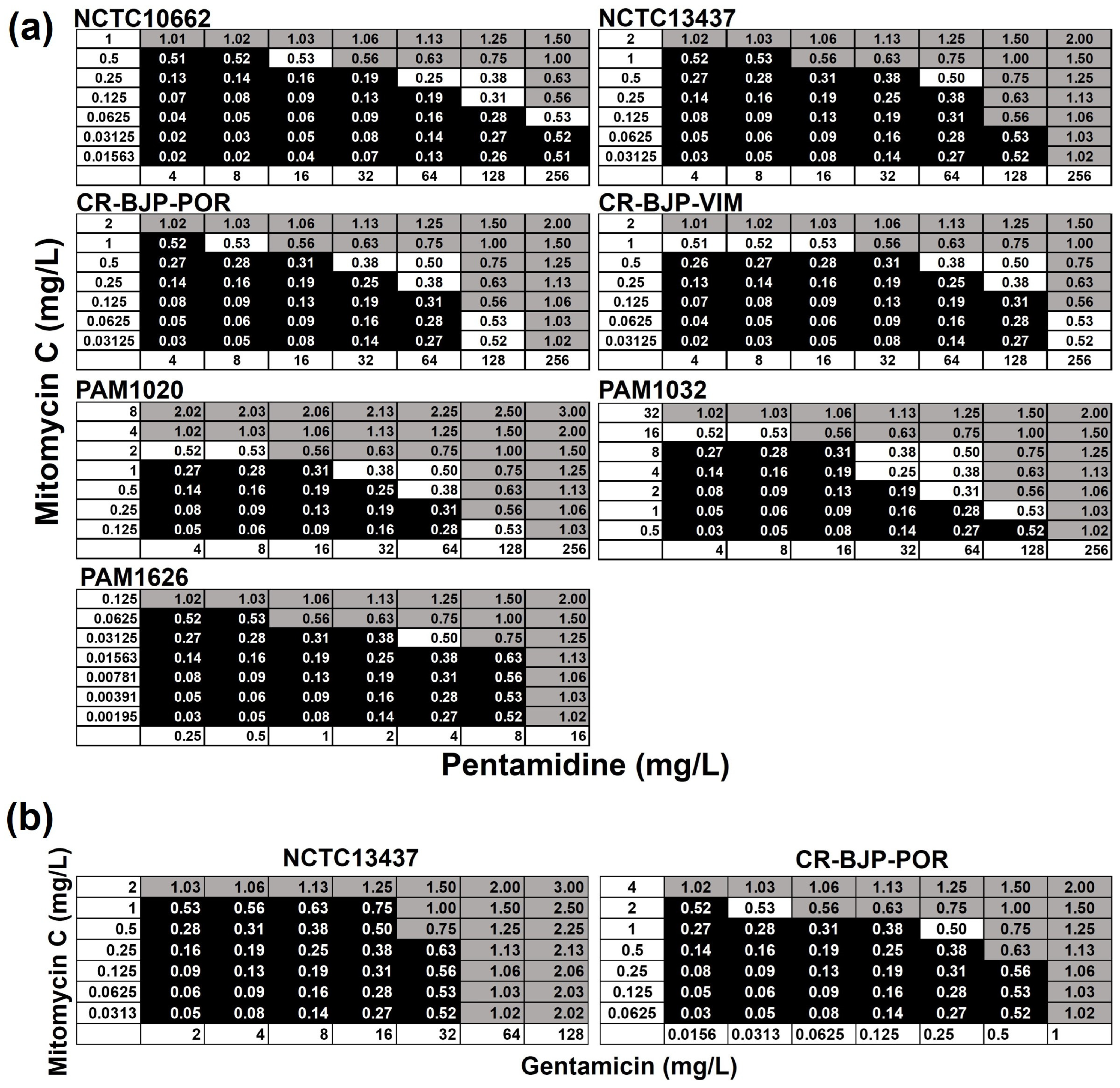
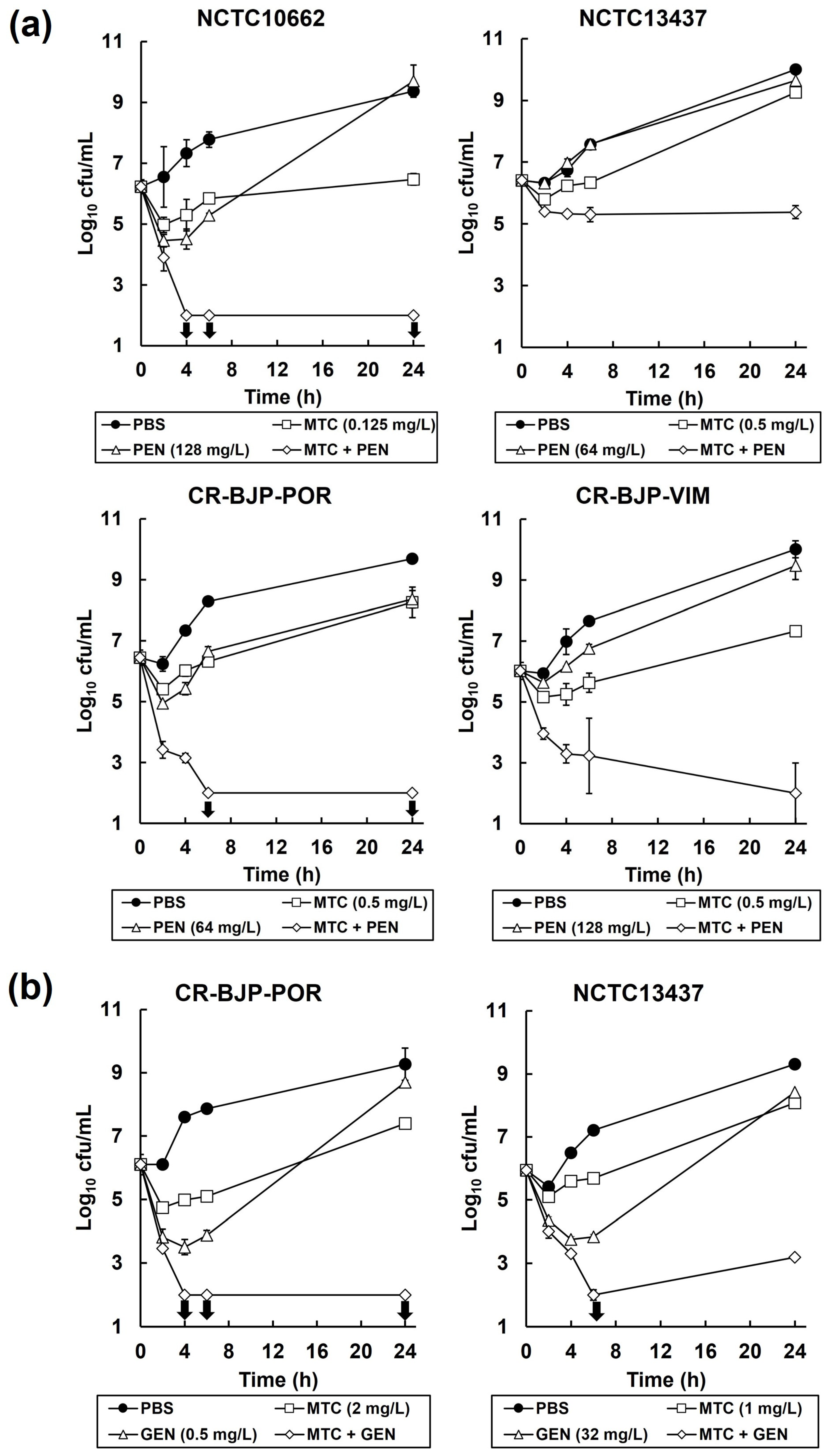
2.3. Combinations of Mitomycin C with Pentamidine, or Gentamicin, In Vitro Result in Synergistic, Bactericidal Inhibition of P. aeruginosa
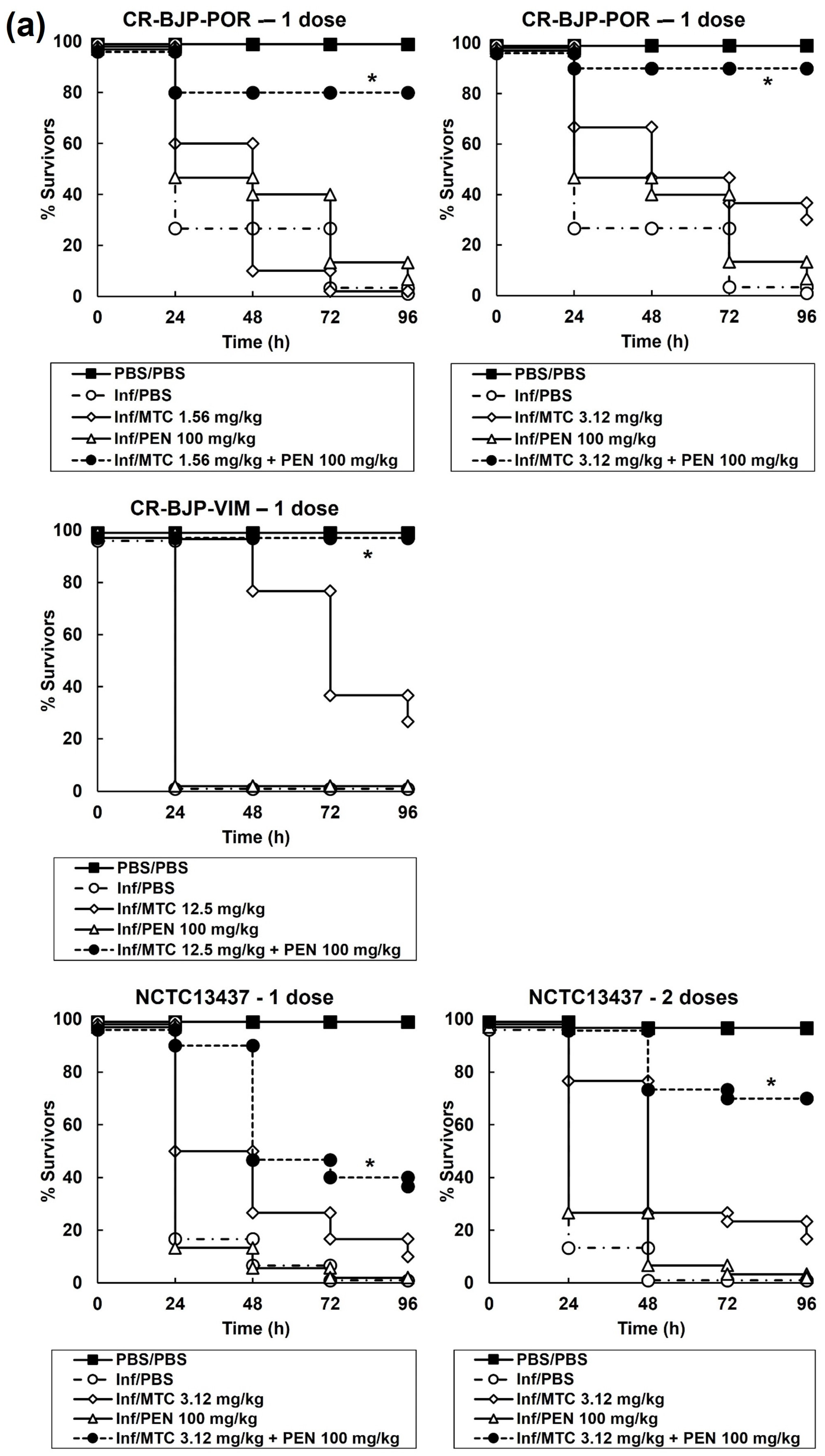
2.4. Combination Therapy with Mitomycin C and Pentamidine or Gentamicin of G. mellonella Larvae Infected with P. aeruginosa Results in Enhanced Efficacy Compared to Monotherapies
3. Discussion
4. Materials and Methods
4.1. Bacteria and Growth Media
4.2. Reagents and G. mellonella Larvae
4.3. Antimicrobial Susceptibility and Checkerboard Assay
4.4. Time–Kill Assay
4.5. G. mellonella Infection Model
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hirsch, E.B.; Tam, V.H. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev. Pharmacoecon. Outcomes Res. 2010, 10, 441–451. [Google Scholar] [CrossRef]
- Sader, H.S.; Castanheira, M.; Mendes, R.E.; Flamm, R.K. Frequency and antimicrobial susceptibility of Gram-negative bacteria isolated from patients with pneumonia hospitalized in ICUs of US medical centres (2015–2017). J. Antimicrob. Chemother. 2018, 73, 3053–3059. [Google Scholar] [CrossRef] [PubMed]
- Palavutitotai, N.; Jitmuang, A.; Tongsai, S.; Kiratisin, P.; Angkasekwinai, N. Epidemiology and risk factors of extensively drug-resistant Pseudomonas aeruginosa infections. PLoS ONE 2018, 13, e0193431. [Google Scholar] [CrossRef] [PubMed]
- Farha, M.A.; Brown, E.D. Drug repurposing for antimicrobial discovery. Nat. Microbiol. 2019, 4, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Barbarossa, A.; Rosato, A.; Corbo, F.; Clodoveo, M.L.; Fracchiolla, G.; Carrieri, A.; Carocci, A. Non-antibiotic drug repositioning as an alternative antimicrobial approach. Antibiotics 2022, 11, 816. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Xia, L.; Huang, W.; Xu, Y.; Gu, Y.; Liu, C.; Ji, L.; Li, W.; Wu, Y.; Zhou, K.; et al. Pentamidine sensitizes FDA-approved non-antibiotics for the inhibition of multidrug-resistant Gram-negative pathogens. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1771–1779. [Google Scholar] [CrossRef] [PubMed]
- Quezada, H.; Martínez-Vázquez, M.; López-Jácome, E.; González-Pedrajo, B.; Andrade, Á.; Fernández-Presas, A.M.; Tovar-García, A.; García-Contreras, R. Repurposed anti-cancer drugs: The future for anti-infective therapy? Expert Rev. Anti-Infect. Ther. 2020, 18, 609–612. [Google Scholar] [CrossRef] [PubMed]
- Soo, V.; Kwan, B.; Quezada, H.; Castillo-Juárez, I.; Pérez-Eretza, B.; García-Contreras, S.; Martínez-Vázquez, M.; Wood, T.; García-Contreras, R. Repurposing of Anticancer Drugs for the Treatment of Bacterial Infections. Curr. Top. Med. Chem. 2016, 17, 1157–1176. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Muñiz, M.Y.; López-Jacome, L.E.; Hernández-Durán, M.; Franco-Cendejas, R.; Licona-Limón, P.; Ramos-Balderas, J.L.; Martinéz-Vázquez, M.; Belmont-Díaz, J.A.; Wood, T.K.; García-Contreras, R. Repurposing the anticancer drug mitomycin C for the treatment of persistent Acinetobacter baumannii infections. Int. J. Antimicrob. Agents 2017, 49, 88–92. [Google Scholar] [CrossRef]
- Kwan, B.W.; Chowdhury, N.; Wood, T.K. Combatting bacterial infections by killing persister cells with mitomycin C. Environ. Microbiol. 2015, 17, 4406–4414. [Google Scholar] [CrossRef]
- Verweij, J.; Pinedo, H. Mitomycin C: Mechanism of action, usefulness and limitations. Anti-Cancer Drugs 1990, 1, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Bradner, W. Mitomycin C: A clinical update. Cancer Treat. Rev. 2001, 27, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Hafiz, S.; Kyriakopoulos, C. Pentamidine. In StatPearls; StatPearls Publishing LLC: Tampa, FL, USA, 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK557586/ (accessed on 17 October 2023).
- Stokes, J.M.; MacNair, C.R.; Ilyas, B.; French, S.; Côté, J.-P.; Bouwman, C.; Farha, M.A.; Sieron, A.O.; Whitfield, C.; Coombes, B.K.; et al. Pentamidine sensitizes Gram-negative pathogens to antibiotics and overcomes acquired colistin resistance. Nat. Microbiol. 2017, 2, 17028. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Espejo, S.; Cebrero-Cangueiro, T.; Labrador-Herrera, G.; Pachón, J.; Pachón-Ibáńez, M.E.; Álvarez-Marín, R. In vitro activity of pentamidine alone and in combination with antibiotics against multidrug-resistant clinical Pseudomonas aeruginosa strains. Antibiotics 2020, 9, 885. [Google Scholar] [CrossRef]
- Woodford, N.; Zhang, J.; Kaufmann, M.E.; Yarde, S.; del Mar Tomas, M.; Faris, C.; Vardhan, M.S.; Dawson, S.; Cotterill, S.L.; Livermore, D.M. Detection of Pseudomonas aeruginosa isolates producing VEB-type extended-spectrum β-lactamases in the United Kingdom. J. Antimic. Chemother. 2008, 62, 1265–1268. [Google Scholar] [CrossRef]
- Lomovskaya, O.; Lee, A.; Hoshino, K.; Ishida, H.; Mistry, A.; Warren, M.S.; Boyer, E.; Chamberland, S.; Lee, V.J. Use of a genetic approach to evaluate the consequences of inhibition of efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1999, 43, 1340–1346. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 13.0. 2023. Available online: http://www.eucast.org (accessed on 17 October 2023).
- Domalaon, R.; Ammeter, D.; Brizuela, M.; Gorityala, B.; Zhanel, G.; Schweizer, F. Repurposed antimicrobial therapy: Tobramycin-ciprofloxacin hybrid augments activity of the anticancer drug mitomycin C against multidrug-resistant Gram-negative bacteria. Front. Microbiol. 2019, 10, 1556. [Google Scholar] [CrossRef] [PubMed]
- Huben, R. Dosage and schedule of mitomycin in clinical setting. Urology 1992, 40, 23–27. [Google Scholar] [CrossRef]
- Pierce, V.M.; Simner, P.J.; Lonsway, D.R.; Roe-Carpenter, D.E.; Johnson, J.K.; Brasso, W.B.; Bobenchik, A.M.; Lockett, Z.C.; Charnot-Katsikas, A.; Ferraro, M.J.; et al. Modified carbapenem inactivation method for phenotypic detection of carbapenemase production among Enterobacteriaceae. J. Clin. Microbiol. 2017, 55, 2321–2333. [Google Scholar] [CrossRef]
- Hill, L.; Veli, N.; Coote, P.J. Evaluation of Galleria mellonella larvae for measuring the efficacy and pharmacokinetics of antibiotic therapies against Pseudomonas aeruginosa infection. Int. J. Antimicrob. Agents 2014, 43, 254–261. [Google Scholar] [CrossRef]
- Eliopoulous, G.; Moellering, R. Antimicrobial combinations. In Antibiotics in Laboratory Medicine 3; Lorian, V., Ed.; Williams and Wilkins: Baltimore, MD, USA, 1996; pp. 330–396. [Google Scholar]
- Bland, J.M.; Altman, D.G. Survival probabilities (the Kaplan-Meier method). Brit. Med. J. 1998, 317, 1572. [Google Scholar] [CrossRef] [PubMed]
- Bland, J.M. The logrank test. Brit. Med. J. 2004, 328, 1073. [Google Scholar] [CrossRef] [PubMed]
- Krezdorn, J.; Adams, S.; Coote, P.J. A Galleria mellonella infection model reveals double and triple antibiotic combination therapies with enhanced efficacy versus a multidrug-resistant strain of Pseudomonas aeruginosa. J. Med. Microbiol. 2014, 63, 945–955. [Google Scholar] [CrossRef] [PubMed]
- Adamson, D.H.; Krikstopaityte, V.; Coote, P.J. Enhanced efficacy of putative efflux pump inhibitor/antibiotic combination treatments versus MDR strains of Pseudomonas aeruginosa in a Galleria mellonella in vivo infection model. J. Antimicrob. Chemother. 2015, 70, 2271–2278. [Google Scholar] [CrossRef]



| Strain | Genotype | Phenotype | Reference |
|---|---|---|---|
| NCTC10662 | Clinical isolate | Antibiotic susceptible control strain | |
| NCTC13437 | Clinical isolate producing VEB-1; VIM-10 β-lactamases | Resistant to β-lactams and fluoroquinolones by an unknown mechanism | [16] |
| CR-BJP-POR | Clinical isolate | Resistant to β-lactams via enhanced efflux or porin loss | Clinical isolate |
| CR-BJP-VIM | Clinical isolate producing a VIM β-lactamase | Resistant to β-lactams, aminoglycosides, fluoroquinolones | Clinical isolate |
| PAM1020 | PA01 prototroph | Wild-type parent strain | [17] |
| PAM1032 | nalB-type mutation | mexAB-oprM over-expressed | [17] |
| PAM1626 | ΔmexAB-oprM::Cm; ΔmexCD-oprJ::Gm; ΔmexEF-oprN::ΩHg | mexAB-oprM; mexCD-oprJ; and mexEF-oprN deleted | [17] |
| Strain | Phenotype | MIC (mg/L) | ||||
|---|---|---|---|---|---|---|
| MTC | PEN | GEN | MEM | CIP | ||
| NCTC10662 | Antibiotic susceptible | 0.5–1 | 512 | - | - | - |
| NCTC13437 | Resistant to β-lactams and fluoroquinolones | 2 | 256 | 64–128 | 64–128 | 32 |
| CR-BJP-POR | Resistant to β-lactams | 2–4 | 256 | 1 | 8 | 0.25 |
| CR-BJP-VIM | Resistant to β-lactams, aminoglycosides, fluoroquinolones | 2 | 512 | - | - | - |
| PAM1020 | Isogenic parent strain of efflux pump mutants | 4–8 | 256 | - | - | - |
| PAM1032 | Over-expression of MexAB-OprM | 16–32 | 256 | - | - | - |
| PAM1626 | Triple deletion of MexAB-OprM, MexCD-OprJ, and MexEF-OprN | 0.125 | 16 | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Svedholm, E.; Bruce, B.; Parcell, B.J.; Coote, P.J. Repurposing Mitomycin C in Combination with Pentamidine or Gentamicin to Treat Infections with Multi-Drug-Resistant (MDR) Pseudomonas aeruginosa. Antibiotics 2024, 13, 177. https://doi.org/10.3390/antibiotics13020177
Svedholm E, Bruce B, Parcell BJ, Coote PJ. Repurposing Mitomycin C in Combination with Pentamidine or Gentamicin to Treat Infections with Multi-Drug-Resistant (MDR) Pseudomonas aeruginosa. Antibiotics. 2024; 13(2):177. https://doi.org/10.3390/antibiotics13020177
Chicago/Turabian StyleSvedholm, Elin, Benjamin Bruce, Benjamin J. Parcell, and Peter J. Coote. 2024. "Repurposing Mitomycin C in Combination with Pentamidine or Gentamicin to Treat Infections with Multi-Drug-Resistant (MDR) Pseudomonas aeruginosa" Antibiotics 13, no. 2: 177. https://doi.org/10.3390/antibiotics13020177
APA StyleSvedholm, E., Bruce, B., Parcell, B. J., & Coote, P. J. (2024). Repurposing Mitomycin C in Combination with Pentamidine or Gentamicin to Treat Infections with Multi-Drug-Resistant (MDR) Pseudomonas aeruginosa. Antibiotics, 13(2), 177. https://doi.org/10.3390/antibiotics13020177






