Emergence of Carbapenemase Genes in Gram-Negative Bacteria Isolated from the Wastewater Treatment Plant in A Coruña, Spain
Abstract
1. Introduction
2. Results
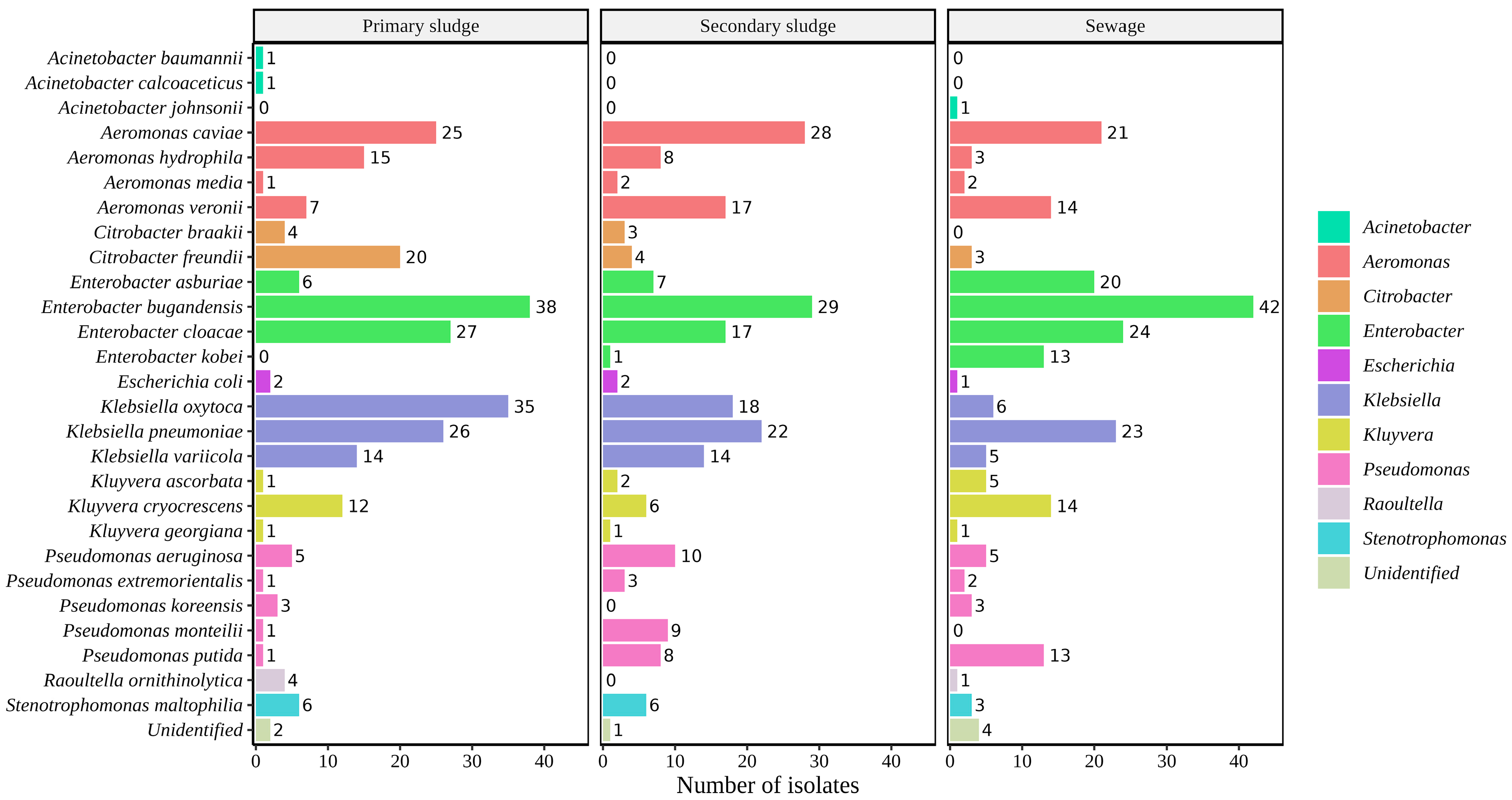
2.1. Identification of Bacterial Isolates
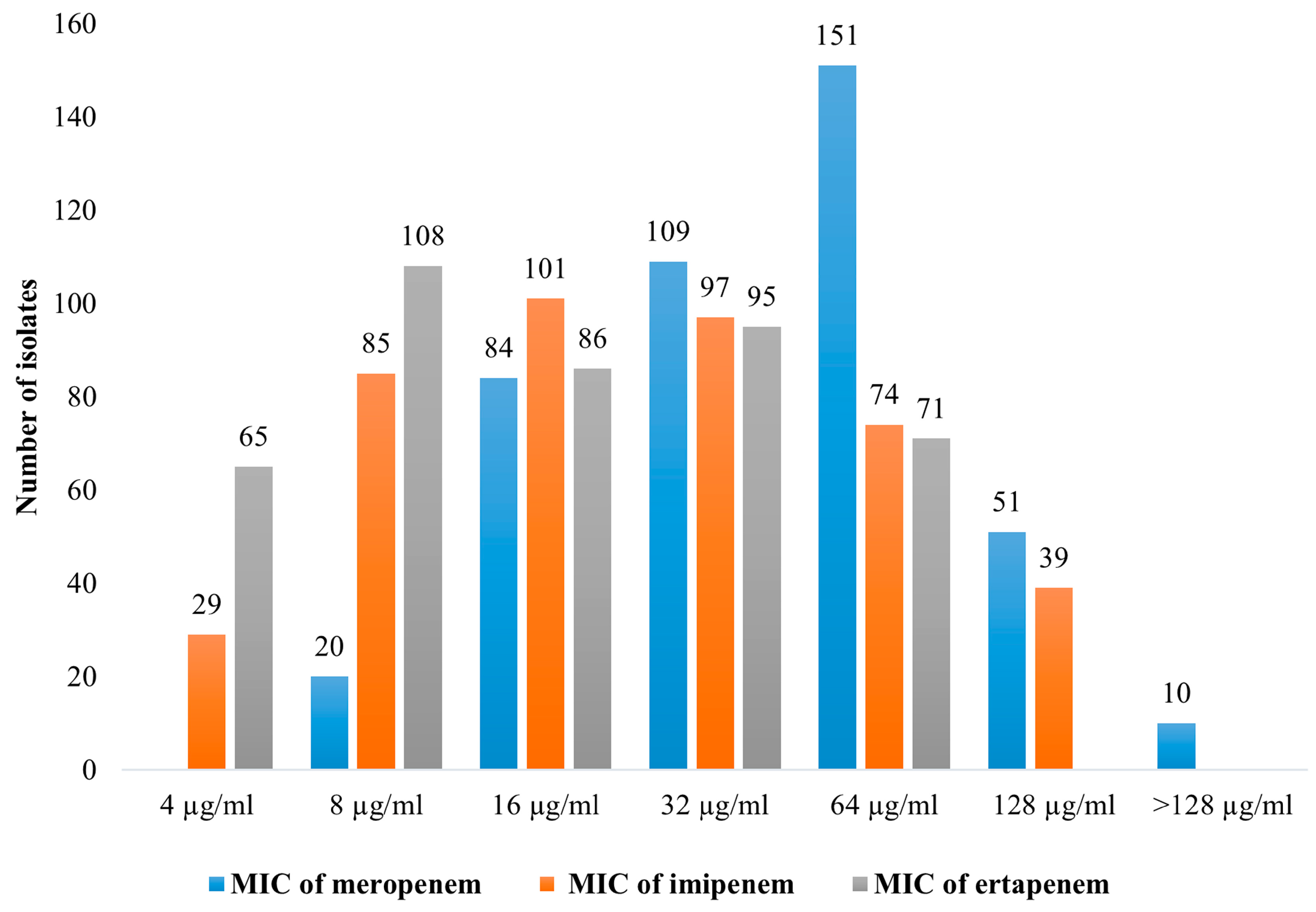
2.2. Antimicrobial Susceptibility Testing
2.3. Carbapenemases Multiplex PCR Detection
2.4. Whole-Genome Sequencing
2.5. Presence of Carbapenemase Genes in Patients from CHUAC
3. Discussion
4. Materials and Methods
4.1. Sampling Site
4.2. Sampling, Isolation, and Bacterial Identification
4.3. Antimicrobial Susceptibility Testing
4.4. Carbapenemases Detection by Multiplex PCR
4.5. Whole-Genome Sequencing
4.6. Bioinformatics
4.7. Clinical Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- O’Neill, J. AMR Review.Org. Available online: https://amr-review.org/home.html (accessed on 31 May 2023).
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef]
- Willyard, C. The Drug-Resistant Bacteria That Pose the Greatest Health Threats. Nature 2017, 543, 15. [Google Scholar] [CrossRef]
- Centers for Disease Control Prevention (CDC). Antibiotic Resistance Threats in the United States, 2019; CDC: Atlanta, GA, USA, 2019.
- Elshamy, A.A.; Aboshanab, K.M. A Review on Bacterial Resistance to Carbapenems: Epidemiology, Detection and Treatment Options. Future Sci. OA 2020, 6, FSO438. [Google Scholar] [CrossRef]
- Codjoe, F.S.; Donkor, E.S. Carbapenem Resistance: A Review. Med. Sci. 2018, 6, 1. [Google Scholar] [CrossRef]
- Papp-Wallace, K.M.; Endimiani, A.; Taracila, M.A.; Bonomo, R.A. Carbapenems: Past, Present, and Future. Antimicrob. Agents Chemother. 2011, 55, 4943–4960. [Google Scholar] [CrossRef]
- Ofer-Friedman, H.; Shefler, C.; Sharma, S.; Tirosh, A.; Tal-Jasper, R.; Kandipalli, D.; Sharma, S.; Bathina, P.; Kaplansky, T.; Maskit, M.; et al. Carbapenems Versus Piperacillin-Tazobactam for Bloodstream Infections of Nonurinary Source Caused by Extended-Spectrum Beta-Lactamase–Producing Enterobacteriaceae. Infect. Control Hosp. Epidemiol. 2015, 36, 981–985. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Izquierdo, M.; Láinez-Ramos-Bossini, A.J.; Rivera-Izquierdo, C.; López-Gómez, J.; Fernández-Martínez, N.F.; Redruello-Guerrero, P.; Martín-delosReyes, L.M.; Martínez-Ruiz, V.; Moreno-Roldán, E.; Jiménez-Mejías, E. OXA-48 Carbapenemase-Producing Enterobacterales in Spanish Hospitals: An Updated Comprehensive Review on a Rising Antimicrobial Resistance. Antibiotics 2021, 10, 89. [Google Scholar] [CrossRef] [PubMed]
- Aurilio, C.; Sansone, P.; Barbarisi, M.; Pota, V.; Giaccari, L.G.; Coppolino, F.; Barbarisi, A.; Passavanti, M.B.; Pace, M.C. Mechanisms of Action of Carbapenem Resistance. Antibiotics 2022, 11, 421. [Google Scholar] [CrossRef]
- Meletis, G. Carbapenem Resistance: Overview of the Problem and Future Perspectives. Ther. Adv. Infect. 2016, 3, 15–21. [Google Scholar] [CrossRef]
- Grundmann, H.; Glasner, C.; Albiger, B.; Aanensen, D.M.; Tomlinson, C.T.; Andrasević, A.T.; Cantón, R.; Carmeli, Y.; Friedrich, A.W.; Giske, C.G.; et al. Occurrence of Carbapenemase-Producing Klebsiella Pneumoniae and Escherichia Coli in the European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE): A Prospective, Multinational Study. Lancet Infect. Dis. 2017, 17, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Sims, N.; Kasprzyk-Hordern, B. Future Perspectives of Wastewater-Based Epidemiology: Monitoring Infectious Disease Spread and Resistance to the Community Level. Environ. Int. 2020, 139, 105689. [Google Scholar] [CrossRef]
- Centers for Disease Control Prevention (CDC). Understanding Antibiotic Resistance in Water: A One Health Approach. Available online: https://www.cdc.gov/onehealth/in-action/understanding-antibiotic-resistance-in-water.html (accessed on 2 June 2023).
- Karkman, A.; Do, T.T.; Walsh, F.; Virta, M.P.J. Antibiotic-Resistance Genes in Waste Water. Trends Microbiol. 2018, 26, 220–228. [Google Scholar] [CrossRef]
- Uluseker, C.; Kaster, K.M.; Thorsen, K.; Basiry, D.; Shobana, S.; Jain, M.; Kumar, G.; Kommedal, R.; Pala-Ozkok, I. A Review on Occurrence and Spread of Antibiotic Resistance in Wastewaters and in Wastewater Treatment Plants: Mechanisms and Perspectives. Front. Microbiol. 2021, 12, 717809. [Google Scholar] [CrossRef]
- Marathe, N.P.; Berglund, F.; Razavi, M.; Pal, C.; Dröge, J.; Samant, S.; Kristiansson, E.; Larsson, D.G.J. Sewage Effluent from an Indian Hospital Harbors Novel Carbapenemases and Integron-Borne Antibiotic Resistance Genes. Microbiome 2019, 7, 97. [Google Scholar] [CrossRef]
- Zhang, L.; Calvo-Bado, L.; Murray, A.K.; Amos, G.C.A.; Hawkey, P.M.; Wellington, E.M.; Gaze, W.H. Novel Clinically Relevant Antibiotic Resistance Genes Associated with Sewage Sludge and Industrial Waste Streams Revealed by Functional Metagenomic Screening. Environ. Int. 2019, 132, 105120. [Google Scholar] [CrossRef]
- Uyaguari, M.I.; Fichot, E.B.; Scott, G.I.; Norman, R.S. Characterization and Quantitation of a Novel β-Lactamase Gene Found in a Wastewater Treatment Facility and the Surrounding Coastal Ecosystem. Appl. Environ. Microbiol. 2011, 77, 8226–8233. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhang, X.-X.; Ye, L. Plasmid Metagenome Reveals High Levels of Antibiotic Resistance Genes and Mobile Genetic Elements in Activated Sludge. PLoS ONE 2011, 6, e26041. [Google Scholar] [CrossRef] [PubMed]
- Mahfouz, N.; Caucci, S.; Achatz, E.; Semmler, T.; Guenther, S.; Berendonk, T.U.; Schroeder, M. High Genomic Diversity of Multi-Drug Resistant Wastewater Escherichia Coli. Sci. Rep. 2018, 8, 8928. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Lazaro-Perona, F.; Falgenhauer, L.; Valverde, A.; Imirzalioglu, C.; Dominguez, L.; Cantón, R.; Mingorance, J.; Chakraborty, T. Insights into a Novel blaKPC-2-Encoding IncP-6 Plasmid Reveal Carbapenem-Resistance Circulation in Several Enterobacteriaceae Species from Wastewater and a Hospital Source in Spain. Front. Microbiol. 2017, 8, 1143. [Google Scholar] [CrossRef] [PubMed]
- Li, A.-D.; Li, L.-G.; Zhang, T. Exploring Antibiotic Resistance Genes and Metal Resistance Genes in Plasmid Metagenomes from Wastewater Treatment Plants. Front. Microbiol. 2015, 6, 1025. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Paakkanen, J.; Österblad, M.; Kirveskari, J.; Hendriksen, R.S.; Heikinheimo, A. Wastewater Surveillance Detected Carbapenemase Enzymes in Clinically Relevant Gram-Negative Bacteria in Helsinki, Finland; 2011–2012. Front. Microbiol. 2022, 13, 887888. [Google Scholar] [CrossRef]
- Gomi, R.; Matsuda, T.; Yamamoto, M.; Chou, P.-H.; Tanaka, M.; Ichiyama, S.; Yoneda, M.; Matsumura, Y. Characteristics of Carbapenemase-Producing Enterobacteriaceae in Wastewater Revealed by Genomic Analysis. Antimicrob. Agents Chemother. 2018, 62, e02501-17. [Google Scholar] [CrossRef]
- Delgado-Blas, J.F.; Valenzuela Agüi, C.; Marin Rodriguez, E.; Serna, C.; Montero, N.; Saba, C.K.S.; Gonzalez-Zorn, B. Dissemination Routes of Carbapenem and Pan-Aminoglycoside Resistance Mechanisms in Hospital and Urban Wastewater Canalizations of Ghana. mSystems 2022, 7, e01019-21. [Google Scholar] [CrossRef]
- Oliveira, M.; Leonardo, I.C.; Nunes, M.; Silva, A.F.; Barreto Crespo, M.T. Environmental and Pathogenic Carbapenem Resistant Bacteria Isolated from a Wastewater Treatment Plant Harbour Distinct Antibiotic Resistance Mechanisms. Antibiotics 2021, 10, 1118. [Google Scholar] [CrossRef]
- Teban-Man, A.; Szekeres, E.; Fang, P.; Klümper, U.; Hegedus, A.; Baricz, A.; Berendonk, T.U.; Pârvu, M.; Coman, C. Municipal Wastewaters Carry Important Carbapenemase Genes Independent of Hospital Input and Can Mirror Clinical Resistance Patterns. Microbiol. Spectr. 2022, 10, e02711-21. [Google Scholar] [CrossRef]
- Yang, F.; Mao, D.; Zhou, H.; Luo, Y. Prevalence and Fate of Carbapenemase Genes in a Wastewater Treatment Plant in Northern China. PLoS ONE 2016, 11, e0156383. [Google Scholar] [CrossRef]
- Zurfluh, K.; Bagutti, C.; Brodmann, P.; Alt, M.; Schulze, J.; Fanning, S.; Stephan, R.; Nüesch-Inderbinen, M. Wastewater Is a Reservoir for Clinically Relevant Carbapenemase- and 16s rRNA Methylase-Producing Enterobacteriaceae. Int. J. Antimicrob. Agents 2017, 50, 436–440. [Google Scholar] [CrossRef]
- Luo, Y.; Yang, F.; Mathieu, J.; Mao, D.; Wang, Q.; Alvarez, P.J.J. Proliferation of Multidrug-Resistant New Delhi Metallo-β-Lactamase Genes in Municipal Wastewater Treatment Plants in Northern China. Environ. Sci. Technol. Lett. 2014, 1, 26–30. [Google Scholar] [CrossRef]
- Logan, L.K.; Weinstein, R.A. The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The Impact and Evolution of a Global Menace. J. Infect. Dis. 2017, 215, S28–S36. [Google Scholar] [CrossRef]
- Oteo, J.; Ortega, A.; Bartolomé, R.; Bou, G.; Conejo, C.; Fernández-Martínez, M.; González-López, J.J.; Martínez-García, L.; Martínez-Martínez, L.; Merino, M.; et al. Prospective Multicenter Study of Carbapenemase-Producing Enterobacteriaceae from 83 Hospitals in Spain Reveals High In Vitro Susceptibility to Colistin and Meropenem. Antimicrob. Agents Chemother. 2015, 59, 3406–3412. [Google Scholar] [CrossRef]
- Cañada-García, J.E.; Moure, Z.; Sola-Campoy, P.J.; Delgado-Valverde, M.; Cano, M.E.; Gijón, D.; González, M.; Gracia-Ahufinger, I.; Larrosa, N.; Mulet, X.; et al. CARB-ES-19 Multicenter Study of Carbapenemase-Producing Klebsiella Pneumoniae and Escherichia Coli From All Spanish Provinces Reveals Interregional Spread of High-Risk Clones Such as ST307/OXA-48 and ST512/KPC-3. Front. Microbiol. 2022, 13, 918362. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Ucha, J.C.; Seoane-Estévez, A.; Rodiño-Janeiro, B.K.; González-Bardanca, M.; Conde-Pérez, K.; Martínez-Guitián, M.; Alvarez-Fraga, L.; Arca-Suárez, J.; Lasarte-Monterrubio, C.; Gut, M.; et al. Activity of Imipenem/Relebactam against a Spanish Nationwide Collection of Carbapenemase-Producing Enterobacterales. J. Antimicrob. Chemother. 2021, 76, 1498–1510. [Google Scholar] [CrossRef]
- Boczek, L.A.; Rice, E.W.; Johnston, B.; Johnson, J.R. Occurrence of Antibiotic-Resistant Uropathogenic Escherichia Coli Clonal Group A in Wastewater Effluents. Appl. Environ. Microbiol. 2007, 73, 4180–4184. [Google Scholar] [CrossRef]
- Tiwari, A.; Kurittu, P.; Al-Mustapha, A.I.; Heljanko, V.; Johansson, V.; Thakali, O.; Mishra, S.K.; Lehto, K.-M.; Lipponen, A.; Oikarinen, S.; et al. Wastewater Surveillance of Antibiotic-Resistant Bacterial Pathogens: A Systematic Review. Front. Microbiol. 2022, 13, 977106. [Google Scholar] [CrossRef]
- Sekizuka, T.; Yatsu, K.; Inamine, Y.; Segawa, T.; Nishio, M.; Kishi, N.; Kuroda, M. Complete Genome Sequence of a blaKPC-2-Positive Klebsiella Pneumoniae Strain Isolated from the Effluent of an Urban Sewage Treatment Plant in Japan. mSphere 2018, 3, e00314-18. [Google Scholar] [CrossRef]
- Sekizuka, T.; Inamine, Y.; Segawa, T.; Hashino, M.; Yatsu, K.; Kuroda, M. Potential KPC-2 Carbapenemase Reservoir of Environmental Aeromonas Hydrophila and Aeromonas Caviae Isolates from the Effluent of an Urban Wastewater Treatment Plant in Japan. Environ. Microbiol. Rep. 2019, 11, 589–597. [Google Scholar] [CrossRef]
- Mantilla-Calderon, D.; Jumat, M.R.; Wang, T.; Ganesan, P.; Al-Jassim, N.; Hong, P.-Y. Isolation and Characterization of NDM-Positive Escherichia Coli from Municipal Wastewater in Jeddah, Saudi Arabia. Antimicrob. Agents Chemother. 2016, 60, 5223–5231. [Google Scholar] [CrossRef]
- Dong, N.; Yang, X.; Chan, E.W.-C.; Zhang, R.; Chen, S. Klebsiella Species: Taxonomy, Hypervirulence and Multidrug Resistance. eBioMedicine 2022, 79, 103998. [Google Scholar] [CrossRef]
- Gorrie, C.L.; Mirčeta, M.; Wick, R.R.; Judd, L.M.; Lam, M.M.C.; Gomi, R.; Abbott, I.J.; Thomson, N.R.; Strugnell, R.A.; Pratt, N.F.; et al. Genomic Dissection of Klebsiella Pneumoniae Infections in Hospital Patients Reveals Insights into an Opportunistic Pathogen. Nat. Commun. 2022, 13, 3017. [Google Scholar] [CrossRef]
- Hernández-García, M.; Pérez-Viso, B.; Navarro-San Francisco, C.; Baquero, F.; Morosini, M.I.; Ruiz-Garbajosa, P.; Cantón, R. Intestinal Co-Colonization with Different Carbapenemase-Producing Enterobacterales Isolates Is Not a Rare Event in an OXA-48 Endemic Area. EClinicalMedicine 2019, 15, 72–79. [Google Scholar] [CrossRef]
- David, S.; Reuter, S.; Harris, S.R.; Glasner, C.; Feltwell, T.; Argimon, S.; Abudahab, K.; Goater, R.; Giani, T.; Errico, G.; et al. Epidemic of Carbapenem-Resistant Klebsiella Pneumoniae in Europe Is Driven by Nosocomial Spread. Nat. Microbiol. 2019, 4, 1919–1929. [Google Scholar] [CrossRef]
- Oteo, J.; Saez, D.; Bautista, V.; Fernández-Romero, S.; Hernández-Molina, J.M.; Pérez-Vázquez, M.; Aracil, B.; Campos, J.; the Spanish Collaborating Group for the Antibiotic Resistance Surveillance Program. Carbapenemase-Producing Enterobacteriaceae in Spain in 2012. Antimicrob. Agents Chemother. 2013, 57, 6344–6347. [Google Scholar] [CrossRef] [PubMed]
- Peirano, G.; Chen, L.; Kreiswirth, B.N.; Pitout, J.D.D. Emerging Antimicrobial-Resistant High-Risk Klebsiella Pneumoniae Clones ST307 and ST147. Antimicrob. Agents Chemother. 2020, 64, e01148-20. [Google Scholar] [CrossRef] [PubMed]
- Ito, R.; Shindo, Y.; Kobayashi, D.; Ando, M.; Jin, W.; Wachino, J.; Yamada, K.; Kimura, K.; Yagi, T.; Hasegawa, Y.; et al. Molecular Epidemiological Characteristics of Klebsiella Pneumoniae Associated with Bacteremia among Patients with Pneumonia. J. Clin. Microbiol. 2015, 53, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Hawkey, J.; Wyres, K.L.; Judd, L.M.; Harshegyi, T.; Blakeway, L.; Wick, R.R.; Jenney, A.W.J.; Holt, K.E. ESBL Plasmids in Klebsiella Pneumoniae: Diversity, Transmission and Contribution to Infection Burden in the Hospital Setting. Genome Med. 2022, 14, 97. [Google Scholar] [CrossRef] [PubMed]
- Mezzatesta, M.L.; Gona, F.; Stefani, S. Enterobacter Cloacae Complex: Clinical Impact and Emerging Antibiotic Resistance. Future Microbiol. 2012, 7, 887–902. [Google Scholar] [CrossRef] [PubMed]
- Sutton, G.G.; Brinkac, L.M.; Clarke, T.H.; Fouts, D.E. Enterobacterhormaechei Subsp. Hoffmannii Subsp. Nov., Enterobacter Hormaechei Subsp. Xiangfangensis Comb. Nov., Enterobacter Roggenkampii Sp. Nov., and Enterobacter Muelleri Is a Later Heterotypic Synonym of Enterobacter Asburiae Based on Computational Analysis of Sequenced Enterobacter Genomes. F1000Res 2018, 7, 521. [Google Scholar] [CrossRef] [PubMed]
- Annavajhala, M.K.; Gomez-Simmonds, A.; Uhlemann, A.-C. Multidrug-Resistant Enterobacter Cloacae Complex Emerging as a Global, Diversifying Threat. Front. Microbiol. 2019, 10, 44. [Google Scholar] [CrossRef]
- Wu, W.; Wang, J.; Zhang, P.; Wang, N.; Yuan, Q.; Shi, W.; Zhang, X.; Li, X.; Qu, T. Emergence of Carbapenem-Resistant Enterobacter Hormaechei ST93 Plasmids Co-Harbouring blaNDM-1, blaKPC-2, and Mcr-9 in Bloodstream Infection. J. Glob. Antimicrob. Resist. 2023, 34, 67–73. [Google Scholar] [CrossRef]
- Takizawa, S.; Soga, E.; Hayashi, W.; Sakaguchi, K.; Koide, S.; Tanabe, M.; Denda, T.; Sugawara, Y.; Yu, L.; Kayama, S.; et al. Genomic Landscape of blaGES-5- and blaGES-24-Harboring Gram-Negative Bacteria from Hospital Wastewater: Emergence of Class 3 Integron-Associated blaGES-24 Genes. J. Glob. Antimicrob. Resist. 2022, 31, 196–206. [Google Scholar] [CrossRef]
- Falgenhauer, L.; Schwengers, O.; Schmiedel, J.; Baars, C.; Lambrecht, O.; Heß, S.; Berendonk, T.U.; Falgenhauer, J.; Chakraborty, T.; Imirzalioglu, C. Multidrug-Resistant and Clinically Relevant Gram-Negative Bacteria Are Present in German Surface Waters. Front. Microbiol. 2019, 10, 2779. [Google Scholar] [CrossRef] [PubMed]
- Rada, A.M.; De La Cadena, E.; Agudelo, C.; Capataz, C.; Orozco, N.; Pallares, C.; Dinh, A.Q.; Panesso, D.; Ríos, R.; Diaz, L.; et al. Dynamics of blaKPC-2 Dissemination from Non-CG258 Klebsiella Pneumoniae to Other Enterobacterales via IncN Plasmids in an Area of High Endemicity. Antimicrob. Agents Chemother. 2020, 64, e01743-20. [Google Scholar] [CrossRef] [PubMed]
- Izdebski, R.; Biedrzycka, M.; Urbanowicz, P.; Żabicka, D.; Gniadkowski, M. Genome-Based Epidemiologic Analysis of VIM/IMP Carbapenemase-Producing Enterobacter Spp., Poland. Emerg. Infect. Dis. 2023, 29, 1618–1626. [Google Scholar] [CrossRef] [PubMed]
- Balero de Paula, S.; Cayô, R.; Streling, A.P.; Silva Nodari, C.; Pereira Matos, A.; Eches Perugini, M.R.; Gales, A.C.; Carrara-Marroni, F.E.; Yamada-Ogatta, S.F. Detection of blaVIM-7 in an Extensively Drug-Resistant Pseudomonas Aeruginosa Isolate Belonging to ST1284 in Brazil. Diagn. Microbiol. Infect. Dis. 2017, 89, 80–82. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Qi, Y.; Li, G.; Wang, Y.; Lou, Z.; Jiang, Y. Characterization of the Genetic Environment of blaKPC in Escherichia Coli Isolates from Hospitals in China. FEMS Microbiol. Lett. 2020, 367, fnaa064. [Google Scholar] [CrossRef] [PubMed]
- Edar Bens SA—Empresa Pública de Depuración de Aguas Residuals—Galicia. Available online: https://edarbens.es/ (accessed on 1 June 2023).
- Trigo-Tasende, N.; Vallejo, J.A.; Rumbo-Feal, S.; Conde-Pérez, K.; Vaamonde, M.; López-Oriona, Á.; Barbeito, I.; Nasser-Ali, M.; Reif, R.; Rodiño-Janeiro, B.K.; et al. Wastewater Early Warning System for SARS-CoV-2 Outbreaks and Variants in a Coruña, Spain. Environ. Sci. Pollut. Res. Int. 2023, 30, 79315–79334. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Quintanilla, M.; Poirel, L.; Nordmann, P. CHROMagar mSuperCARBA and RAPIDEC® Carba NP Test for Detection of Carbapenemase-Producing Enterobacteriaceae. Diagn. Microbiol. Infect. Dis. 2018, 90, 77–80. [Google Scholar] [CrossRef]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 11th ed.; CLSI standard M07; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 31st ed.; CLSI supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2021. [Google Scholar]
- Mudzana, R.; Mavenyengwa, R.T.; Gudza-Mugabe, M. Analysis of Virulence Factors and Antibiotic Resistance Genes in Group B Streptococcus from Clinical Samples. BMC Infect. Dis. 2021, 21, 125. [Google Scholar] [CrossRef]
- Ribeiro Junior, J.C.; Tamanini, R.; Soares, B.F.; Oliveira, A.M.D.; Silva, F.D.G.; Silva, F.F.D.; Augusto, N.A.; Beloti, V. Efficiency of Boiling and Four Other Methods for Genomic DNA Extraction of Deteriorating Spore-Forming Bacteria from Milk. Semin. Ciências Agrárias 2016, 37, 3069. [Google Scholar] [CrossRef]
- Poirel, L.; Walsh, T.R.; Cuvillier, V.; Nordmann, P. Multiplex PCR for Detection of Acquired Carbapenemase Genes. Diagn. Microbiol. Infect. Dis. 2011, 70, 119–123. [Google Scholar] [CrossRef]
- Bogaerts, P.; Rezende de Castro, R.; de Mendonça, R.; Huang, T.-D.; Denis, O.; Glupczynski, Y. Validation of Carbapenemase and Extended-Spectrum β-Lactamase Multiplex Endpoint PCR Assays According to ISO 15189. J. Antimicrob. Chemother. 2013, 68, 1576–1582. [Google Scholar] [CrossRef]
- Nanoporetech/Duplex-Tools: Splitting of Sequence Reads by Internal Adapter Sequence Search. Available online: https://github.com/nanoporetech/duplex-tools (accessed on 12 September 2023).
- Rrwick/Filtlong: Quality Filtering Tool for Long Reads. Available online: https://github.com/rrwick/Filtlong (accessed on 12 September 2023).
- De Coster, W.; Rademakers, R. NanoPack2: Population-Scale Evaluation of Long-Read Sequencing Data. Bioinformatics 2023, 39, btad311. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of Long, Error-Prone Reads Using Repeat Graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Nanoporetech/Medaka: Sequence Correction Provided by ONT Research. Available online: https://github.com/nanoporetech/medaka (accessed on 12 September 2023).
- Huang, Y.-T.; Liu, P.-Y.; Shih, P.-W. Homopolish: A Method for the Removal of Systematic Errors in Nanopore Sequencing by Homologous Polishing. Genome Biol. 2021, 22, 95. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- KmerFinder Genomic Epidemiology/Kmerfinder—Bitbucket. Available online: https://bitbucket.org/genomicepidemiology/kmerfinder/src/master/ (accessed on 12 September 2023).
- Brown, C.T.; Irber, L. Sourmash: A Library for MinHash Sketching of DNA. J. Open Source Softw. 2016, 1, 27. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus Sequence Typing of Total-Genome-Sequenced Bacteria. J. Clin. Microbiol. 2020, 50, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and Standardized Annotation of Bacterial Genomes via Alignment-Free Sequence Identification. Microb. Genom. 2021, 7, 000685. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.; Nash, J.H.E. MOB-Suite: Software Tools for Clustering, Reconstruction and Typing of Plasmids from Draft Assemblies. Microb. Genom. 2018, 4, e000206. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Lam, M.M.C.; Wick, R.R.; Watts, S.C.; Cerdeira, L.T.; Wyres, K.L.; Holt, K.E. A Genomic Surveillance Framework and Genotyping Tool for Klebsiella Pneumoniae and Its Related Species Complex. Nat. Commun. 2021, 12, 4188. [Google Scholar] [CrossRef] [PubMed]
- Bayliss, S.C.; Thorpe, H.A.; Coyle, N.M.; Sheppard, S.K.; Feil, E.J. PIRATE: A Fast and Scalable Pangenomics Toolbox for Clustering Diverged Orthologues in Bacteria. GigaScience 2019, 8, giz119. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.-Y. Ggtree: An r Package for Visualization and Annotation of Phylogenetic Trees with Their Covariates and Other Associated Data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]



| Gene | Oligonucleotide Name | Sequence (5′-3′) | Product Size (bp) | References |
|---|---|---|---|---|
| blaKPC | KPC-Fm KPC-Rm | CGTCTAGTTCTGCTGTCTTG CTTGTCATCCTTGTTAGGCG | 798 232 | [66] |
| blaOXA-48 | OXA-F OXA-R | GCGTGGTTAAGGATGAACAC CATCAAGTTCAACCCAACCG | 438 | [66] |
| blaVIM | VIM-F VIM-R | GATGGTGTTTGGTCGCATA CGAATGCGCAGCACCAG | 390 | [66] |
| blaIMP | IMP-F IMP-R | GGAATAGAGTGGCTTAAYTCTC GGTTTAAYAAAACAACCACC | 232 | [66] |
| blaNDM | NDM-F NDM-R | GGTTTGGCGATCTGGTTTTC CGGAATGGCTCATCACGATC | 621 | [66] |
| blaGES | GES-F GES-R | CTGGCAGGGATCGCTCACTC TTC CGATCAGCCACCTCTCA | 600 | [67] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nasser-Ali, M.; Aja-Macaya, P.; Conde-Pérez, K.; Trigo-Tasende, N.; Rumbo-Feal, S.; Fernández-González, A.; Bou, G.; Poza, M.; Vallejo, J.A. Emergence of Carbapenemase Genes in Gram-Negative Bacteria Isolated from the Wastewater Treatment Plant in A Coruña, Spain. Antibiotics 2024, 13, 194. https://doi.org/10.3390/antibiotics13020194
Nasser-Ali M, Aja-Macaya P, Conde-Pérez K, Trigo-Tasende N, Rumbo-Feal S, Fernández-González A, Bou G, Poza M, Vallejo JA. Emergence of Carbapenemase Genes in Gram-Negative Bacteria Isolated from the Wastewater Treatment Plant in A Coruña, Spain. Antibiotics. 2024; 13(2):194. https://doi.org/10.3390/antibiotics13020194
Chicago/Turabian StyleNasser-Ali, Mohammed, Pablo Aja-Macaya, Kelly Conde-Pérez, Noelia Trigo-Tasende, Soraya Rumbo-Feal, Ana Fernández-González, Germán Bou, Margarita Poza, and Juan A. Vallejo. 2024. "Emergence of Carbapenemase Genes in Gram-Negative Bacteria Isolated from the Wastewater Treatment Plant in A Coruña, Spain" Antibiotics 13, no. 2: 194. https://doi.org/10.3390/antibiotics13020194
APA StyleNasser-Ali, M., Aja-Macaya, P., Conde-Pérez, K., Trigo-Tasende, N., Rumbo-Feal, S., Fernández-González, A., Bou, G., Poza, M., & Vallejo, J. A. (2024). Emergence of Carbapenemase Genes in Gram-Negative Bacteria Isolated from the Wastewater Treatment Plant in A Coruña, Spain. Antibiotics, 13(2), 194. https://doi.org/10.3390/antibiotics13020194






